Abstract
The development of matrix metalloproteinase (MMP) inhibitors has often been frustrated by a lack of specificity and subsequent off-target effects. More recently, inhibitor design has considered secondary binding sites (exosites) to improve specificity. Small molecules and peptides have been developed that bind exosites in the catalytic (CAT) domain of MMP-13, the CAT or hemopexin-like (HPX) domain of MT1-MMP, and the collagen binding domain (CBD) of MMP-2 and MMP-9. Antibody-based approaches have resulted in selective inhibitors for MMP-9 and MT1-MMP that target CAT domain exosites. Triple-helical “mini-proteins” have taken advantage of collagen binding exosites, producing a family of novel probes. A variety of non-traditional approaches that incorporate exosite binding into the design process has yielded inhibitors with desirable selectivities within the MMP family.
Keywords: Matrix metalloproteinase (MMP), Exosite, Collagen, Inhibitor, Triple-helical peptide
Introduction
Matrix metalloproteinases (MMPs) have long been recognized as potential targets for a variety of pathologies, including tumor angiogenesis and metastasis, osteoarthritis (OA), inflammation, periodontitis, vascular diseases, post-myocardial infarction remodeling, neurodegenerative diseases, and neuropsychiatric disorders [1–7]. The development of MMP inhibitors has typically proceeded along the path of active site Zn2+ inhibition. The most common zinc-binding group used for this purpose is hydroxamic acid [8,9]. However, one reason why hydroxamic acid--based inhibitors have not been successful in clinic trials is their lack of selectivity [9,10]. The low selectivity originated from the fact that inhibitors targeting the enzyme active sites face the challenge of very similar chemistry and configuration of these sites across the MMPs [11]. In addition, under certain circumstances, hydroxamic acids may chelate zinc in a non-selective fashion [9,10]. An often observed side effect of hydroxamic acid-based MMP inhibitors has been musculoskeletal syndrome (MSS). MSS has been attributed to inhibition of MMP-1 and ADAM17/TACE [12,13]. A pyrimidine-2,4,6-trione derivative that inhibits MT1-MMP, MMP-2, and MMP-9 is not associated with MSS, and thus demonstrates that better selectivity has the potential to create therapeutically useful MMP inhibitors [14]. Similarly, MMP-13 inhibition does not induce MSS in rat models [15].
More recent strategies for developing inhibitors with greater selectivity consider secondary binding sites (exosites) [16–19]. Also referred to as regulatory sites, unique exosites have been proposed to be present in all MMPs [20]. Considerable prior work has utilized phage display or combinatorial peptide libraries to find peptide-based inhibitors of MMPs [21]. Although these inhibitors may target exosites, the actual binding sites have often not been identified. The following discussion focuses on probes that interact with distinct secondary binding sites of MMPs, and in some cases utilize non-traditional zinc-interaction motifs.
MMP-13 specificity pockets within the catalytic domain
Aventis discovered a pyrimidine dicarboxamide that had low micromolar potency for MMP-13 and no activity against other MMPs when tested at 100 μM [22]. The potency of this compound was further improved to a low nanomolar compound (N4,N6-bis(4-fluoro-3-methylbenzyl)pyrimidine-4,6-dicarboxamide) without losing selectivity [22]. The Aventis molecule binds within a “specificity loop” (subsite S1′) of the MMP-13 catalytic (CAT) domain, which is recognized as an exosite (Fig. 1) [22,23]. Pfizer reported discovery of highly selective nanomolar range MMP-13 inhibitors based on pyrimidinedione and quinazolinone scaffolds acting via binding to the same S1′ exosite [24,25]. Furthermore, pyrimidinedione derivatives were efficacious and safe in rabbit and dog models of OA [25,26] and mouse models of rheumatoid arthritis [27]. Similarly, Alantos Pharmaceuticals identified a new class of highly selective non-Zn2+-binding MMP-13 inhibitors [15,28,29]. ALS 1-0635 provided histologic and clinical efficacy without muscoskeletal toxicity. Binding studies of ALS 1-0635 to the MMP-13 CAT domain indicated non-competitive, reversible MMP-13 inhibition and non-exclusive binding when tested against a non-specific Zn2+ chelator. The compound displayed bovine and human articular cartilage protection at sub-micromolar concentrations in vitro. It also provided chondroprotection in the in vivo rat model of acute and chronic OA at reasonable concentrations. Furthermore, no MSS was observed in ALS 1-0635-treated animals, even at a 200-fold greater concentration than that of marimastat known to induce this condition [15].
Fig. 1.
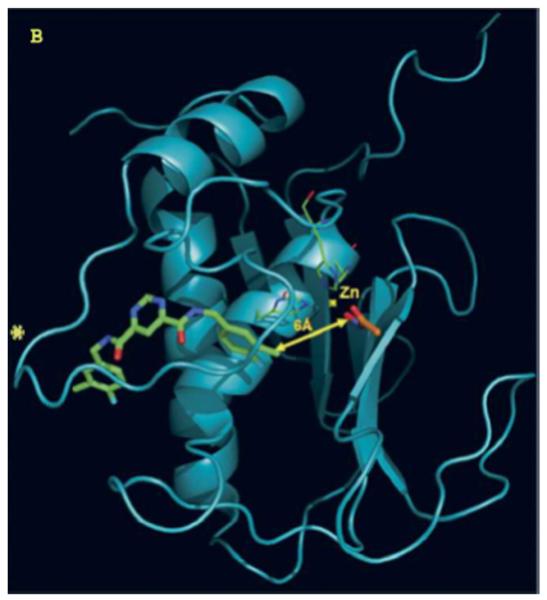
Docked structure of MMP-13 CAT domain with pyrimidine dicarboxamide (green) and acetohydroxamate (orange). The two docked structures are 6 Å apart. The “selectivity loop” is denoted by an *.
Reprinted from [23] with permission.
