Abstract
Humans and mice detect pain, itch, temperature, pressure, stretch and limb position via signaling from peripheral sensory neurons. These neurons are divided into three functional classes (nociceptors/pruritoceptors, mechanoreceptors and proprioceptors) that are distinguished by their selective expression of TrkA, TrkB or TrkC receptors, respectively. We found that transiently coexpressing Brn3a with either Ngn1 or Ngn2 selectively reprogrammed human and mouse fibroblasts to acquire key properties of these three classes of sensory neurons. These induced sensory neurons (iSNs) were electrically active, exhibited distinct sensory neuron morphologies and matched the characteristic gene expression patterns of endogenous sensory neurons, including selective expression of Trk receptors. In addition, we found that calcium-imaging assays could identify subsets of iSNs that selectively responded to diverse ligands known to activate itch- and pain-sensing neurons. These results offer a simple and rapid means for producing genetically diverse human sensory neurons suitable for drug screening and mechanistic studies.
Pain, itch and other disorders affecting peripheral sensory neurons pose a large unmet medical need1,2. Diverse subtypes of sensory neurons in the dorsal root ganglia (DRG) or the trigeminal nerve are responsible for mediating acute and chronic pain, histamine-dependent and histamine-independent itch, and unique subtypes are also affected in neurologic disorders such as Friedreich’s ataxia and diabetic neuropathy3. More recently, TrpV1 expressing nociceptors have also been linked to immune responses and implicated in the regulation of aging and metabolism4. In addition, rare human mutations in nociceptor-specific genes have been shown to underlie familial pain disorders. For example, inactivating mutations in the ion channel NaV1.7 result in dangerous insensitivity to pain, whereas activating mutations in the same channel cause chronic neuropathic pain in affected individuals5. Similarly, more common inherited genetic variants are likely to affect sensory neuron function, though the mechanisms governing this in human sensory neurons remain largely uncharacterized2.
Peripheral sensory neurons comprise three distinct subtypes, which are distinguished by their function as nociceptors/pruritoceptors, mechanoceptors and proprioceptors. These subclasses are characterized by their selective expression of one member of the Trk receptor gene family: TrkA, TrkB and TrkC6. In a subclass, individual neurons exhibit further diversity. For example, different subsets of TrkA positive neurons respond to distinct stimuli via expression of Trp receptor proteins7. Neurons expressing TrpV1 respond to heat and to capsaicin8, whereas cold-sensitive neurons express TrpM8 and respond to menthol9,10. Neurons expressing TrpA1 can respond to noxious compounds, including allyl isothiocyanate11. Subsets of TrkA neurons also respond to histamine, chloroquine and other pruritogenic compounds based, at least partly, on the expression of the histamine receptors (H1 and H4) and the Mrgpr family proteins, which are quite divergent between mouse, rat and human12–15. Understanding the molecular basis for these forms of sensation would benefit from improved methods for identifying, studying and manipulating peripheral sensory neurons in vitro.
Traditional methods for studying sensory neurons have largely relied on animal models given the difficulty of obtaining and manipulating human neurons2,16. Although animal models are useful, as they can recapitulate the endogenous circuitry of this system, human mechanisms of pain and itch perception can differ from those of rodents and inter-individual genetic differences may also influence responses to nociceptive and pruritogenic compounds17,18. These factors may help to explain high rates of attrition during clinical trials for promising analgesics19. Although cadaveric peripheral sensory neurons are available for research, these cells cannot be genetically altered and have limited availability. Thus, a means to produce functionally responsive, yet genetically diverse, human sensory neurons in sufficient numbers for mechanistic studies or drug screening would enable research into human specific aspects of pain, itch and other disorders that affect this class of neurons.
One means to produce genetically diverse human or mouse neurons of a specific subtype is to develop protocols to differentiate them from human pluripotent stem cells (hPSCs). Encouragingly, hPSCs have been differentiated into nociceptive populations of neurons that respond to P2XR3 agonists used to mimic inflammatory pain, some of which also responded to capsaicin. However, other clinically relevant sensory neuronal subtypes have not been efficiently produced directly from hPSCs20. An alternative method to produce desired human neuronal subtypes is to generate neurons directly from fibroblasts via transcription factor overexpression. This direct reprogramming approach offers a potentially more rapid and selective method to produce defined functional classes of neurons21–28.
Most established direct reprogramming methods require the neurogenic basic helix-loop-helix (bHLH) transcription factor Ascl1 (also known as Mash1)21,22,24–28. However, peripheral sensory neurons arise from precursors that express alternative neurogenic bHLH proteins Neurogenin 1 (Ngn1) and/or Neurogenin 2 (Ngn2)29, but not Ascl1. These precursors subsequently differentiate into mature sensory neurons that express the transcription factor Brn3a (also called Pou4f1), but no longer express Ngn1 or Ngn2. Mouse developmental studies have shown that Ngn1, Ngn2 and Brn3a are required for appropriate differentiation of these neurons and have indicated that, although Ngn1 and Ngn2 have distinct roles in generating the TrkA versus TrkB/TrkC subsets of sensory neurons, they can also partly compensate for one another29–32.
We found that expressing Brn3a with either Ngn1 or Ngn2 was sufficient to selectively convert both human and mouse fibroblasts into induced neurons that exhibited key molecular, morphological and electrophysiological characteristics of endogenous peripheral sensory neurons. Notably, nearly all (90%) of the induced neurons produced by this method exhibited hallmarks of one of the three subclasses of sensory neurons. Calcium imaging and electrophysiology confirmed that these induced neurons were sufficiently mature to permit detection of responses to diverse pain- and itch-inducing compounds. These results indicate that developmentally relevant transcription factors can be employed to rapidly and selectively produce neurons of a desired subtype that may be used to study neuronal responses in vitro.
RESULTS
Transient coexpression of Brn3a with either Ngn1 or Ngn2 reprograms fibroblasts into mature neurons
Previous reports of neural direct reprogramming using transcription factors have included Ascl1, either in combination with additional factors, such as Brn2, or more recently as a single factor21–28. However, in vivo, peripheral sensory neurons express Brn3a, but not Brn2, and arise from precursors that express Ngn1 or Ngn2, but not Ascl1. Thus, we tested whether pairing Ngn1 or Ngn2 with Brn3a could produce neurons with features similar to peripheral sensory neurons. To achieve transient expression of these factors, we cloned cDNAs for each gene into doxycycline (dox)-inducible lentiviral vectors (Supplementary Fig. 1a). Next, we transduced mouse embryonic fibroblasts (MEFs) with either Brn3a and Ngn1 (BN1) or Brn3a and Ngn2 (BN2). After 8 d of induction, dox was removed and the cells were maintained in the absence of dox for an additional 6 d (termed day 14 of reprogramming). To our surprise, in both the BN1 and BN2 conditions, we observed numerous Tuj1-positive cells exhibiting stereotypical neuronal morphology, the majority (~90%) of which also coexpressed Map2, a marker of more mature neurons (Fig. 1a,b and Supplementary Fig. 1b). Control experiments using individual transcription factors (Ngn1, Ngn2 or Brn3a) failed to induce Map2 expression, although rare Tuj1-positive cells with predominantly non-neuronal morphologies were observed (Supplementary Fig. 1d). No Tuj1- or Map2-positive cells were found in untreated MEFs (Supplementary Fig. 1c,d). Finally, to establish the optimal induction time window, we performed time course experiments and found that 7–9 d of dox produced the greatest number of Map2-positive cells (Supplementary Fig. 1e). Subsequent experiments were performed using 8 d of induction, followed by six or more days of maturation in the absence of dox, which reproducibly produced Map2-, Tuj1- positive neuronal-like cells (Supplementary Fig. 1f).
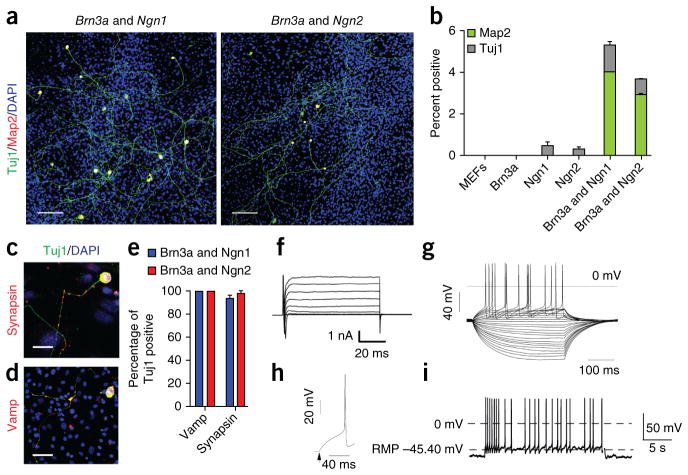
Figure 1.
Transient coexpression of two developmentally relevant transcription factors stably reprograms fibroblasts to attain properties of functionally mature neurons. (a) Transient coexpression of Brn3a with either Ngn1 or Ngn2 for 8 d in fibroblasts induced cells with neural morphology that stained for the pan-neural markers Map2 (red) and Tuj1 (green) Cells were immunostained on day 14. Scale bars represent 100 μm. (b) Coexpression of Brn3a with Ngn1 or Ngn2 was required for induction of Map2 and Tuj1 double-positive cells. Transcription factors were induced for 8 d and immunostained on day 14. Data are presented as means ± s.e.m. from three independent replicates. n = 50,000 cells per genotype. (c) Synapsin expression (red) in BN2 neural cells. Cells were counterstained for Tuj1 (green). (d) Vamp (red) expression in BN1 neural cells. Scale bars in c and d represent 25 μm. (e) The majority of neurons induced with BN1 and BN2 expressed synaptic markers indicative of mature neurons (Vamp, (BN1 = 100%, n = 52, BN2 = 100%, n = 44; Synapsin, BN1 = 93.4%, n = 47, BN2 = 97.9%, n = 44). Data are presented as means ± s.e.m. from two independent experiments. (f) Whole-cell currents recorded in voltage-clamp mode. Inward fast-inactivating Na+ and outward currents were observed. (g) Representative subthreshold voltage responses and action potentials evoked from neural cells after 14 d in culture. Current pulses of 350-ms starting at −145 pA. (h) Representative single voltage spike isolated from trace in g measured at +95-pA current level. (i) Representative train of spontaneous action potentials observed in 14-d culture.