Although selective MMP-13 inhibitors have been described by Alantos, Aventis, Boehringer, Pfizer, and Wyeth, important pharmacokinetic (PK) and/or other data have not been reported for many of these compounds, and no clinical studies have appeared. For example, no PK or MSS data has been reported for the Aventis and Wyeth compounds [22,30]. The first series of Pfizer compounds, while exhibiting good PK and MSS data, were tested against a limited number of MMPs [31–33]. In similar fashion, the Boehringer compounds exhibited good PK data but were tested against a limited number of MMPs, and not at all in a MSS model [34,35]. The Alantos compounds exhibited excellent MMP selectivity and good PK data, but were not tested in a MSS model [28,29]. Only the second series of Pfizer compounds were reported to exhibit excellent MMP selectivity and good PK and MSS data [24,25,27]. However, as mentioned above, no clinical studies have been reported for the Pfizer compounds. In our hands, we found the primary Pfizer compound (E)-4-((1-methyl-2,4-dioxo-6-(3-phenylprop-1-enyl)-1,2-dihydroquinazolin-3(4H)-yl)methyl)-benzoic acid (Fig. 2) to have low solubility (it could only be tested at a maximal concentration of 2.5 μM), and it inhibited cytochrome P450 1A2 [36].
Fig. 2.
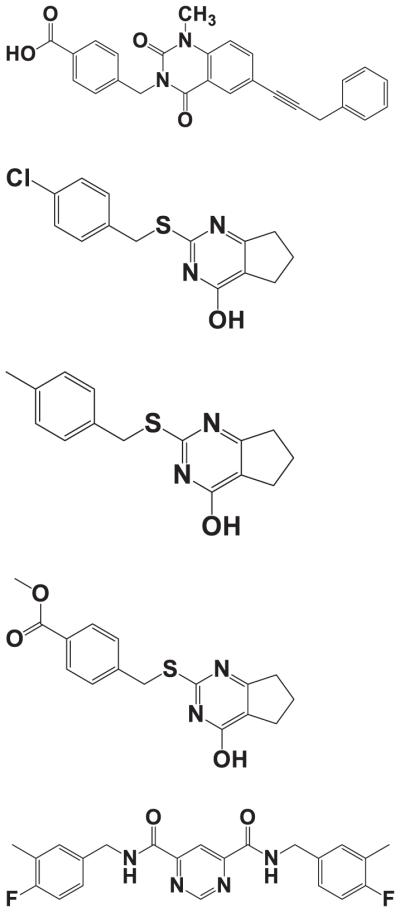
Structures, from top to bottom, of (E)-4-((1-methyl-2,4-dioxo-6-(3-phenylprop-1-enyl)-1,2-dihydroqui-nazolin-3(4H)-yl)methyl)benzoic acid, reported by Pfizer as Compound 2 [24], compound Q/4 (2-[(4-chlorobenzyl)thio]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one) [37], compound Q1/20 (2-[(4-methylphenyl)methyl sulfanyl]-1,5,6,7-tetrahydrocyclopenta[d]pyrimidin-4-one) [38], compound Q2/24 (methyl 4-[(4-oxo-1,5,6,7-tetrahydrocyclo penta[d]pyrimidin-2-yl)sulfanylmethyl] benzoate) [38], and N4,N6-bis(4-fluoro-3-methylbenzyl)pyrimidine-4,6-dicarboxamide, reported by Aventis as Compound 4 [22].
As a result of high-throughput screening and structure–activity relationship studies, we identified a novel, highly selective class of MMP-13 inhibitors (Fig. 2) [37]. Medicinal chemistry characterization of the compound Q/4 2-(arylmethylthio)-cyclopentapyr-imidin-4-one scaffold led to two compounds (2-[(4-methylphenyl)methyl sulfanyl]-1,5,6,7-tetrahydrocy-clopenta[d]pyrimidin-4-one; Q1/20 and methyl 4-[(4-oxo-1,5,6,7-tetrahydrocyclo penta[d]pyrimidin-2-yl) sulfanylmethyl] benzoate; Q2/24) that demonstrated improved potency (as measured by Ki) and selectivity compared to compound 4 [38]. Most significantly, compound 20 did not inhibit MMP-8, whereas compound 4 did. In addition, compounds 4, 20, and 24 did not inhibit MMP-1 or TACE [38], and MSS has been attributed to inhibition of MMP-1 and ADAM17/TACE [13].
Mechanistic characterization revealed a noncompetitive nature of these inhibitors with binding constants in the low μM range. Surprisingly, compound Q/4 exhibited non-mutually exclusive binding (positive cooperativity) when co-tested with the Aventis molecule N4,N6-bis(4-fluoro-3-methylbenzyl)pyrimidine-4,6-dicarboxamide (see Fig. 2 for structure) [36]. Similarly, compound Q/4 binding to MMP-13 was synergistic with the Pfizer molecule (Fig. 2). These results indicate that compound Q/4 has the potential to work through a different mechanism than previously identified inhibitors. Crystallographic analyses revealed two binding modes for compound Q1/20 in the MMP-13 S1′ subsite and in a S1/S2* subsite [36]. Type II collagen- and cartilage-protective effects exhibited by compounds Q/4, Q1/20, and Q2/24 suggested that these compounds might be efficacious in future in vivo studies. Finally, these compounds were also highly selective when tested against a panel of 30 proteases, which, in combination with a good cytochrome P450 inhibition profile, suggested low off-target toxicity and drug–drug interactions in humans [36].
Compounds Q/4, Q1/20, and Q2/24 have 10–100 times lower affinity than the primary Pfizer and Aventis compounds. However, compounds Q/4, Q1/20, and Q2/24 possess a smaller molecular scaffold than previously described MMP-13 inhibitors (Fig. 2), suggesting that greater affinity can be achieved by increasing the size of future analogs of Q/4, Q1/20, and Q2/24. A similar approach was recently described using a thienol[2,3-d]pyrimidine scaffold and extension into the S1′′ subsite of MMP-13 [39]. Targeting multiple structural water molecules within the S1′ subsite as binding partners has also been suggested as a way to improve the potency of MMP-13 inhibitors [40].
MT-MMP loop in the CAT domain
The need for proper interaction of MT1-MMP with cell surface partners is exemplified in studies of the 163–170 loop region within MT1-MMP CAT domain. This MT (membrane type)-loop region is present in the CAT domain of MT1-MT6-MMPs, but is absent in all other MMPs. Deletion of this loop results in the mislocalization of MT1-MMP relative to the β1 integrin adhesion complexes and subsequent decrease of activity towards collagen films [41].
Monoclonal antibody (mAb) 9E8 inhibits MT1-MMP activation of proMMP-2, but not other MT1-MMP catalytic activities [42]. Analysis of mAb 9E8 interaction with MT1-MMP determined that the antibody bound to the Pro163 to Gln174 loop in the CAT domain [43]. Binding of mAb 9E8 to the loop interferes with TIMP-2 binding, preventing formation of the MT1-MMP· TIMP-2·proMMP-2 complex required for proMMP-2 activation [43].
An MT1-MMP near infrared probe was designed based on a peptide sequence identified in a phage display substrate library [44]. To enhance selectivity, the phage display library was screened against the MT1-MMP loop sequence (Arg160 to Gln174, designated MT1-160p) rather than the complete sequence of MT1-MMP or its CAT domain. The non-substrate peptide MT1-AF7p (His-Trp-Lys-His-Leu-His-Asn-Thr-Lys-Thr-Phe-Leu) displayed the highest affinity towards MT1-160p (KD = 0.075 nM). MT1-AF7p was labeled with Cy5.5 (Cy5.5-MT1-AF7p) and chosen for further validation in vivo [44]. The evaluation was performed in mice carrying MDA-MB-435 breast cancer xenografts (expressing high levels of MT1-MMP) or A549 xenografts (low MT1-MMP levels). MDA-MB-435 xenografts had significantly higher signal accumulation and better tumor contrast than the A549 xenografts. However, more precise quantitative data on tumor uptake and PK will be needed to determine the further utility of this probe.
The HPX domain
Diversity in the HPX domains among MMPs makes this exodomain a good target in the search for selective MMP inhibitors. A hurdle is the typically low affinity of compounds that disrupt protein–protein interactions.
The outer blade regions of the MT1-MMP HPX domain have non-homologous loop sequences compared to other members of the MMP family. Our laboratory synthesized peptide models of the 5 MT1-MMP HPX domain loops (blade I strand 4, blade II strand 2, blade II strands 3–4, blade III strand 1, and blade IV strand 4), and examined their inhibitory activity for MT1-MMP processing of a triple-helical substrate [21]. Two peptides were micromolar inhibitors of MT1-MMP but did not inhibit MMP-1. Val-Phe-Asp-Glu-Ala-Ser-Leu-Glu-Pro-NH2, from blade II strands 3–4, had the best IC50 value, 238 μM. This peptide may directly or indirectly inhibit MT1-MMP dimerization or interaction of MT1-MMP with the substrate triple-helix. Val-Arg-Asn-Asn-Gln-Val-Nle-Asp-Gly-Tyr-Pro-Nle-Pro-NH2, which is an N-terminal extension and modified version of the blade IV strand 4 peptide, was more effective than the parent peptide for MT1-MMP inhibition of triple-helical peptidase activity (IC50 = 670 μM).
A small molecule MT1-MMP HPX domain inhibitor was identified using virtual ligand screening of the NCI/NIH Developmental Therapeutics Program ~275,000 compound library [45]. Compound NSC405020 [3,4-dichloro-N-(1-methylbutyl)benzamide] inhibited MT1-MMP homodimerization but not catalytic activity towards a fluorogenic peptide substrate or proMMP-2 activation. NSC405020 was shown to reduce the collagenolytic activity of MCF7-β3/MT1-MMP cells. The compound was effective in vivo, as intratumoral injections reduced tumor size significantly.
Collagen binding exosites
Using a combinatorial one peptide one bead library, Xu et al. determined that the MMP-2 collagen binding domain (CBD) binds a short segment of the α1(I) collagen chain [46]. More specifically, Cys-Gly-Ala-Hyp-Gly-Ala-Hyp-Gly-Ser-Gln-Gly-Ala (designated P713, where Hyp = 4-hydroxyproline) was identified as an inhibitor of MMP-2 activity. P713 inhibited 90% of MMP-2 gelatin cleavage (IC50 of ~ 30 μM), but less than 20% of the MMP-2 activity on a peptide substrate which did not require the CBD for binding. To examine the specificity towards MMP-2, comparative inhibition assays were performed with MMP-8, with no alteration in MMP-8 activities observed upon P713 treatment [46].
Based on the single-stranded peptide model of the α1(I)715-721 collagen sequence identified above as a ligand for the MMP-2 CBD, our group assembled a triple-helical version of this ligand [α1(I)715-721 THP; (Gly-Pro-Hyp)4-Gly-Ala-Hyp-Gly-Ala-Hyp-Gly-Ser-Gln-Gly-Ala-Hyp-(Gly-Pro-Hyp)3-GPY-NH2] [47]. α1(I)715–721 THP inhibited MMP-2 and MMP-9 hydrolysis of a triple-helical peptide model of type V collagen, but did not inhibit MMP-2 or MMP-9 hydrolysis of a short, single-stranded substrate or a triple-helical peptide model of types I–III collagen. To our knowledge, this demonstrated the first use of an exosite binder to selectively inhibit one collagen-based MMP activity (type V) but not another (types I–III).
Incorporation of a zinc binding group within a collagen-model, triple-helical construct would add sequence diversity and eliminate off target interactions with non-collagenolytic proteases. Phosphinic peptides [Ψ{PO2H–CH2}] have been shown to behave as transition state analog inhibitors of MMPs [48]. Phosphinate triple-helical MMP inhibitors have several potential advantages over other inhibitor constructs. These analogs allow incorporation of specificity elements for both the S and S′ subsites of the enzyme. Although binding to the non-primed region of the active site is generally weaker than the primed site to prevent product inhibition [49], it does add sequence diversity and potential selectivity. The triple-helical structure allows for interaction with both the active site and exosites [50–52]. Triple-helical probes are less susceptible to general proteolysis than peptides and other folded proteins and thus offer favorable PK [53–56]. We have found that selective, high affinity inhibitors can be developed for MMPs based on triple-helical structure.