More than 98% of Map2, Tuj1 double-positive cells induced by BN1 or BN2 expressed synapsin and synaptobrevin (also called VAMP), which, for endogenous neurons, suggests that they are capable of forming synapses and releasing synaptic vesicles (Fig. 1c–e). To assess the electrophysiological properties of the induced neurons, we performed whole-cell patch-clamp recordings on 12 cells with neuronal morphology that were identified using a genetically encoded reporter that labels endogenous DRG sensory neurons as well as induced neurons generated using our protocols (Pchd21::CRE × Ai9)33 (Supplementary Fig. 2a) Their resting membrane potentials ranged between −23 and −72 mV, with an average of −42.5 mV (s.e.m. = 14.6, n = 9 individual cells), which is similar to resting membrane potentials reported for other induced neuron21,22,26,27. Depolarizing voltage steps elicited fast inward currents followed by slow outward currents, consistent with the opening of voltage-dependent sodium and potassium channels, respectively (Fig. 1f and Supplementary Fig. 2b,c). Of the 12 recorded cells, 11 cells (91.6%) fired action potentials in response to depolarizing current steps and 3 cells exhibited spontaneous firing (Fig. 1g–i and Supplementary Fig. 2a,b). In addition, at least one neuron exhibited voltage sags consistent with the presence of inward-rectifying Ih currents (Supplementary Fig. 2a). Taken together, these data indicate that transient expression of BN1 or BN2 converts fibroblasts into neuron-like cells exhibiting electrophysiological properties consistent with a mature neuronal identity.
Neurons induced with BN1 or BN2 exhibit molecular and morphological hallmarks of peripheral sensory neurons
Peripheral sensory neurons are characterized by the expression of a number of relatively specific molecular markers, including neurofilaments, neurotransmitters and neuropeptides (Supplementary Fig. 3a). The intermediate neurofilament peripherin, which is primarily found in the peripheral nervous system, and the heavy neurofilament NF200 are selectively expressed in two distinct populations of sensory neurons, the C-fiber and A-fiber neurons, respectively34,35. We observe that, in both BN1 and BN2, populations of induced neurons expressed peripherin (BN1 = 64.17 ± 9.67%, BN2 = 51.93 ± 9.12%; Fig. 2a,b) and NF200 (BN1 = 29.3 ± 1.46%, BN2 = 30.60 ± 3.11%; Fig. 2a,c). Peripheral sensory neurons are excitatory glutamatergic neurons that express three vesicular glutamate transporters 1, 2 and 3 (also known as vGlut1 (Slc17a7), vGlut2 (slc17a6) and vGlut3 (slc17a8), respectively), but not GABA6,36–38. Similarly, BN1- and BN2-induced neurons expressed all three vesicular glutamate transporters, but not GABA (Fig. 2a and Supplementary Fig. 3b). A subset of nociceptive neurons also expresses the neuropeptide CGRP7,36. Approximately 10% of BN1- and BN2-induced neurons expressed CGRP (Fig. 2a and Supplementary Fig. 3b). These data indicate that neurons induced with BN1 and BN2 express characteristic neurofilaments, neurotransmitters and peptides found in peripheral sensory neurons.
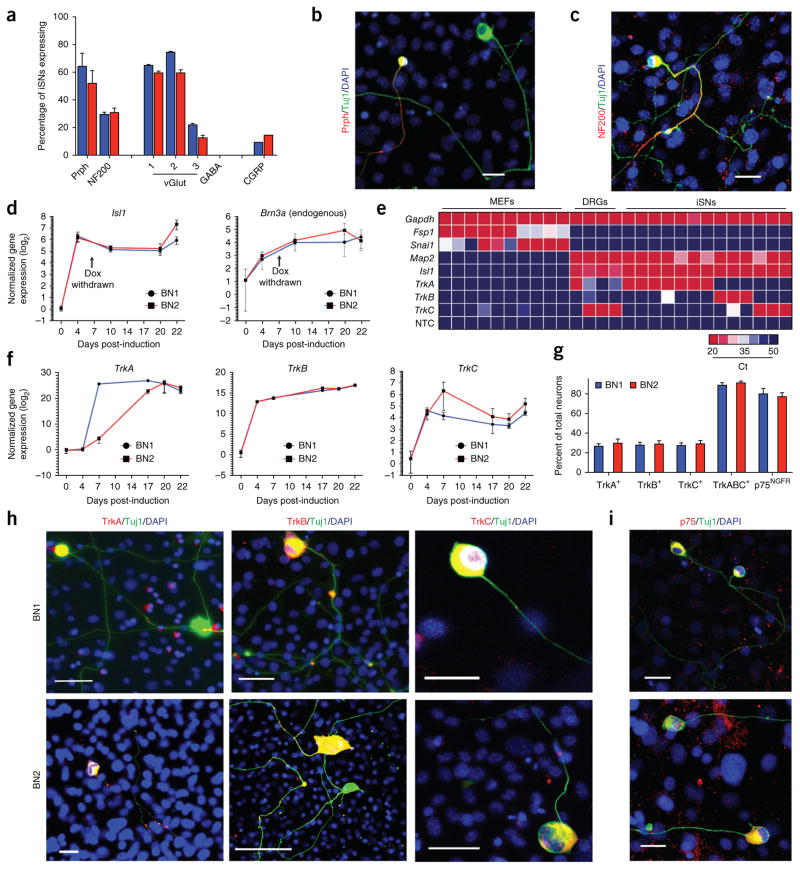
Figure 2.
Neurons induced with BN1 or BN2 exhibit molecular hallmarks of the peripheral sensory neural lineage. (a) Quantification of neurofilament, neurotransmitter and neuropeptide expression in BN1 and BN2 neurons. Expression was assessed 20 d post induction via immunostaining (Prph, BN1 = 64.2%, n = 253, BN2 = 51.9%, n = 204; NF200, BN1 = 29.4%, n = 353, BN2 = 30.5%, n = 307; data are presented as means ± s.e.m. from four independent experiments; vGlut1, BN1 = 64.8%, n = 57, BN2 = 59.5%, n = 52; vGlut2, BN1 = 74.5%, n = 129, BN2 = 59.4%, n = 123; vGlut3, BN1 = 21.9%, n = 138, BN2 = 12.4%, n = 158; GABA, BN1 = 0%, n = 238, BN2 = 0%, n = 243; CGRP, BN1 = 9.1%, n = 99, BN2 = 14.3%, n = 91; data are presented as means ± s.e.m. from two independent experiments). (b) A subset of neurons induced via BN1 or BN2 expressed the Prph (red). Scale bar represents 25 μm. (c) A subset of neurons induced via BN1 or BN2 expressed the peripheral neurofilament NF200 (red). Scale bar represents 25 μm. (d) Time course of quantitative RT-PCR analysis of Isl1 and endogenous Brn3a expression following induction of Brn3a and Ngn1 (blue) or Brn3a and Ngn2 (red). Fold induction was calculated as the increase in expression from un-induced fibroblasts cultured for the same duration in the same conditions for each time point. Dox was withdrawn permanently 8 d post-induction as indicated by the arrows. (e) Single-cell real-time RT-PCR of iSNs on day 20. NTC, no template control. (f) Fold induction of TrkA, TrkB and TrkC following induction of Brn3a and Ngn1 (blue) or Brn3a and Ngn2 (red). Fold induction was calculated as the increase in expression from un-induced fibroblasts cultured for the same duration in the same conditions for each time point. (g) Quantification of neurons expressing p75Ngf and each of the three Trk receptors individually or combined (TrkA, BN1 = 26.8%, n = 72, BN2 = 29.6%, n = 100; TrkB, BN1 = 27.6%, n = 146, BN2 = 29.0%, n = 178; TrkC, BN1 = 27.3%, n = 102, BN2 = 28.9%, n = 113; TrkABC, BN1 = 88.9%, n = 182, BN2 = 91.9%, n = 156; p75NGFR, BN1 = 80.3%, n = 86, BN2 = 79.9%, n = 117). Data are presented as means ± s.e.m. from two independent experiments. (h) Representative immunostaining for TrkA, TrkB and TrkC 20 d post-induction. (i) p75Ngf immunostaining 20 d post induction. Scale bars in h and i represent 25 μm.
Complete reprogramming involves the stabilization of neuronal transcriptional programs and the concomitant loss of genes expressed in the parental fibroblast. Mature peripheral sensory neurons express Isl1 and Brn3a, but no longer express the developmental neurogenic factors Ngn1 or Ngn2 (refs. 29,39,40). RT-PCR analysis of bulk populations revealed that both Isl1 and endogenous Brn3a expression were strongly upregulated during reprogramming with either BN1 or BN2, and their expression persisted after removal of dox (Fig. 2d). In contrast, endogenous Ngn1 and Ngn2 were initially upregulated during reprogramming, but their expression was not maintained after dox removal (Supplementary Fig. 3c). Neurons generated with BN1 or BN2 did not express detectable levels of markers of other neuronal subtypes such as Satb2, Ctip2, Brn2, Tbr2 and Tbx21 (data not shown). To assess whether induced neurons had downregulated fibroblast genes, we performed single-cell RT-PCR. These studies confirmed that neurons induced with BN1 and BN2 upregulated expression of Map2 and Isl1 and downregulated the fibroblast genes Snai1 and Fsp1 (Fig. 2e and Supplementary Fig. 3d). These data indicate that transient expression of BN1 and BN2 stably reprograms fibroblasts to establish a neuronal identity consistent with that of endogenous peripheral sensory neurons.