We have produced a series of THPIs based on GlyΨ{PO2H–CH2}Leu, GlyΨ{PO2H–CH2}Val, and GlyΨ{PO2 H–CH2}Ile transition state analogs (Table 1) [16,47,50,57,58]. The GlyΨ{PO2H–CH2}Leu inhibitor [C6-Gly-Pro-Flp-(Gly-Pro-Hyp)4-Gly-Pro-Gln-GlyΨ{PO2H-CH2}(R,S)Leu-Ala-Gly-Gln-Arg-Gly-Ile-Arg-(Gly-Pro-Hyp)4-Gly-Pro-Flp-NH2], developed based on the MMP cleavage site in types I–III collagen [47], was effective for all collagenolytic MMPs tested, but offered a substantial range of activities (Table 1).
Table 1.
Inhibition of MMPs by GlyΨ{PO2H–CH2}Xxx THPIs
| Enzyme | Inhibitor | Inhibitor Tm (°C) | Ki (app) (nM) |
|---|---|---|---|
| MMP-1 | GlyΨ{PO2H–CH2}Leu | 30 | 7.83 ± 1.03a |
| MMP-1(R291A) | GlyΨ{PO2H–CH2}Leu | 30 | 12.76 ± 1.60a |
| MMP-1(I290A,R291A) | GlyΨ{PO2H–CH2}Leu | 30 | 23.66 ± 0.03a |
| MMP-2 | GlyΨ{PO2H–CH2}Leu | 30 | 0.18 ± 0.00a |
| MMP-9 | GlyΨ{PO2H–CH2}Leu | 30 | 0.02 ± 0.01a |
| MT1-MMP | GlyΨ{PO2H–CH2}Leu | 30 | 122.5 ± 22.3a |
| MMP-2 | GlyΨ{PO2H–CH2}Val | 25 | 4.14 ± 0.47a |
| MMP-9 | GlyΨ{PO2H–CH2}Val | 25 | 1.76 ± 0.05a |
| MMP-2 | Stabilized GlyΨ{PO2H–CH2}Val | 43 | 189.1 ± 26.54 |
| MMP-9 | Stabilized GlyΨ{PO2H–CH2}Val | 43 | 90.6 ± 6.67 |
| MMP-8 | GlyΨ{PO2H–CH2}Ile-His-Lys | <5 | 124.6 ± 6.9 |
| MT1-MMP | GlyΨ{PO2H–CH2}Ile-His-Lys | <5 | 4704 ± 708.4 |
| MMP-1 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 110.59 ± 29.8 |
| MMP-2 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 17.82 ± 1.9 |
| MMP-3 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 13600.33 ± 5160.7 |
| MMP-8 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 62.1 ± 2.5 |
| MMP-9 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 0.03 ± 0.02 |
| MMP-13 | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 77.13 ± 14.4 |
| MT1-MMP | GlyΨ{PO2H–CH2}Ile-Tyr-Phe | 40 | 46.15 ± 4.7 |
Assay performed at 10 °C.
Two mutants of MMP-1, MMP-1(R291A) and MMP-1(I290A,R291A), where Ile290 and Arg291 were identified as two residues within the HPX domain that facilitate interactions between collagenolytic MMPs (MMP-1) and triple-helical structures, were tested with GlyΨ{PO2H–CH2}Leu THPI [50]. Low nM Ki values were observed for inhibition of MMP-1 activity by the THPI (Table 1). The Ki values for MMP-1(R291A) and MMP-1(I290A,R291A) were ~ 2-times and ~ 4-times higher, respectively, than wild type MMP-1 (Table 1). Thus, mutation of Ile290 and Arg291 decreased the affinity of MMP-1 for the THPI, indicating interaction of inhibitor with the HPX domain.
The GlyΨ{PO2H–CH2}Val inhibitor [C6-(Gly-Pro-Hyp)4-Gly-Pro-Pro-GlyΨ{PO2H–CH2}(R,S)Val-Val-Gly-Glu-Gln-Gly-Glu-Gln-Gly-Pro-Pro-(Gly-Pro-Hyp)4-NH2], based on the cleavage site in type V collagen by MMP-9 [16], was selective for MMP-2 and MMP-9 [16]. As was the case for the GlyΨ{PO2H–CH2}Leu THPI, the thermal stability of the GlyΨ{PO2H–CH2}Val THPI was greatly reduced compared to the parent substrate (Table 1) [16]. We synthesized a stabilized version of the α1(V)GlyΨ{PO2H–CH2}Val THPI, designated α1(V)GlyΨ{PO2H–CH2}Val [mep14,32,Flp15,33] THPI. This THPI was tested with MMP-2 and MMP-9, and the Ki values were 189.1 and 90.6 nM, respectively (Table 1).
α1(V)GlyΨ{PO2H–CH2}Val [mep14,32,Flp15,33] THPI has been applied in vivo. Citrate synthase (CS) was identified as an intracellular substrate of MMP-9 in a mice model of post-myocardial infarction (MI) [59]. Increased CS processing was abolished with the inhibition of MMP-9 by α1(V)GlyΨ{PO2H–CH2}Val [mep14,32,Flp15,33] THPI [59]. Thus, MMP-9 inhibition improved mitochondrial function post-MI.
GlyΨ{PO2H–CH2}Ile inhibitors were synthesized based on prior results with single-stranded, hydroxamate-based inhibitors that were selective between MMP-8 and MT1-MMP [60]. Unfortunately, neither of the two GlyΨ{PO2H–CH2}Ile inhibitors were selective for MT1-MMP over MMP-8 (Table 1). This result was consistent with our prior study in which selectivity found in single-stranded peptides is not applicable to the same sequences within a triple-helix [61]. Nonetheless, the GlyΨ{PO2H–CH2}Ile-Tyr-Phe THPI did offer a promising lead for an MT1-MMP probe.