In vivo, selective expression of one of the three Trk receptors, TrkA, TrkB and TrkC, is critical for the execution of molecular programs that distinguish the three functional classes of sensory neurons6,41,42. Quantitative RT-PCR revealed that mRNA for TrkA, TrkB and TrkC was detectable as early as 4 d post-induction in both the BN1 and BN2 conditions. Trk receptor mRNA levels plateaued after 7 d, but were maintained for at least 22 d, indicating that conversion was stable and independent of the inducing factors (Fig. 2f). TrkA, TrkB and TrkC proteins were present in the soma and along the axons of neurons induced with either BN1 or BN2 (Fig. 2g,h). Each of the Trk receptors was found in approximately 25–30% of the induced neurons, suggesting that these cells comprise three distinct populations (Fig. 2e). To confirm this, we performed simultaneous staining with all three Trk antibodies, as well as with each pairwise combination. Co-staining for all three Trk receptors labeled approximately 90% of induced neurons (Fig. 2g), and each pair-wise combination of TrkA, TrkB and TrkC stained approximately 60% of induced neurons (Supplementary Fig. 3f). In addition, single cell RT-PCR on individual induced neurons revealed non-overlapping TrkA, TrkB and TrkC expression (Fig. 2e and Supplementary Fig. 3f). These data demonstrate that TrkA-, TrkB- and TrkC-positive cells represent three distinct populations that constitute the majority of neurons induced by BN1 or BN2. Sensory neurons also express the low-affinity nerve growth factor receptor p75 (ref. 43). Encouragingly, immunostaining showed that approximately 80% of neurons induced by BN1 and BN2 expressed p75 (Fig. 2g,i). Because neurons induced by BN1 or BN2 coexpress specific combinations of transcription factors, neurofilaments, neurotransmitters, neuropeptides and key characteristic of peripheral sensory neurons while selectively expressing one of the three Trk receptors, we referred to them as induced sensory neurons (iSNs).
In vivo, the size of sensory neuron cell bodies correlates with Trk receptor expression. TrkA-positive neurons have the smallest cell bodies, whereas TrkB-positive neurons are intermediate and TrkC-positive neurons are the largest44. TrkA-, TrkB- and TrkC-expressing iSNs generated by both BN1 and BN2 formed populations with distinct size distributions that were significantly different from each other (P < 0.0001) except for BN1-induced TrkB- and TrkC-expressing iSNs (P = 0.072) (Fig. 3a and Supplementary Fig. 4a–d). Sensory neurons have a single neurite that bifurcates distal to the cell body. Although this pseudounipolar morphology is lost when endogenous sensory neurons are cultured, the majority of induced neurons exhibited a pseudounipolar morphology in vitro (BN1 = 92.0 ± 0.4%, BN2 = 84.7 ± 1.4%; Fig. 3b,c and Supplementary Fig. 5). The remaining induced neurons were either unipolar without observable bifurcation (BN1 = 6.1 ± 0.4%, BN2 = 11.2 ± 1.4%) or bipolar (BN1 = 1.9 ± 0.02%, BN2 = 3.4 ± 0.1%) (Fig. 3b,c). In contrast, neurons generated using Brn2, Ascl1 and Zic1 (BAZ) were primarily multipolar26; less than 10% of neurons observed using these factors were pseudounipolar (Fig. 3c). These findings demonstrate that direct reprogramming can establish distinct neuronal morphologies in the absence of exogenous cues derived from the endogenous environment or appropriate synaptic partners.
Figure 3.
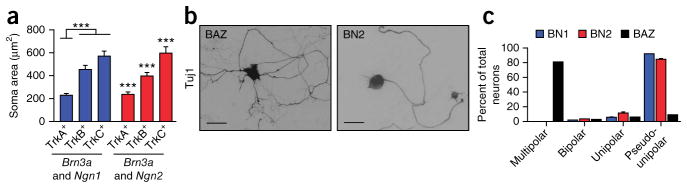
Reprogramming induces peripheral sensory neural morphology. (a) TrkA-, TrkB- and TrkC-immunoreactive neurons had distinct distribution of soma size. Graph depicts mean soma areas by Trk expression (TrkA, BN1 = 231 μm2, n = 56, BN2 = 238 μm2, n = 49; TrkB, BN1 = 456 μm2, n = 40, BN2 = 397 μm2, n = 47; TrkC, BN1 = 569 μm2, n = 40, BN2 = 598 μm2, n = 36). Error bars represent ±s.e.m. ***P < 0.001 (Kruskal-Wallis test) from two independent experiments (BN1: TrkA to TrkB, P = 1.8 × 10−8; TrkA to TrkC, P = 2.67 × 10−10; TrkB to TrkC, P = 0.072; BN2: TrkA to TrkB, P = 7.03 × 10−7; TrkA to TrkC, P = 5.39 × 10−10; TrkB to TrkC, P = 0.000916). (b) Typical morphology of pseudounipolar cells induced by BAZ and BN1 and BN2. Scale bars represent 100 μm. (c) The majority of Map2-, Tuj1-positive cells induced via Brn3a and Ngn1 or Brn3a and Ngn2 were pseudounipolar. Shown is the quantification of the neural morphologies observed in a representative experiments 14 d post-induction (multipolar, BN1 = 0%, BN2 = 0%, BAZ = 81.3%; bipolar, BN1 = 1.9%, BN2 = 3.4%, BAZ = 6.2%; unipolar, BN1 = 6.1%, BN2 = 11.2%, BAZ = 3.1%; pseudounipolar, BN1 = 92.0%, BN2 = 84.7%, BAZ = 9.3%). BN1, n = 212; BN2, n = 206; BAZ, n = 132. Bars represent means from two independent experiments. Error bars represent s.e.m.
Notably, these assays did not identify any major differences between neurons produced with BN1 or BN2, which contrasts with some results from mouse knockout studies29. One explanation for this could be that, during reprogramming, these factors have a general ‘proneural’ role. To address this question, we replaced Ngn1 and Ngn2 with the alternate proneural bHLH factor Ascl1 (Mash1). Expression of Ascl1with Brn3a induced cells that expressed Tuj1 and Map2 with similar efficiencies as with Ngn1 or Ngn2 with Brn3a (Supplementary Fig. 6a). Neurons induced by cooperative expression of Brn3a and Ascl1 exhibited primarily unipolar morphologies, consistent with either an immature phenotype or similarity with iSNs (Supplementary Fig. 6c,d). However, Brn3a-induced and Ascl1-induced neuronal cells did not express TrkA, TrkB or TrkC, indicating that they were not fully specified sensory neurons (Supplementary Fig. 6b). These data argue that, although Ngn1 and Ngn2 appear to be interchangeable in vitro, not every bHLH proneural factor can produce this neural subclass.
iSNs include cells that functionally resemble nociceptive sensory neurons
The utility of iSNs for mechanistic and screening studies depends on the ability of these neurons to recapitulate functional properties of biomedically relevant neural subtypes. Assays for distinguishing functional properties of sensory neurons are predominately limited to the nociceptive sub-lineage because the proprioceptive function requires innervation of Golgi tendon organs and skeletal muscle, and the mechanoceptive sub-lineage has mechanical responses that are similar to those of fibroblasts. We therefore focused on physiologically characterizing the TrkA lineage of nociceptive neurons. We first assessed expression of receptor ion channels Trpv1, Trpm8 and Trpa1, which detect heat, cold and noxious chemicals, respectively7. Trp channel expression was not detectable in uninfected MEFs or in neurons induced with the proneural cocktail Brn2, Ascl1 and Zic1 (BAZ; Fig. 4a and Supplementary Fig. 7a). However, iSNs induced with either BN1 or BN2 robustly expressed Trpa1, Trpm8 and Trpv1 (Fig. 4a and Supplementary Fig. 7a). In addition, BN1 or BN2 iSNs selectively expressed the voltage-gated sodium channel NaV1.7 (also known as Scn9a), which has a critical role in the generation and conductance of action potentials in nociceptors (Fig. 4a and Supplementary Fig. 7a).
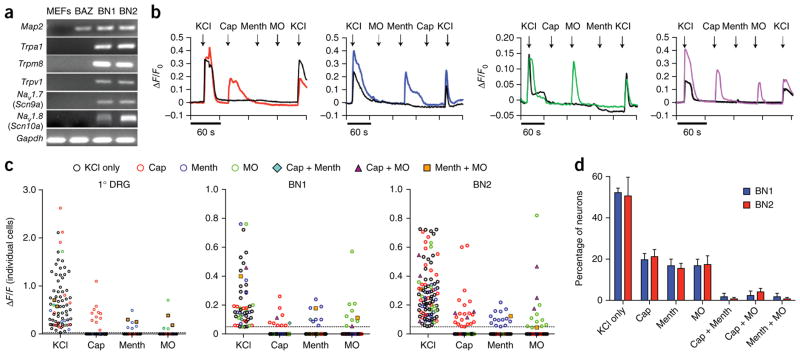
Figure 4.
iSNs exhibit functional properties of sensory neurons. (a) RT-PCR analysis of MEFs, neurons induced with BAZ and iSNs generated with BN1 or BN2. Trpa1, Trpm8, Trpv1 and NaV1.7 were detected in BN1 and BN2 conditions, but not in MEFs or BAZ. Full-length images are shown in Supplementary Figure 12. (b) Representative calcium responses for 10 μM capsaicin (Cap), 100 μM menthol (Menth) and 100 μM mustard oil (MO). Populations of BN1 and BN2 iSNs responded to Cap (BN1, 19.7 ± 3.0%; BN2, 21.2 ± 3.5%), Menth (BN1, 15.4 ± 2.6%; BN2, 17.4 ± 4.3%) and MO (BN1, 12.9 ± 3.8%; BN2, 12.5 ± 3.4%). A smaller population responded to two ligands sequentially: Cap and Menth (BN1, 1.7 ± 1.7%; BN2, 0.7 ± 0.7%), Cap and MO (BN1, 2.3 ± 2.3%; BN2, 4.1 ± 1.7%), and Menth and MO (BN1, 1.7 ± 1.7%; BN2, 0.7 ± 0.7%). Calcium transients were measured using Map2::GCAMP5.G. Calcium responses were calculated as the change in fluorescence (ΔF) over the initial fluorescence (Fo). Depolarization with 25 mM KCl was used at the beginning and end of each experiment to confirm neural identity and sustained functional capacity. (c) ΔF/F0 intensity plot showing the response of individual cells to each ligand for primary DRG neurons (1° DRG, n = 78), BN1 (n = 50) and BN2 (n = 139) iSNs. Each cell is represented in each column. Cells responded to either KCl only (black circle), KCl plus one other ligand (colored circle) or KCl plus two other ligands (blue diamond, purple triangle or orange square). (d) Distribution of KCl responders that responded to either KCl only, KCl plus one other ligand or KCl with two other ligands. Data are presented as means ± s.e.m. from at least four experiments.
Endogenous sensory neurons exhibit functional diversity in their responses to different Trp channel agonists. To determine whether iSNs exhibit similar diversity, we performed calcium imaging. We transduced iSNs with lentivirus encoding the fluorescent calcium reporter protein GCAMP5.G under the control of a Map2 promoter. To ensure that cells retained functional viability throughout the experiments, we transiently washed cells with 2.5 mM KCl at the beginning and end of each experiment. Only cells that responded in both cases were included in analysis. We serially exposed iSNs to menthol, allyl isothiocyanate (mustard oil) and capsaicin in a randomized order. These compounds selectively activate TrpM8, TrpA1and TrpV1, respectively8–11,20. Both BN1 and BN2 iSNs exhibited rapid transient calcium fluctuations in response to capsaicin, menthol and mustard oil (Fig. 4b–d). The majority of iSNs responded to only one ligand and a small subpopulation responded to two ligands sequentially. We did not observe any iSNs responding to all three sensory ligands. The responses of the iSNs were very similar to those of primary mouse DRG neurons, but were different from those of induced neurons generated with the BAZ factors, which did not respond to menthol or mustard oil (Fig. 4a,c and Supplementary Fig. 7b,c). We did, however, observe one capsaicin-responsive BAZ-induced iSN of the 51 neurons tested. Consistent with a minimal nociceptive capacity we failed to detect Trp channel expression in neurons induced with BAZ (Fig. 4a and Supplementary Fig. 7a).
To confirm that the calcium transients observed with GCAMP reflect cellular depolarization, we also measured sodium transients using the sodium dye CoroNa-Green. Depolarization with KCl increased the intracellular concentration of sodium in the majority (~90%) of Map2-positive iSNs (Supplementary Fig. 7d–f). Menthol or capsaicin elicited an increase in intracellular sodium in smaller subpopulations of Map2-positive iSNs (Supplementary Fig. 7d–f). Lidocaine, an inhibitor of voltage-gated sodium channels, blocked intracellular sodium increase, establishing the presence of functional voltage-gated sodium channels (Supplementary Fig. 7d–f). Collectively, these results demonstrate that iSNs are comprised of functionally diverse nociceptive neurons that express the primary receptors and signaling pathways required to selectively respond to biomedically relevant sensory stimuli.
Reprogramming fibroblasts to peripheral sensory neurons does not require proliferating or specialized embryonic precursors
In our reprogramming experiments, the three major subsets of sensory neurons (TrkA, TrkB and TrkC) reproducibly arose in roughly equivalent numbers. One mechanism for this could involve a multipotent proliferating precursor that differentiates into these three subtypes. To address this possibility, we performed two experiments. First, MEFs were reprogrammed in the presence of the proliferation marker EdU (5-ethynyl-2′-deoxyuridine, a click-chemistry BrdU analog) starting on the third day of induction. After 14 d of reprogramming, less than 0.5% of the Map2-positive cells were labeled with EdU (Fig. 5a,b). In a complimentary experiment, we inhibited mitosis with arabinofuranosyl cytidine (AraC). Mitotic inhibition from day 3 did not significantly alter the number of neurons produced during reprogramming (P > 0.05; Fig. 5c,d). These results indicate that iSNs are not derived from a continuously dividing progenitor and predict that the diversification of neurons may involve stochastic cell fate choices.
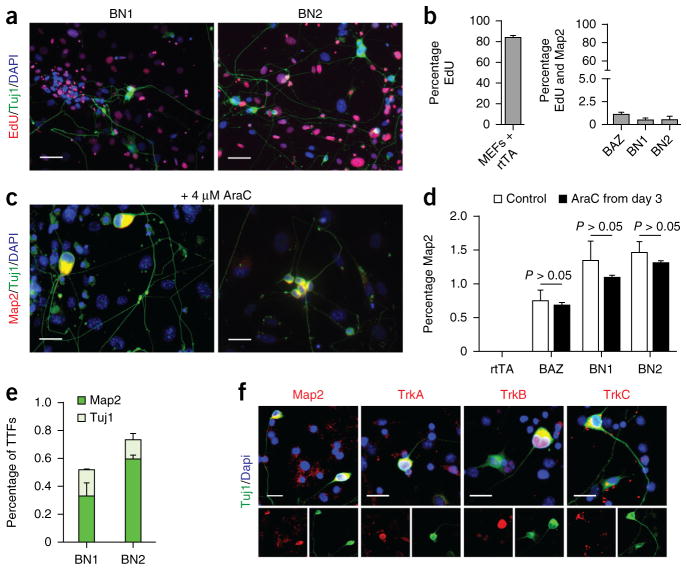
Figure 5.
iSN reprogramming does not require proliferating or specialized embryonic precursor. (a) EdU and Map2 staining 14 d post induction. Scale bars represent 25 μm. (b) Quantification of the number of Map2-positive cells that co-stained for the mitotic indicator EdU. rtTA indicates MEFs infected with reverse tetracycline trans-activator. BAZ indicates MEFs infected with Brn2, Mash1 (also known as Ascl1) and Zic1, a previously reported transcriptional cocktail for the direct conversion of MEFs to neurons26. Bar are means from two separate experiments. Error bars are s.e.m. For each condition, 50,000 cells were counted in three biological replicates. (c) BN1 and BN2 generated neurons in the presence of the mitotic inhibitor AraC. AraC was applied from 3 d post induction until the end of the experiment at 4 μM, a concentration empirically determined to inhibit >90% proliferative cells. Scale bars represent 25 μm. (d) No significant difference was observed in the number of Map2-positive neurons generated in the presence or absence of the mitotic inhibitor AraC (BAZ, P = 0.19; BN1, P = 0.49; BN2, P = 0.40). Bars are means of two independent experiments. Error bars are s.e.m. For each condition, 50,000 cells were counted in three biological replicates. (e) Quantification of Map2-, Tuj1-positive cells induced from tail-tip fibroblasts. n = 50,000 cells per genotype from two independent experiments. Error bars represent s.e.m. for each condition. (f) BN1 and BN2 induced neural cells from TTFs that stain for peripheral sensory markers. Scale bars represent 25 μm.
To examine whether iSN reprogramming may require the presence of embryonic precursor that is ‘pre-committed’ to this lineage, we characterized the parental MEFs by RT-PCR. MEFs expressed the fibroblast genes Fsp1 and Snai1, whereas the neural crest and neural progenitor markers Sox10, Nestin, Pax6, Pax3, Isl1 and Prph were not detectable (Supplementary Fig. 8). However, despite removal of neural tissue during MEF derivation, a small population of contaminating p75-positive cells was present (Supplementary Fig. 8a,b). To examine whether the p75-positive population was critical for reprogramming with BN1 or BN2, we purified p75-negative MEFs via FACS and subsequently reprogrammed them with BN1 or BN2. BN1 and BN2 reprogramming efficiency of unsorted versus p75-negative MEFs did not vary significantly (P > 0.05; Supplementary Fig. 8c,d). This suggests that the p75 population is not required for efficient reprogramming. To determine whether an embryonic cell type is required for BN1 or BN2 reprogramming, we derived tail-tip fibroblasts (TTFs) from 5-d-old pups. When we infected TTFs with BN1 or BN2 and reprogrammed via our protocol, Map2-, Tuj1-positive neural cells arose (Fig. 5e). As previously observed, these cells exhibited pseudounipolar morphologies and expressed the TrkA, TrkB and TrkC receptors (Fig. 5f).
BN1 and BN2 convert human fibroblasts into peripheral sensory neurons
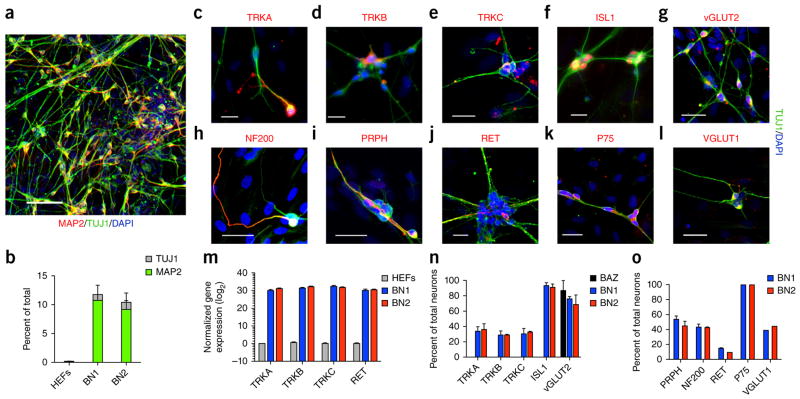
Next, we sought to determine whether this method could be applied to human cells. We produced human embryonic fibroblasts (HEFs) from induced pluripotent stem cells (iPSCs) using established methods22. We transduced the HEFs with inducible lentiviral vectors encoding either BN1 or BN2 and the dox-dependent activator rtTA as described in the mouse experiments. Transcription factor expression was induced for 8 d, followed by 7 d of culture in the absence of dox. On day 15 of reprogramming, we observed MAP2-, TUJ1-postive cells with neuronal morphologies in the BN1 and BN2 conditions, but not in control uninfected HEFs or in HEFs treated with single transcription factors (Fig. 6a,b).