Antibodies and minibodies
Preparation of mouse mAbs using MMP-9 as antigen resulted in the identification of REGA-3G12, a selective inhibitor of MMP-9 [62]. REGA-3G12 recognizes the Trp116 to Lys214 region of MMP-9, located in CAT domain but not part of the Zn2+ binding site [63]. REGA-3G12 bound to MMP-9 with KD = 2.1 nM and inhibited MMP-9 catabolism of type II gelatin [62]. A single chain variable fragment derived from REGA-3G12 selectively inhibited MMP-9 compared to MMP-2 [64]. Gelatin hydrolysis was inhibited 44% at a single chain variable fragment concentration of 5 μM [64]. REGA-3G12 was effective in vivo, preventing interleukin-8-induced mobilization of hematopoietic progenitor cells in rhesus monkeys [65].
A selective, fully human MT1-MMP inhibitory antibody (DX-2400; IC50 ~ 1–5 nM) was developed using a human Fab displaying phage library [66,67]. DX-2400 was found to inhibit tumoral MT1-MMP activity, which in turn inhibited MDA-MB-231 primary tumor growth but not MCF-7 tumor growth in xenograft mouse models [66]. DX-2400 also inhibited metastasis [66]. Unfortunately, antibodies are subject to proteolysis and may be removed from circulation rapidly.
Targeting Abs directly to the metal complex in the MMP active site would be expected to increase binding affinity. In addition, as the enzymes' active site is obligatory, it can rarely develop drug resistance through genetic or epigenetic alternations. Achieving the ultimate goal of Ab superior potency and selectivity will also require additional enzyme surface epitope binding. These proposed novel function blocking Abs targeting the MMP active site (designated metallobodies) would exhibit superior properties over classical Abs by mimicking molecular recognition aspects of MMPs endogenous inhibitors, TIMPs, while offering higher selectivity [68].
To generate specificity against the Zn2+–protein complex, mice are immunized with rationally designed synthetic organic ligands bound to metal ion (Zinc-Tripod), which mimic structural and chemical motifs of the relatively exposed catalytic Zn2+-His machinery in the active MMP. This stimulates a first immune response against the small synthetic mimicry antigen and is followed by subsequent immunization with the full-length MMP to induce in vivo affinity maturation towards the native conformation of the catalytic site and additional surface epitopes presented in the whole enzyme active form. The Sagi laboratory demonstrated that this immunization procedure yields function blocking metallobodies directed at the catalytic Zn2+ and enzyme surface epitopes in activated MMP-9 [68]. Metallobody SDS4 selectively bound and inhibited MMP-2/-9 with Ki = 40 nM, and was shown to have therapeutic potential in an inflammatory bowl disease animal model [68]. Noteworthy, MMPs neutralizing mAbs raised by conventional methods (against native proteins or protein fragments) generally interact with surface loops rather then the zinc ion in the active form of the enzyme [63,66].
Overview
The application of structural biology approaches has yielded detailed information on the secondary binding sites/exosites utilized by MMPs for interaction with their substrates and inhibitors [18,69]. In turn, biomolecules designed for specific exosite targeting have provided selective MMP probes. Thus, exosite-based inhibitors appear to have significant advantages over prior active site-based inhibitors, achieving a level of selectively ultimately desired for potential MMP therapeutic agents.
Acknowledgments
I gratefully acknowledge the NIH (CA98799, AR063795, GM106469, and NHLBI contract 268201000036C) and the Multiple Sclerosis National Research Institute for the support of my laboratory's research on MMPs.
Abbreviations
- CAT
catalytic
- CBD
collagen-binding domain
- HPX
hemopexin-like
- mAb
monoclonal antibody
- MI
myocardial infarction
- MMP
matrix metalloproteinase
- MSS
musculoskeletal syndrome
- MT
membrane type
- OA
osteoarthritis
- PK
pharmacokinetic
- THP
triple-helical peptide
- THPI
triple-helical peptide inhibitor
References
- [1].Rosenberg GA. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet. 2009;8:205–16. doi: 10.1016/S1474-4422(09)70016-X. [DOI] [PubMed] [Google Scholar]
- [2].Hu J, Van den Steen PE, Sang Q-XA, Opdenakker G. Matrix metalloproteinase inhibitors as therapy for inflammatory and vascular diseases. Nat Rev Drug Discov. 2007;6:480–98. doi: 10.1038/nrd2308. [DOI] [PubMed] [Google Scholar]
- [3].Rivera S, Khrestchatisky M, Kaczmarek L, Rosenberg GA, Jaworski DM. Metzincin proteases and their inhibitors: foes or friends in nervous system physiology? J Neurosci. 2010;30:15337–57. doi: 10.1523/JNEUROSCI.3467-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Troeberg L, Nagase H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim Biophys Acta. 2012;1824:133–45. doi: 10.1016/j.bbapap.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sbardella D, Faciglione GF, Gioia M, Ciaccio C, Tundo GR, Marini S, et al. Human matrix metalloproteinases: an ubiquitarian class of enzymes involved in several pathological processes. Mol Aspects Med. 2012;33:119–208. doi: 10.1016/j.mam.2011.10.015. [DOI] [PubMed] [Google Scholar]
- [6].Newby AC. Matrix metalloproteinase inhibition therapy for vascular diseases. Vascul Pharmacol. 2012;56:232–44. doi: 10.1016/j.vph.2012.01.007. [DOI] [PubMed] [Google Scholar]
- [7].Lindsey ML, Zamilpa R. Temporal and spatial expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases following myocardial infarction. Cardiovasc Ther. 2012;30:31–41. doi: 10.1111/j.1755-5922.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Whittaker M, Floyd CD, Brown P, Gearing AJH. Design and therapeutic application of matrix metalloproteinase inhibitors. Chem Rev. 1999;99:2735–76. doi: 10.1021/cr9804543. [DOI] [PubMed] [Google Scholar]
- [9].Jacobsen JA, Major Jourden JL, Miller MT, Cohen SM. To bind zinc or not to bind zinc: an examination of innovative approaches to improved metalloproteinase inhibition. Biochim Biophys Acta. 2010;1803:72–94. doi: 10.1016/j.bbamcr.2009.08.006. [DOI] [PubMed] [Google Scholar]
- [10].Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101:10000–5. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–72. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- [12].Peterson JT. The importance of estimating the therapeutic index in the development of matrix metalloproteinase inhibitors. Cardiovasc Res. 2006;69:677–87. doi: 10.1016/j.cardiores.2005.11.032. [DOI] [PubMed] [Google Scholar]
- [13].Becker DP, Barta TE, Bedell LJ, Boehm TL, Bond BR, Carroll J, et al. Orally active MMP-1 sparing α-tetrahydropyranyl and α-piperidinyl sulfone matrix metalloproteinase (MMP) inhibitors with efficacy in cancer, arthritis, and cardiovascular disease. J Med Chem. 2010;53:6653–80. doi: 10.1021/jm100669j. [DOI] [PubMed] [Google Scholar]
- [14].Devy L, Dransfield DT. New strategies for the next generation of matrix-metalloproteinase inhibitors: selectively targeting membrane-anchored MMPs with therapeutic antibodies. Biochem Res Int. 2011;2011:191670. doi: 10.1155/2011/191670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baragi VM, Becher G, Bendele AM, Biesinger R, Bluhm H, Boer J, et al. A new class of potent matrix metalloproteinase 13 inhibitors for potential treatment of osteoarthritis: evidence of histologic and clinical efficacy without musculoskeletal toxicity in rat models. Arthritis Rheum. 2009;60:2008–18. doi: 10.1002/art.24629. [DOI] [PubMed] [Google Scholar]
- [16].Lauer-Fields JL, Brew K, Whitehead JK, Li S, Hammer RP, Fields GB. Triple-helical transition-state analogs: a new class of selective matrix metalloproteinase inhibitors. J Am Chem Soc. 2007;129:10408–17. doi: 10.1021/ja0715849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morrison CJ, Butler GS, Rodríguez D, Overall CM. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21:645–53. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- [18].Sela-Passwell N, Trahtenherts A, Krüger A, Sagi I. New opportunities in drug design of metalloproteinase inhibitors: combination between structure-function experimental approaches and systems biology. Expert Opin Drug Discovery. 2011;6:527–42. doi: 10.1517/17460441.2011.560936. [DOI] [PubMed] [Google Scholar]
- [19].Sela-Passwell N, Rosenblum G, Shoham T, Sagi I. Structural and functional bases for allosteric control of MMP activities: can it pave the path for selective inhibition? Biochim Biophys Acta. 2010;1803:29–38. doi: 10.1016/j.bbamcr.2009.04.010. [DOI] [PubMed] [Google Scholar]
- [20].Udi Y, Fragai M, Grossman M, Mitternacht S, Arad-Yellin R, Calderone V, et al. Unraveling hidden regulatory sites in structurally homologous metalloproteases. J Mol Biol. 2013;425:2330–46. doi: 10.1016/j.jmb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- [21].Ndinguri MW, Bhowmick M, Tokmina-Roszyk D, Robichaud TK, Fields GB. Peptide-based selective inhibitors of matrix metalloproteinase-mediated activities. Molecules. 2012;17:14230–48. doi: 10.3390/molecules171214230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Engel CK, Pirard B, Schimanski S, Kirsch R, Habermann J, Klingler O, et al. Structural basis for the highly selective inhibition of MMP-13. Chem Biol. 2005;12:181–9. doi: 10.1016/j.chembiol.2004.11.014. [DOI] [PubMed] [Google Scholar]
- [23].Gooljarsingh LT, Lakdawala A, Coppo F, Luo L, Fields GB, Tummino PJ, et al. Characterization of an exosite binding inhibitor of matrix metalloprotease 13. Protein Sci. 2008;17:66–71. doi: 10.1110/ps.073130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Johnson AR, Pavlovsky AG, Ortwine DF, Prior F, Man C-F, Bornemeier DA, et al. Discovery and characterization of a novel inhibitor of matrix metalloprotease-13 that reduces cartilage damage in vivo without joint fibroplasia side effects. J Biol Chem. 2007;282:27781–91. doi: 10.1074/jbc.M703286200. [DOI] [PubMed] [Google Scholar]
- [25].Li JJ, Nahra J, Johnson AR, Bunker A, O'Brien P, Yue W-S, et al. Quinazolinones and pyrido[3,4-d]pyrimidin-4-ones as orally active and specific matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. J Med Chem. 2008;51:835–41. doi: 10.1021/jm701274v. [DOI] [PubMed] [Google Scholar]
- [26].Settle S, Vickery L, Nemirovskiy O, Vidmar T, Bendele A, Messing D, et al. Cartilage degradation biomarkers predict efficacy of a novel, highly selective matrix metalloproteinase 13 inhibitor in a dog model of osteoarthritis: confirmation by multivariate analysis that modulation of type II collagen and aggrecan degradation peptides parallels pathologic changes. Arthritis Rheum. 2010;62:3006–15. doi: 10.1002/art.27596. [DOI] [PubMed] [Google Scholar]
- [27].Jüngel A, Ospelt C, Lesch M, Thiel M, Sunyer T, Schorr O, et al. Effect of the oral application of a highly selective MMP-13 inhibitor in three different animal models of rheumatoid arthritis. Ann Rheum Dis. 2010;69:898–902. doi: 10.1136/ard.2008.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Piecha D, Weik J, Kheil H, Becher G, Timmermann A, Jaworski A, et al. Novel selective MMP-13 inhibitors reduce collagen degradation in bovine articular and human osteoarthritis cartilage explants. Inflamm Res. 2010;59:379–89. doi: 10.1007/s00011-009-0112-9. [DOI] [PubMed] [Google Scholar]