Figure 6.
Human iSNs are generated using BN1 and BN2. (a) Expression of BN1 and BN2 converted HEFs into MAP2 and TUJ1 double-positive cells with neuronal morphologies 14 d after induction; dox was removed on day 8. Scale bar represents 100 μm. (b) Percentage of TUJ1-positive (gray) and MAP2 and TUJ1 double-positive (green) cells generated from HEFs in BN1, BN2 and rtTA control conditions. n = 50,000 cells per genotype from two independent experiments. (c–l) Neurons induced with BN1 and BN2 expressed peripheral sensory markers. Scale bars represent 25 μm. (m) Quantitative RT-PCR of TRK receptors in MEFs and iSNs generated with BN1 or BN2. Expression levels are normalized to MAP2. Bars and error bars represent means and s.e.m. from two independent biological replicates. (n) Quantification of TUJ1-positive cells expressing TRKA, TRKB, TRKC, vGLUT2 and ISL1 in BN1, BN2, BAZ and rtTA control conditions (TRKA, BN1 = 33.6%, n = 109, BN2 = 35.8%, n = 131, BAZ = 0%, n = 52; TRKB, BN1 = 28.6%, n = 138, BN2 = 29.0%, n = 135, BAZ = 0%, n = 109; TRKC, BN1 = 30.0%, n = 44, BN2 = 23.3%, n = 98, BAZ = 0%, n = 101; ISL1, BN1 = 93.2%, n = 278, BN2 = 90.5%, n = 27, BAZ = 0%, n = 181; vGLUT2, BN1 = 75.9%, n = 135, BN2 = 68.6%, n = 166, BAZ = 86.7%, n = 121). Bars represent means and error bars represent s.e.m. from three independent experiments. (o) Quantification of TUJ1-positive cells expressing NF200, PRPH and RET in BN1 and BN2 (PRPH, BN1 = 53.4%, n = 141, BN2 = 44.5%, n = 169; NF200, BN1 = 42.8%, n = 70, BN2 = 42.1%, n = 126; RET, BN1 = 14.3%, n = 223, BN2 = 9.1%, n = 154; P75, BN1 = 100%, n = 120, BN2 = 100%, n = 120; VGLUT1, BN1 = 38.5%, n = 122, BN2 = 44.0%, n = 184). Error bars represent s.e.m. from two independent experiments.
As seen with the mouse iSNs, human BN1- and BN2-induced neurons expressed TrkA, TrkB and TrkC, along with other characteristic markers of peripheral sensory neurons, as determined by immu-nostaining and RT-PCR assays (Fig. 6c–o). Each TRK receptor was expressed in approximately 30% of the MAP2-, TUJ1-positive cells (Fig. 6c–e,n), whereas the majority of neurons expressed the characteristic transcription factor ISL1 (BN1 = 93.15 ± 4.14%, BN2 = 90.5 ± 5.14%; Fig. 6f,n). In contrast, neurons derived using the pan-neural cocktail Brn2, Ascl1 and Zic1 (BAZ) did not express these markers (Fig. 6n). However, both BN1 and BN2 and BAZ neurons were predominantly glutamatergic, as measured by immunostaining with vGLUT2 (Fig. 6g,n). BN1 and BN2 iSNs also expressed VGLUT1 (Fig. 6l,o), but not GABA (data not shown). BN1 and BN2 iSNs also expressed p75 in nearly all iSNs (Fig. 6k,o), whereas the GDNF receptor c-RET was expressed in a smaller subset (Fig. 6j,o). Finally, iSNs expressed the characteristic neurofilaments peripherin (PRPH) and NF200 that are associated with C- and A-fiber neurons, respectively (Fig. 6h,i,o). This pattern of coordinated gene expression and subtype diversity suggests that a sensory neuron phenotype was selectively imparted to human fibroblasts through transient ectopic expression of Brn3a with either Ngn1 or Ngn2, as we previously observed with mouse cells.
To produce iSNs more rapidly, we would find it useful to generate them directly from adult human cells, without involving an iPSC intermediate. We therefore tested whether reprogramming with BN1 or BN2 could convert adult human dermal fibroblasts to neurons. BN1 and BN2 were transiently expressed in fibroblasts for 8 d and subsequently silenced via removal of dox. After 14 d of reprogramming, MAP2-, TUJ1-positive cells with neuronal morphologies were observed in approximately 1% of starting fibroblasts (Supplementary Fig. 9a,b). In addition, more than 90% of the neurons induced from human dermal fibroblasts expressed ISL1 (Supplementary Fig. 9c) and approximately 30% expressed either TRKA, TRKB or TRKC receptor (Supplementary Fig. 9d,e). These results indicate that generation of human iSNs does not require embryonic fibroblasts and establish a second method for producing patient-specific sensory neurons.
Human iSNs exhibit molecular and physiological properties of mature sensory neurons
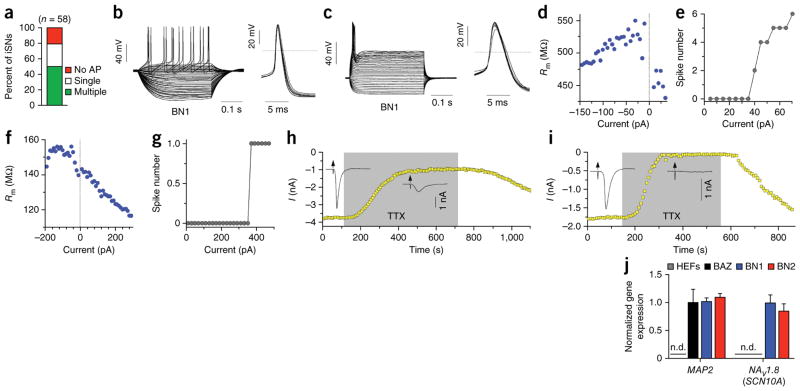
To determine whether human iSNs display membrane and physiological properties of mature neurons, we performed patch-clamp recordings on cells with neuronal morphologies, as identified by phase-contrast microscopy (Supplementary Fig. 10a). In total, we recorded from 58 cells, of which 46 (79%) displayed a healthy resting membrane potential and fired action potentials in response to depolarizing current pulses (Fig. 7a). On the basis of responses to current steps, we classified cells as either single-spiking or multiple-spiking iSNs, which were observed in both BN1 (n = 36) and BN2 (n = 10) neurons (Fig. 7a–c and Supplementary Fig. 10b,c). The multiple-spiking iSNs increased their firing rate to increasing depolarizing current steps (Fig. 7b,d,e and Supplementary Fig. 10b,d,e). The single-firing iSNs exhibited a single action potential in response to suprathreshold depolarizing currents, even at levels greater than 50 pA above their rheobase (average = 77.5 pA; Fig. 7c,f,g and Supplementary Fig. 10c,f,g). The average resting membrane potential for the single-spiking and multiple-spiking types were similar at −54.0 and −55.0 mV, respectively (n = 17 and n = 29 individual cells; Supplementary Fig. 10h). The mean amplitude of first action potentials in single spiking iSNs and the slope of their input-output relationship (I-O gain) was significantly lower than those is regular multiple-spiking iSNs (Supplementary Fig. 10h). Notably, most iSNs displayed a high input resistance at rest and little to no sag in response to strongly hyperpolarizing current steps. Depolarizing voltage steps elicited fast inward currents followed by slow outward currents, consistent with the opening of voltage-activated sodium and potassium channels, respectively (Fig. 7h). These data demonstrate that human iSNs possess membrane and electrophysiological properties that are characteristic of functional neurons.
Figure 7.
Human iSNs display physiological properties of mature sensory neurons. (a) The observed frequency of iSNs types in all cells patched. (b,c) Representative traces from whole-cell patch-clamp recordings showing multiple-spiking (b) and single-spiking (c) type iSNs, with single action potentials shown to the right. (d,f) The input resistance of multiple-spiking (d) and single-spiking (f) iSNs plotted as a function of the injected current. (e,g) The number of action potentials fired at increasing levels of current for multiple-spiking (e) and single- spiking (g) iSNs. (h,i) Time course of inward current during perfusion with 100 nM TTX (gray area) and during wash out (white area). Insets are representative current traces before and during the TTX application. The effect was reversible in both cells. (j) iSNs selectively expressed the TTX-resistant sodium channel SCN10A. Expression of MAP2 and SCN10A in HEFs, BAZ iSNs and iSNs generated by BN1 and BN2 was measured by real-time RT-PCR from two independent experiments. Gene expression was normalized to BN1 iSNs. n.d. signifies undetectable expression of that gene. Error bars represent s.e.m. from three experiments.
An additional characteristic of some nociceptive neurons is resistance to TTX. TTX resistance is predominantly mediated by SCN10A (also known as NaV1.8). To examine whether a population of iSNs are TTX resistant, we stimulated the iSNs in 8-s intervals using −20-mV command steps of 50-ms duration and applied 100 or 300 nM TTX in the bath. In all experiments, TTX blocked Na+ currents reversibly, but the degree of the blocking effect varied across cells (Fig. 7h,i). In the presence of 100 nM TTX under steady state conditions, ~36% of iSNs (n = 11) exhibited residual Na+ currents greater than 15% of the original inward Na+ current. Similarly, even in the presence of 300 nM TTX, 25% of iSNs (n = 8) exhibited residual Na+ currents over 12% of the original current. In addition, we found that SCN10A was expressed in iSNs derived with BN1 and BN2, suggesting that the TTX resistance that we observed could derive from this channel, as it does in vivo (Fig. 7j).