- [29].Gege C, Bao B, Bluhm H, Boer J, Gallagher BM, Korniski B, et al. Discovery and evaluation of a non-Zn chelating, selective matrix metalloproteinase 13 (MMP-13) inhibitor for potential intra-articular treatment of osteoarthritis. J Med Chem. 2012;55:709–16. doi: 10.1021/jm201152u. [DOI] [PubMed] [Google Scholar]
- [30].Chen JM, Nelson FC, Levin JI, Mobilio D, Moy FJ, Nilakantan R, et al. Structure-based design of a novel, potent, and selective inhibitor for MMP-13 utilizing NMR spectroscopy and computer-aided molecular design. J Am Chem Soc. 2000;122:9648–54. [Google Scholar]
- [31].Blagg JA, Noe MC, Wolf-Gouveia LA, Reiter LA, Laird ER, Chang SP, et al. Potent pyrimidinetrione-based inhibitors of MMP-13 with enhanced selectivity over MMP-14. Bioorg Med Chem Lett. 2005;15:1807–10. doi: 10.1016/j.bmcl.2005.02.038. [DOI] [PubMed] [Google Scholar]
- [32].Reiter LA, Freeman-Cook KD, Jones CS, Martinelli GJ, Antipas AS, Berliner MA, et al. Potent, selective pyrimidinetrione-based inhibitors of MMP-13. Bioorg Med Chem Lett. 2006;16:5822–6. doi: 10.1016/j.bmcl.2006.08.066. [DOI] [PubMed] [Google Scholar]
- [33].Shah M, Huang D, Blick T, Connor A, Reiter LA, Hardink JR, et al. An MMP13-selective inhibitor delays primary tumor growth and the onset of tumor-associated osteolytic lesions in experimental models of breast cancer. PLoS One. 2012;7:e29615. doi: 10.1371/journal.pone.0029615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Heim-Riether A, Taylor SJ, Liang S, Gao DA, Xiong Z, August EM, et al. Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors. Bioorg Med Chem Lett. 2009;19:5321–4. doi: 10.1016/j.bmcl.2009.07.151. [DOI] [PubMed] [Google Scholar]
- [35].Gao DA, Xiong Z, Heim-Riether A, Amodeo L, August EM, Cao X, et al. SAR studies of non-zinc-chelating MMP-13 inhibitors: improving selectivity and metabolic stability. Bioorg Med Chem Lett. 2010;20:5039–43. doi: 10.1016/j.bmcl.2010.07.036. [DOI] [PubMed] [Google Scholar]
- [36].Spicer TP, Jiang J, Taylor AB, Hart PJ, Roush WR, Fields GB, et al. Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro. J Med Chem. 2014;57:9598–611. doi: 10.1021/jm501284e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lauer-Fields JL, Minond D, Chase PS, Baillargeon PE, Saldanha SA, Stawikowska R, et al. High throughput screening of potentially selective MMP-13 exosite inhibitors utilizing a triple-helical FRET substrate. Bioorg Med Chem. 2009;17:990–1005. doi: 10.1016/j.bmc.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Roth J, Minond D, Darout E, Liu Q, Lauer J, Hodder P, et al. Identification of novel, exosite-binding matrix metalloproteinase-13 inhibitor scaffolds. Bioorg Med Chem Lett. 2011;21:7180–4. doi: 10.1016/j.bmcl.2011.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nara H, Sato K, Naito T, Mototani H, Oki H, Yamamoto Y, et al. Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxy-benzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1″ binding site. Bioorg Med Chem. 2014;22:5487–505. doi: 10.1016/j.bmc.2014.07.025. [DOI] [PubMed] [Google Scholar]
- [40].Fischer T, Riedl R. Strategic targeting of multiple water-mediated interactions: a concise and rational structure-based design approach to potent and selective MMP-13 inhibitors. ChemMedChem. 2013;8:1457–61. doi: 10.1002/cmdc.201300278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Woskowicz AM, Weaver SA, Shitomi Y, Ito N, Itoh Y. MT-LOOP-dependent localization of membrane type I matrix metalloproteinase (MT1-MMP) to the cell adhesion complexes promotes cancer cell invasion. J Biol Chem. 2013;288:35126–37. doi: 10.1074/jbc.M113.496067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ingvarsen S, Porse A, Erpicum C, Maertens L, Jürgensen HJ, Madsen DH, et al. Targeting a single function of the multifunctional matrix metalloproteinase MT1-MMP: impact on lymphangiogenesis. J Biol Chem. 2013;288:10195–204. doi: 10.1074/jbc.M112.447169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shiryaev SA, Remacle AG, Golubkov VS, Ingvarsen S, Porse A, Behrendt N, et al. A monoclonal antibody interferes with TIMP-2 binding and incapacitates the MMP-2-activating function of multifunctional, pro-tumorigenic MMP-14/MT1-MMP. Oncog. 2013;2:e80. doi: 10.1038/oncsis.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhu L, Wang H, Wang L, Wang Y, Jiang K, Li C, et al. High-affinity peptide against MT1-MMP for in vivo tumor imaging. J Control Release. 2011;150:248–55. doi: 10.1016/j.jconrel.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Remacle AG, Golubkov VS, Shiryaev SA, Dahl R, Stebbins JL, Chernov AV, et al. Novel MT1-MMP small-molecule inhibitors based on insights into hemopexin domain function in tumor growth. Cancer Res. 2012;72:2339–49. doi: 10.1158/0008-5472.CAN-11-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu X, Chen Z, Wang Y, Bonewald L, Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem J. 2007;406:147–55. doi: 10.1042/BJ20070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lauer-Fields JL, Whitehead JK, Li S, Hammer RP, Brew K, Fields GB. Selective modulation of matrix metalloproteinase 9 (MMP-9) functions via exosite inhibition. J Biol Chem. 2008;283:20087–95. doi: 10.1074/jbc.M801438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gall AL, Ruff M, Kannan R, Cuniasse P, Yiotakis A, Dive V, et al. Crystal structure of the stromelysin-3 (MMP-11) catalytic domain complexed with a phosphinic inhibitor mimicking the transition-state. J Mol Biol. 2001;307:577–86. doi: 10.1006/jmbi.2001.4493. [DOI] [PubMed] [Google Scholar]