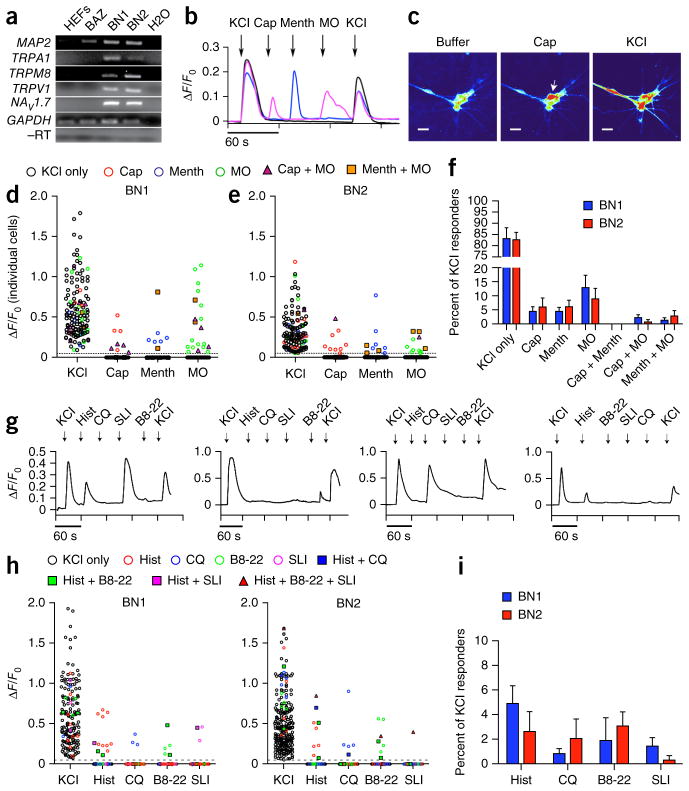
As in the mouse, human nociceptive neurons express TRPA1, TRPM8, TRPV1 and NaV1.7. Human iSNs generated with both BN1 and BN2 exhibited robust expression of the TRP channels and NaV1.7, whereas we detected no expression in HEFs or in induced neurons generated with the BAZ factors (Fig. 8a). To determine whether the TRP channels were functional, we performed calcium-imaging studies, as described for the mouse iSNs. To further validate the GCAMP analysis, we immunostained human iSNs for MAP2 and TUJ1 following addition of KCl. We found that GCAMP signals occurred in iSNs and that the vast majority (~90%) of iSNs were calcium responsive (Supplementary Fig. 11a–d). Human iSNs generated with either BN1 or BN2 exhibited rapid and selective calcium transients in response to capsaicin, menthol and mustard oil (Fig. 8b–e). A few iSNs responded to two ligands sequentially, including mustard oil with either capsaicin or menthol; however, we did not detect any neurons that simultaneously responded to both capsaicin and menthol, or to all three ligands. These calcium fluxes in response to ligand specific activation suggest that TRPA1, TRPM8 and TRPV1 are functional in human iSNs (Fig. 8d–f).
Figure 8.
Human iSNs possess functional properties of sensory neurons. (a) RT-PCR analysis showing presence of ligand gated calcium channels TRPA1, TRPM8, TRPV1 and the voltage-gated sodium channel NaV1.7 in BN1 and BN2 human iSNs compared with BAZ and uninfected controls (HEFs) at 16 d post induction. BN1, BN2 and BAZ samples expressed MAP2 compared with uninfected controls. TRPA1, TRPM8, TRPV1 and NaV1.7 were present only in BN1 and BN2. GAPDH amplification in samples provided the loading control, and GAPDH used in the reverse transcriptase–negative (−RT) control for contaminating genomic DNA. Full-length images are shown in Supplementary Figure 12. (b) Representative calcium responses for 10 μM capsaicin (Cap), 100 μM menthol (Menth), and 100 μM mustard oil (MO). Calcium transients were measured using Map2::GCAMP5.G. Calcium responses were calculated as the change in fluorescence (ΔF) over the initial fluorescence (Fo). Depolarization with 25 mM KCl was used at the beginning and end of each experiment to confirm neural identity and sustained functional capacity. (c) Screenshot images of GCAMP fluorescence in BN2 iSNs in response to the addition of buffer, 10 μM capsaicin (cap) and 25 mM KCl. Scale bars represent 25 μm. (d,e) ΔF/F0 intensity plot showing the response of individual cells to each ligand. Each cell is represented in each column. Cells respond to KCl only (black circle), KCl plus one other ligand (colored circle) or KCl plus two other ligands (triangle or square). (f) Distribution of ligand response in BN1 (n = 146) and BN2 (n = 138). Populations of iSNs responded to Cap (BN1, 4.4 ± 1.7%; BN2, 5.9 ± 3.3%), Menth (BN1, 4.4 ± 1.4%; BN2, 6.0 ± 2.3%) and MO (BN1, 12.8 ± 4.5%; BN2, 8.8 ± 3.7%), with small subpopulations responding to two ligands sequentially. iSNs responded to two ligands sequentially: Cap and MO (BN1, 3.2 ± 0.7%; BN2, 1.2 ± 1.2%), Menth and MO (BN1, 1.9 ± 1.9%; BN2, 1.1 ± 1.1%). The majority of KCl responsive iSNs did not respond to any of the three ligands (BN1, 83.1 ± 4.9%; BN2, 82.6 ± 3.4%). Bars represent means and error bars represent s.e.m. from seven independent experiments. (g) Representative calcium responses for 100 μM histamine (Hist), 100 μM chloroquine (CQ), 10 μM BAM8-22 (B8-22), 10 μM SLI-GRL (SLI). (h) ΔF/F0 intensity plots of three separate ligand combination regimes showing the response of individual cells to each ligand. Each cell is represented in each column. Cells responded to KCl only (black circle), KCl plus one other ligand (colored circle) or KCl plus two other ligands (triangle or square). (i) Distribution of ligand response in BN1 and BN2 to Hist (BN1, 4.8 ± 1.4%; BN2, 2.6 ± 1.6%), CQ (BN1, 0.8 ± 0.4%; BN2, 2.0 ± 1.6%), B8-22 (BN1, 1.8 ± 1.8%; BN2, 3.1 ± 1.1%), SLI (BN1, 1.5 ± 0.7%; BN2, 0.3 ± 0.3%), Hist and CQ (BN1, n = 288; BN2, n = 302), and B8-22 and SLI (BN1, n = 183; BN2, n = 234). Bars represent means and error bars represent s.e.m. from three independent experiments.
In addition to detecting pain and temperature, subsets of peripheral sensory neurons also mediate itch. Although the pathways regulating human and mouse itch are not well defined and appear to differ between species, numerous compounds that promote itch are known to act on pruritoceptors12–15 We tested whether subsets of human iSNs respond to known pruritogenic compounds, such as histamine, chloroquine, and the peptides BAM 8-22 (B8-22) and SLIGRL (SLI). Indeed, distinct subsets of human iSNs responded selectively to histamine, chloroquine, B8-22 and SLI (Fig. 8g–i). Collectively, these results indicate that BN1 or BN2 can reprogram human fibroblasts to iSNs that are capable of responding to pruritogens in addition to exhibiting numerous other features of endogenous sensory neurons.
DISCUSSION
We found that two different combinations of developmentally relevant transcription factors were sufficient to convert mouse and human fibroblasts into electrically active induced neurons that expressed key markers of peripheral sensory neurons, recapitulated aspects of sensory neuron morphology, and exhibited selective responses to chemical mimics of pain, temperature and itch. Thus, our results provide several lines of evidence that iSNs are functional phenocopies of peripheral sensory neurons and, as a result, may have utility for the study of sensory neuron biology, disease and drug screening.
A key question raised by these findings is the extent to which iSNs resemble their endogenous counterparts. iSNs are specified in an environment that is markedly different from the developing spinal cord. However, the vast majority of iSNs exhibit multiple characteristics of endogenous sensory neurons that persist after reprogramming factor withdrawal. Notably, several of these features, such as pseudounipolar morphology and selective responses to ligands for the Trp and Mrgpr receptor families, are rarely observed in other neuronal subtypes. Furthermore, all of the molecular markers that we tested were present in iSNs, with one exception: mouse iSNs lacked a Ret-positive population. In contrast, a subpopulation of human iSNs expressed RET, suggesting that variation in donor cells can influence the outcomes of reprogramming. Functionally, the majority of iSNs fired action potentials and a subset of iSNs exhibited signatures of TTX resistant channels, but we did not detect signs of presynaptic input. This, perhaps, is not surprising, as we did not supply iSNs with appropriate presynaptic neural subtypes. Given the extent to which iSNs resemble their endogenous counterparts in the absence of the appropriate environment, it is tempting to speculate that BN1 and BN2 engage an evolutionarily conserved feedforward transcriptional cascade that induces cell autonomous features of endogenous sensory neurons. However, it is also possible that deficiencies in iSNs reflect incomplete reprogramming by BN1 and BN2 or inherent limitations of reprogramming methods.
Another similarity between iSNs and endogenous sensory neurons is their extensive cell type diversity. Endogenous sensory neurons exhibit diversity in their expression of Trk receptors, neurofilaments and neuropeptides, and in their responses to pain- and itch-inducing ligands. We found that iSNs also exhibited this diversity. However, in contrast with endogenous development, diversity among iSNs arose without transit through a dividing intermediate or exposure to patterning cues found in the developing nervous system. How does reprogramming with two transcription factors produce this extensive cellular subtype diversification? Neuronal diversity can arise via determined mechanisms based on extrinsic cues or cell-intrinsic differences, or from stochastic activation of gene expression. Determined mechanisms that could influence the outcome of reprogramming include inherent diversity in the donor cell population or variable expression of transcription factors. However, in the case of Trk receptor expression, notably consistent proportions of the TrkA, TrkB and TrkC were maintained across experiments performed with four different cell populations and multiple viral preparations. This suggests that determined mechanisms based on donor cell type or factor stoichiometry are not major determinants of iSN diversity. Alternatively, Trk receptor expression could arise stochastically, such that each donor cell has a roughly equivalent chance of diversifying into each of the three sublineages. Similar stochastic mechanisms have been shown to produce diversity among olfactory sensory neurons and fly photoreceptors45. Establishing the means by which iSNs diversify will be important to determine the extent to which it is possible to selectively specify desired subsets of sensory neurons using reprogramming.
We found that the iSNs that we generated with either BN1 or BN2 were markedly similar. This contrasts with mouse developmental studies that reported distinct roles for Ngn1 and Ngn2 in sensory neuron specification. In these studies, Ngn1 null mice exhibit a nearly complete loss of TrkA nociceptive neurons29, whereas mice lacking Ngn2 exhibit a reduction in TrkB and TrkC neurons that is later restored by differentiation of Ngn1-dependent precursors into the TrkB and TrkC lineages29. These results predict that BN1 would preferentially generate TrkA neurons, whereas BN2 would produce iSNs biased toward the TrkB and TrkC subtypes. However, we found that both factor combinations reproducibly induced TrkA, TrkB and TrkC expression in equal numbers of neurons, and that other properties that segregate with Trk receptor expression were distributed similarly in iSNs produced with BN1 or BN2. One possible explanation for this is that Ngn1 and Ngn2 transactivate one another during reprogramming, resulting in functionally equivalent expression levels in cells treated with either combination. Alternatively, Ngn1 and Ngn2 may have overlapping or equivalent activities during reprogramming, which is consistent with developmental studies29. Notably, we found that Ascl1 could not substitute for Ngn1 and Ngn2 in generating iSNs, suggesting that Ngn1 and Ngn2 have a specific role in producing iSNs.
Modeling the personalized biology of pain and sensory pathology has long been a major challenge for pharmacotherapy. It is becoming increasingly recognized that genetic factors are a major contributor to personalized pain phenotypes and associated pharmacological responses. Our results suggest a new a method for producing electrically active iSNs from humans of diverse genetic backgrounds. Accordingly, employing iSNs rather than human cadaveric tissue or animal models may allow more accurate modeling of human sensory neuron function and disease. For example, we found that subsets of iSNs responded to ligands that mimic itch. The biology of itch and the underlying mechanisms have only recently been elucidated in rodents12,14,29. In humans, less is known about itch, in part because of divergence of pruritogenic receptors between species and the scarcity of human somatosensory neurons to study in the laboratory14. In addition, we found that iSNs expressed physiologically active TRP ion channels that mediate functional responses to noxious sensory ligands, thereby enabling cell-based studies of pain in diverse patient populations. This method also produces candidate mechanoreceptors and proprioceptors, which may be useful for investigating the molecular basis of touch and to model degenerative diseases that affect subsets of peripheral sensory neurons, such as Friedreich’s ataxia. Finally, given that iSNs may be generated from fibroblasts derived from human pluripotent cells, this method is compatible with genome editing approaches that enable selection of specific subtypes of sensory neurons, allowing more precise analyses of the functional effect of particular genetic variants in the context of the appropriate neuronal subtype.
ONLINE METHODS
Embryonic fibroblasts isolation and derivation
Wild-type CD1 mice were bred at The Scripps Research Institute animal facility. MEFs were isolated from E14.5 embryos under a dissection microscope. The head, internal organs and spinal column containing the dorsal root ganglion was removed and discarded to eliminate cells with neurogenic potential. The remaining tissue was manually disassociated in 0.25% trypsin (vol/vol, Gibco) for 10 min at 37 °C subsequently the digestion solution was diluted and removed via centrifugation and subsequently seeded onto tissue culture plates. MEFs were grown to confluence and passaged at least twice before use. For HEF differentiation, human iPSCs colonies were harvested using 1 mg ml−1 collagenase type IV and differentiated by embryoid bodies (EBs) formation. The EBs were cultured for 7 d in non-adherent suspension culture dishes (Corning), 2 d in 20% KSR medium and the following 5 d in 10% FBS DMEM (vol/vol). On day 8, the EBs were plated onto adherent tissue culture dishes and passaged according to primary fibroblast protocols using trypsin for three passages before the start of experiments.
Molecular cloning, cell culture and lentiviral infection
The cDNAs for human BRN3A (97% homologous to mouse Brn3a peptide) and mouse Ngn1 and Ngn2 were cloned into lentiviral constructs under the control of tetracycline operator (TetO) using the following primers: BRN3A forward and reverse, respectively, 5′-ATGATGTCCATGAACAGCAAGCAG and 5′-TCAGTAAGTGGCAGAGAATTTC. Ngn1 forward and reverse, respectively, 5′-ATGCCTGCCCCTTTGGAGACC and 5′-TTCAGCGAGGGTGCAGCAACC. Ngn2 forward and reverse, respectively, 5′-ATGTTCGTCAAATCTGAGACTCT GG and 5′-AAACCAGAGCTGGTCTCCACC. Replication-incompetent VSVg-coated lentiviral particles were packaged in 293T cells as previously described33. Passage three CD1 MEFs were infected with lentivirus in MEF media (DMEM + 10% FBS and penicillin/streptomycin). After 12–16 h of infection media-containing virus was replaced with fresh MEF media. Transcription factors were induced 48 h post infection media by switching to MEF media supplemented with 5 μg ml−1 dox (Sigma). On day 4, media was changed with N3 media with 5 μg ml−1 dox (Sigma)26. 7 d post induction, dox was withdrawn unless otherwise stated. 10 d post-induction, media was replaced with neural maintenance media, which consisted of a 1:1 mix of DMEM/F12 (Invitrogen) and Neurobasal supplemented with B27, and NGF, BDNF and GDNF, all at 10 ng ml−1. Efficiency of conversion was measured by the number of Map2-positive cells divided by to the initial number of cells plated. In experiments with very low conversion efficiencies, we observed that reprogrammed cells exhibited neuronal morphology and marker expression that differed from the results of typical experiments, perhaps as a result of infrequent coexpression of the reprogramming factors or to other conditions related to cell culture. Thus, only experiments with conversion efficiency greater than 1% of starting population were analyzed.
Immunohistochemistry, and RT-PCR
For immunofluorescence staining, cells were fixed 4% paraformaldehyde (vol/vol) for 10 min at 20–24 °C. Cells were then washed three times with phosphate-buffered saline (PBS) and subsequently permeabilized with 0.1% Triton X-100 (vol/vol, Sigma) in PBS. After washing and permeabilization, cells were blocked in 5% horse serum (vol/vol) for 30 min at 20–24 °C. Primary staining was performed overnight at 4 °C in block. Secondary antibodies were diluted in blocking solution and stained at 20–24 °C for 1 h. EdU staining was performed using Click-it EdU kit (C10337, Invitrogen) following manufacturer’s instructions. The following antibodies and dilutions were used: Ms-βIII-Tubulin (Tuj1) (1:1,000, Covance MMS-435P), Rb-βIII-Tubulin (Tuj1) (1:1,000, Covance MRB-435P), Ms-Map2 (1:500, BD 556320), Ms-VGlut1 (1:100, Millipore MAB5502), Rb-Vglut2 (1:50, abcam ab72310), Gp-VGlut3 (1:1,000, Millipore AB5421), Ms-Brn3a (1:200, Millipore MAB1585), Gt-human Ret (1:100, R&D AF1485), Gt-mouse Ret (1:100, R&D AF482), Ms-Islet1 (1:200, DSHB 40.2D6), Gt-mouse TrkB (1:200, R&D BAF1494), Gt-TrkA (1:200, R&D AF175), Sh-TrkC (1:200, Abcam ab72120), Ms-NF200 (1:200, Millipore MAB5266), Rb-Peripherin (1:200, Millipore AB1530), Rb-Gaba (1:500, Sigma A2052), Rb-Ngn1 (1:500, Abcam 66498), Rb-Ngn2 (1:500, Millipore 5682), Rb-P75 (Abcam 8874), Rb-CGRP (1:500, Neuromics RA24112), Ms-VAMP (1:200, Synaptic systems 104 211), Rb-Synapsin (1:1,000, E028). Secondaries: Life Technologies: antibody to goat (1:1,000, A21447D-G647), antibody to mouse (1:1,000, A10036-DM546), antibody to mouse (1:1,000, A21202-DM488), antibody to rabbit (1:1,000, A10040-DR546), antibody to sheep (1:1,000, A11015-DSh488), antibody to goat (1:1,000, A11056-DGt546), antibody to rabbit (1:1,000, A21206-DRb488), antibody to rat (1:1,000, A21208-DRt488) and antibody to sheep (1:1,000, A21098-DSh546).
For RT-PCR analysis, total RNA was isolated at the time points indicated using Trizol (Invitrogen) following manufacturer’s instructions, treated with DNaseI (Ambion) and 1.0 μg was reverse transcribed with iScript (BioRad). PCRs were performed using TaqMan Gene Expression Assays (Applied Biosystems) or SYBR green. For quantitative RT-PCR from single cells, single cells were grown on glass coverslips from which they were isolated 3 weeks after induction using a using a patch pipet and micromanipulator. Cells were placed in 4 μl of lysis/RT buffer consisting of Superscript III RT buffer (Invitrogen) supplemented with 0.5% (vol/vol) NP-40, 1 mM DTT, and SuperRnase inhibitor (Ambion), and Prime RNase inhibitor (5 Prime). Cells were spun down in a microcentrifuge and flash frozen at −80 °C until further processing. Reverse transcription was performed using SuperScript III (Invitrogen) with 130 nM of each gene-specific 3′ primers. Reverse transcription products were than subjected to 15 cycles of target specific pre-amplification using 15 nM of outside nested primers designed to produce amplicons of 300–400 bp. Quantitative real-time PCR was subsequently performed using SYBR select (Applied Biosystems, 4472918) with internal primers designed to generate amplicons approximately 100 base pairs. To ensure specificity, template titrations were performed and only primers that demonstrated linear amplifications were used. Melt curves were also obtained for single cells and controls to ensure specificity of products.
Statistics
For samples assumed to have a normal distribution, we applied one-way ANOVA with Newman-Keuls multiple comparison test. In Figure 3a, populations were determined not to have a normal distribution using the D’Agostino & Pearson omnibus test. The comparisons between populations without normal distribution used the non-parametric Kruskal-Wallis test. For Figure 5d, we used an unpaired Student’s t test; data distribution was assumed to be normal, but was not formally tested. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications21,22,25,27.