- [49].Babine RE, Bender SL. Molecular recognition of protein-ligand complexes: applications to drug design. Chem Rev. 1997;97:1359–472. doi: 10.1021/cr960370z. [DOI] [PubMed] [Google Scholar]
- [50].Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem. 2009;284:24017–24. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bertini I, Fragai F, Luchinat C, Melikian M, Toccafondi M, Lauer JL, et al. Structural basis for matrix metalloproteinase 1 catalyzed collagenolysis. J Am Chem Soc. 2012;134:2100–10. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, et al. Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc Natl Acad Sci U S A. 2012;109:12461–6. doi: 10.1073/pnas.1204991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lauer-Fields JL, Juska D, Fields GB. Matrix metalloproteinases and collagen catabolism. Biopolymers (Pept Sci) 2002;66:19–32. doi: 10.1002/bip.10201. [DOI] [PubMed] [Google Scholar]
- [54].Fan C-Y, Huang C-C, Chiu W-C, Lai C-C, Liou G-G, Li H-C, et al. Production of multivalent protein binders using a self-trimerizing collagen-like peptide scaffold. FASEB J. 2008;22:3795–804. doi: 10.1096/fj.08-111484. [DOI] [PubMed] [Google Scholar]
- [55].Ndinguri MW, Zheleznyak A, Lauer JL, Anderson CJ, Fields GB. Application of collagen-model triple-helical peptide-amphiphiles for CD44 targeted drug delivery systems. J Drug Deliv. 2012;2012:592602. doi: 10.1155/2012/592602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yasui H, Yamazaki CM, Nose H, Awada C, Takao T, Koide T. Potential of collagen-like triple helical peptides as drug carriers: their in vivo distribution, metabolism, and excretion profiles in rodents. Biopolymers (Pept Sci) 2013;100:705–13. doi: 10.1002/bip.22234. [DOI] [PubMed] [Google Scholar]
- [57].Bhowmick M, Sappidi RR, Fields GB, Lepore SD. Efficient synthesis of fmoc-protected phosphinic pseudodipeptides: building blocks for the synthesis of matrix metalloproteinase inhibitors (MMPIs) Biopolymers (Pept Sci) 2011;96:1–3. doi: 10.1002/bip.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bhowmick M, Fields GB. Synthesis of Fmoc-Gly-Ile phosphinic pseudodipeptide: residue specific conditions for construction of matrix metalloproteinase inhibitor building blocks. Int J Peptide Res & Ther. 2012;18:335–9. doi: 10.1007/s10989-012-9307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].de Castro Brás LE, Cates CA, DeLeon-Pennell KY, Ma Y, Iyer RP, Halade GV, et al. Citrate synthase is a novel in vivo matrix metalloproteinase-9 substrate that regulates mitochondrial function in the post-myocardial infarction left ventricle. Antioxid Redox Signal. 2014;21:1974–85. doi: 10.1089/ars.2013.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Uttamchandani M, Wang J, Li J, Hu M, Sun H, Chen KY-T, et al. Inhibitor fingerprinting of matrix metalloproteases using a combinatorial peptide hydroxamate library. J Am Chem Soc. 2007;129:7848–58. doi: 10.1021/ja070870h. [DOI] [PubMed] [Google Scholar]
- [61].Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, et al. Differentiation of secreted and membrane-type matrix metalloproteinase activities based on substitutions and interruptions of triple-helical sequences. Biochemistry. 2007;46:3724–33. doi: 10.1021/bi062199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Paemen L, Martens E, Masure S, Opdenakker G. Monoclonal antibodies specific for natural human neutrophil gelatinase B used for affinity purification, quantitation by two-site ELISA and inhibition of enzymatic activity. Eur J Biochem. 1995;234:759–65. doi: 10.1111/j.1432-1033.1995.759_a.x. [DOI] [PubMed] [Google Scholar]
- [63].Martens E, Leyssen A, Van Aelst I, Fiten P, Piccard H, Hu J, et al. A monoclonal antibody inhibits gelatinase B/MMP-9 by selective binding to part of the catalytic domain and not to the fibronectin or zinc binding domains. Biochim Biophys Acta. 2007;1770:178–86. doi: 10.1016/j.bbagen.2006.10.012. [DOI] [PubMed] [Google Scholar]
- [64].Hu J, Van den Steen PE, Houde M, Ilenchuk TT, Opdenakker G. Inhibitors of gelatinase B/matrix metalloproteinase-9 activity comparison of a peptidomimetic and polyhistidine with single-chain derivatives of a neutralizing monoclonal antibody. Biochem Pharmacol. 2004;67:1001–9. doi: 10.1016/j.bcp.2003.10.030. [DOI] [PubMed] [Google Scholar]
- [65].Pruijt JF, Fibbe WE, Laterveer L, Pieters RA, Lindley IJ, Paemen L, et al. Prevention of interleukin-8-induced mobilization of hematopoietic progenitor cells in rhesus monkeys by inhibitory antibodies against the metalloproteinase gelatinase B (MMP-9) Proc Natl Acad Sci U S A. 1999;96:10863–8. doi: 10.1073/pnas.96.19.10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, et al. Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 2009;69:1517–26. doi: 10.1158/0008-5472.CAN-08-3255. [DOI] [PubMed] [Google Scholar]
- [67].Zucker S, Cao J. Selective matrix metalloproteinase (MMP) inhibitors in cancer therapy: ready for prime time? Cancer Biol Ther. 2009;8:1–3. doi: 10.4161/cbt.8.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sela-Passwell N, Kikkeri R, Dym O, Rozenberg H, Margalit R, Arad-Yellin R, et al. Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential. Nat Med. 2011;18:143–7. doi: 10.1038/nm.2582. [DOI] [PubMed] [Google Scholar]
- [69].Fields GB. Biophysical studies of matrix metalloproteinase (MMP)/triple-helix complexes. In: Christov C, editor. Advances in protein chemistry and structural biology. Metal-containing enzymesLondon: Elsevier, Inc.; 2014. pp. 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]