Calcium imaging
Calcium imaging was performed on mouse and human iSNs 2–3 weeks post-induction using Map2::GCAMP5.G lentiviral reporter46. Imaging was performed in Tyrode’s solution (145 mM NaCl, 2.5 mM KCl, 10 mM Hepes, NaH2PO4, 2 mM CaCl2, 1 mM MgCl2, 10 mM Glucose, and 0.4 mM ascorbic acid) at a constant flow rate of 250 ml h−1. To monitor calcium response, capsaicin, menthol and mustard oil were added sequentially in randomized orders to the flow chamber at a 10× concentration to deliver a final concentration of 10 μM capsaicin, 100 μM menthol and 100 μM mustard oil. Each tracing experiment was bracketed by an initial and final pulse of 25 mM KCl to confirm neural identity and sustained functional ability. Only cells with neural identity and sustained functional ability were analyzed. Calcium responses were determined by calculating the change in fluorescence over the initial fluorescence (F − F0)/F0, where F = the fluorescence at a given time point and F0 = the mean basal, unstimulated fluorescence of each cell. A typical non-response area was selected for fluorescence bleed normalization and background subtraction. The threshold for a positive calcium response to the addition of a ligand was determined as 1 (F − F0)/F0 greater than 0.03 in a 7.4-s window. The intensity threshold for a positive GCAMP response at (F − F0)/F0 > 0.03 was determined as 5 s.d. above the mean of s.d. from (F − F0)/F0 recordings in BN1 and BN2 neurons during a period of no stimulation (n = 20). The 7.4-s response window was determined as one s.d. above the mean response time from KCl addition to peak GCAMP intensity in BN1 and BN2 neurons (n = 30).
Electrophysiology
Fibroblasts were plated, transduced and cultured on laminin coated thermanoxplastic coverslips (13 mm) as described in cell culture methods. Coverslips were placed in the recording chamber mounted on a Nikon Eclipse FN microscope. Spontaneous activity and evoked responses were recorded at 33 °C under whole-cell patch clamp. Signals were amplified using a MultiClamp700B (Molecular Devices) and acquired at 20 kHz using the data acquisition software DASYLab v.11 (National Instruments). Patch pipettes were pulled from standard wall glass of 1.5-mm OD (Warner Instruments) and had input resistances of 6–8 MOhm. The composition of the bath solution (artificial cerebrospinal fluid) was 125 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, 25 mM glucose. For recording action potentials and voltage-gated currents, patch electrodes were filled with the following solution 120 mM potassium-glutonate, 10 mM KCl, 10 mM HEPES, 10 mM EGTA, 2 mM MgATP, 0.3 mM Na3GTP; pH 7.3. To record voltage responses of the iPSCs, we used incrementing levels of current steps of 350-ms duration. Initial current step level was −50 to −200 pA depending on the observed input resistance of the cell and the steps were incremented by +5 pA. Analysis of the evoked responses was performed in software developed by A. Szücs (IVAnalyzer). Several physiological parameters including the resting membrane potential, rheobase, input resistance at rest, spike amplitude and half-width among others were determined for each cell. To monitor voltage-gated membrane currents, we applied step commands of 50-ms duration and ranging from −60 to +10 mV. In experiments with TTX application, we monitored Na+ currents at −20 mV voltage level using 8-s interstimulus intervals. Outward K+ currents at −20 mV command level were generally minor relative to the inward Na+ component. Leakage correction was performed digitally by using a negative step (−90 mV) preceding each stimulus command. In experiments characterizing TTX resistivity of Na+ currents, we applied TTX at 100–300 nM concentrations in bath and recorded control levels, TTX effect and washout in the same file.
A Supplementary methods checklist is available.
Supplementary Material
Acknowledgments
We would like to thank M. Talantova and D. Zhang for assistance with mouse electrophysiology, K. Spencer for assistance with microscopy, A. Patapoutian, S. Murthy, A. Dubin, S. Ranade and J. Mathur for helpful discussions, and W. Ferguson for technical assistance. This research was supported by the National Institute on Drug Abuse (DA031566 to P.S.), the National Institute on Deafness and other Communication Disorders (DC012592 to K.K.B.), the National Institute of Mental Health (MH102698 to K.K.B.), the California Institute for Regenerative Medicine (RB3-02186 to K.K.B.), the Baxter Family, Norris and Del Webb Foundations (K.K.B.), by Las Patronas and the Dorris Neuroscience Center (K.K.B.), a pre-doctoral fellowship from the California Institute of Regenerative Medicine (J.W.B. and R.K.T.), an NSF Predoctoral Fellowship (R.K.T.), the Andrea Elizabeth Vogt Memorial Award (J.W.B.) and the Scripps Stem Cell Postdoctoral Fellowship (V.L.S.).
Footnotes
AUTHORS CONTRIBUTIONS
K.K.B., J.W.B. and K.T.E. designed and conceived the experiments. K.T.E., J.W.B., V.L.S., R.K.T. and D.W. performed the experiments. A.S. and P.P.S. performed electrophysiology. K.K.B., J.W.B. and K.T.E. wrote and revised the manuscript and all of the authors edited the final drafts.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Finnerup NB, Sindrup SH, Jensen TS. The evidence for pharmacological treatment of neuropathic pain. Pain. 2010;150:573–581. doi: 10.1016/j.pain.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 2.Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci. 2002;5:1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 3.Koeppen AH, et al. The dorsal root ganglion in Friedreich’s ataxia. Acta Neuropathol. 2009;118:763–776. doi: 10.1007/s00401-009-0589-x. [DOI] [PubMed] [Google Scholar]
- 4.Riera CE, et al. TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell. 2014;157:1023–1036. doi: 10.1016/j.cell.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 5.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 6.Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 7.Patapoutian A, Tate S, Woolf CJ. Transient receptor potential channels: targeting pain at the source. Nat Rev Drug Discov. 2009;8:55–68. doi: 10.1038/nrd2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 9.Peier AM, et al. TRP channel that senses cold stimuli and menthol. Cell. 2002;108:705–715. doi: 10.1016/s0092-8674(02)00652-9. [DOI] [PubMed] [Google Scholar]
- 10.McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416:52–58. doi: 10.1038/nature719. [DOI] [PubMed] [Google Scholar]
- 11.Macpherson LJ, et al. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines: Abstract: Nature. Nature. 2007;445:541–545. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- 12.Han L, et al. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci. 2013;16:174–182. doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikand P, Dong X, LaMotte RH. BAM8–22 peptide produces itch and nociceptive sensations in humans independent of histamine release. J Neurosci. 2011;31:7563–7567. doi: 10.1523/JNEUROSCI.1192-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, et al. Sensory neuron–specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell JK, McQueen DS, Rees JL. Involvement of histamine H4 and H1 receptors in scratching induced by histamine receptor agonists in BalbC mice. Br J Pharmacol. 2004;142:374–380. doi: 10.1038/sj.bjp.0705754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132:549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Lanier LH. Variability in the pain threshold. Science. 1943;97:49–50. doi: 10.1126/science.97.2506.49. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen CS, Price DD, Vassend O, Stubhaug A, Harris JR. Characterizing individual differences in heat-pain sensitivity. Pain. 2005;119:65–74. doi: 10.1016/j.pain.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 20.Chambers SM, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 22.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu ML, et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chanda S, et al. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanier J, Dykes IM, Nissen S, Eng SR, Turner EE. Brn3a regulates the transition from neurogenesis to terminal differentiation and represses non-neural gene expression in the trigeminal ganglion. Dev Dyn. 2009;238:3065–3079. doi: 10.1002/dvdy.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Q, Chen Z, Barrantes IDB, Luis de la Pompa J, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 32.McEvilly RJ, et al. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384:574–577. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- 33.Boland MJ, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 34.Ferri GL, et al. Neuronal intermediate filaments in rat dorsal root ganglia: differential distribution of peripherin and neurofilament protein immunoreactivity and effect of capsaicin. Brain Res. 1990;515:331–335. doi: 10.1016/0006-8993(90)90618-l. [DOI] [PubMed] [Google Scholar]
- 35.Fornaro M, et al. Neuronal intermediate filament expression in rat dorsal root ganglia sensory neurons: an in vivo and in vitro study. Neuroscience. 2008;153:1153–1163. doi: 10.1016/j.neuroscience.2008.02.080. [DOI] [PubMed] [Google Scholar]
- 36.Lallemend F, Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–381. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Landry M, Bouali-Benazzouz R, Mestikawy El S, Ravassard P, Nagy FDR. Expression of vesicular glutamate transporters in rat lumbar spinal cord, with a note on dorsal root ganglia. J Comp Neurol. 2004;468:380–394. doi: 10.1002/cne.10988. [DOI] [PubMed] [Google Scholar]
- 38.Li L, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dykes IM, Lanier J, Eng SR, Turner EE. Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev. 2010;5:3. doi: 10.1186/1749-8104-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, et al. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fariñas I, et al. Characterization of neurotrophin and Trk receptor functions in developing sensory ganglia: direct NT-3 activation of TrkB neurons in vivo. Neuron. 1998;21:325–334. doi: 10.1016/s0896-6273(00)80542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moqrich A, et al. Expressing TrkC from the TrkA locus causes a subset of dorsal root ganglia neurons to switch fate. Nat Neurosci. 2004;7:812–818. doi: 10.1038/nn1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett GL, Bartlett PF. The p75 nerve growth factor receptor mediates survival or death depending on the stage of sensory neuron development. Proc Natl Acad Sci USA. 1994;91:6501–6505. doi: 10.1073/pnas.91.14.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon SB, Armanini MP, Ling LH, Phillips HS. Expression and coexpression of Trk receptors in subpopulations of adult primary sensory neurons projecting to identified peripheral targets. Neuron. 1994;12:1161–1171. doi: 10.1016/0896-6273(94)90323-9. [DOI] [PubMed] [Google Scholar]
- 45.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Addis RC, et al. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS ONE. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.