Abstract
Objective
Traditionally, karyotype and fluorescence in situ hybridization (FISH) were used for cytogenetic testing of infants with congenital heart disease who underwent cardiac surgery at our institution. Recently, chromosome microarray analysis (CMA) has been performed in lieu of the traditional tests. A standardized approach to cytogenetic testing does not exist in this population. The purpose of this study was to assess the utility of CMA based on our current ordering practice.
Design
We reviewed the records of all infants (< 1 year old) who underwent cardiac surgery at our institution from January 2010 to June 2013. Data included results of all cytogenetic testing performed. Diagnostic yield was calculated as the percentage of significant abnormal results obtained by each test modality. Patients were grouped by classification of congenital heart disease (CHD).
Results
Two hundred and seventy-five (51%) of 535 infants who underwent cardiac surgery had cytogenetic testing. Of those tested, 154 (56%) had multiple tests performed and at least 18% were redundant or overlapping. The utilization of CMA has increased each year since its implementation. The diagnostic yield for karyotype, FISH and CMA was 10%, 12% and 14% respectively. CMA yield was significantly higher in patients with septal defects (33%, p = 0.01) compared to all other CHD classes. CMA detected abnormalities of unknown clinical significance in 13% of infants tested.
Conclusions
In our center, redundant cytogenetic testing is frequently performed in infants undergoing cardiac surgery. The utilization of chromosome microarray analysis has increased over time and abnormalities of unknown clinical significance are detected in an important subset of patients. A screening algorithm that risk-stratifies based on classification of CHD and clinical suspicion may provide a practical, data-driven approach to genetic testing in this population and limit unnecessary resource utilization.
Keywords: chromosome microarray analysis, congenital heart disease, genetic testing
Introduction
Congenital heart defects are the most common structural birth defect and are estimated to occur in about 4–10 per 1000 live births (1–3). The etiology of most of these defects is generally thought be a multifactorial combination of genetic and environmental influences (4). As our capability to detect chromosomal abnormalities has improved, so has our understanding of the genetic contribution to congenital heart disease (CHD). Estimates of the prevalence of chromosomal abnormalities in patients with CHD have varied widely and ranged anywhere from 8–27% (5–10).
The presence of cytogenetic abnormalities in patients with CHD has important potential ramifications. Many syndromes have extracardiac manifestations that require surveillance and often benefit from early intervention. A genetic diagnosis also has prognostic implications. Several studies have shown increased perioperative complications and poorer outcomes in infants with genetic abnormalities who undergo cardiac surgery (11–14). Lastly, recurrence risk data exists for a many cytogenetic abnormalities and this information may influence future family planning.
In 2007, the AHA released recommendations for genetic testing in patients with CHD (15). While these recommendations still guide the clinician in identifying high-risk populations, they do not specifically address the utilization of chromosome microarray analysis (CMA). Thus, the development and adoption of CMA has increased the complexity of the question: who to test and how to test them?
Conventionally, karyotyping and florescence in situ hybridization (FISH) have been utilized for cytogenetic testing of infants with congenital heart disease. More recently, CMA is being utilized in lieu of the traditional tests. This technology has at least 5-fold increased resolution than traditional karyotyping (6). Not surprisingly, the increased sensitivity of CMA allows it to detect abnormalities that are unable to be identified by karyotyping and FISH. The power of CMA is so robust that chromosomal abnormalities of unknown clinical significance are frequently encountered in patients with CHD (7, 16).
There is significant variation amongst centers with regard to genetic testing practices in patients with CHD (5, 7, 9, 10). In the CMA era, a standardized approach to genetic screening that delineates testing algorithms for specific patient phenotypes does not exist. Furthermore, the diagnostic yield of CMA in infants undergoing cardiac surgery is not well established.
The purpose of this study is to review our institution’s current genetic testing practices and assess the utility of CMA as a screening tool in infants undergoing cardiac surgery. We hypothesized that:
Redundant testing is often performed.
Differences exist in the prevalence of chromosomal abnormalities amongst types of CHD.
CMA detected chromosomal abnormalities of unknown clinical significance are not uncommon.
Methods
This is a single-institution retrospective review and was approved by the Medical University of South Carolina Institutional Review Board. Informed consent to review the charts was waived.
Study population
All infants under 1 year of age who underwent cardiac surgery at our institution from January 1, 2010 to June 30, 2013 were identified using the Society of Thoracic Surgeons (STS) Database. No patients were excluded from review.
Chromosome microarray analysis
Microarray-based chromosome analysis is currently performed at our institution using the I Scan® System with the Infinium® OExPls Cytoconsortium Array BeadChip. This microarray consists of >800,000 genetic markers. The markers provide information on the copy number status of the entire genome and provide single nucleotide polymorphism genotyping that allows for detection of uniparental disomy, loss of heterozygosity and identity by descent. Patient hybridization data is compared to a compilation of information obtained from the HapMap set of 270 control individuals. Criteria for designating a reportable aberration include deletions larger than 200 kb with a minimum of 20 consecutive markers disrupted and duplications larger than 500 kb, unless the area is associated with benign copy number variation. Smaller aberrations are reported only if the regions have a high likely clinical significance. Loss of heterozygosity is reported when the region is greater than 3 Mb. Genomic linear positions are given relative to NCBI build 37. Results are compared with a public database of known, common copy number variations seen in healthy controls, and common population variants were excluded from the secondary analysis. The test was developed and its appropriate performance characteristics determined by the Molecular Diagnostics Laboratory of the Medical University of South Carolina.
Data collection
The STS database was queried to identify infants who underwent cardiac surgery within the study period. This database provided additional diagnostic information regarding each patient’s type of congenital heart disease. This list of patients was then cross-referenced with an institution-based clinical data warehouse to determine which patients had cytogenetic testing performed and corresponding test results. Genetic testing included karyotype, FISH, and CMA. Every FISH test that was performed in the study population was included in the analysis. Results of CMA were interpreted as normal, abnormal or unknown clinical significance. The significance of CMA results was determined based on the interpretation of a molecular pathologist. Additionally, all results deemed abnormal underwent a secondary review by a pediatric geneticist.
Classification of CHD
Patients were grouped into mutually exclusive diagnostic classes according to the type of cardiovascular malformation as outlined in the National Birth Defects Prevention Study (17). CHD classes included: conotruncal, atrioventricular septal defect (AVSD), anomalous pulmonary venous return (APVR), heterotaxy, complex, left ventricular outflow tract obstruction (LVOTO), septal, and right ventricular outflow tract obstruction (RVOTO). For the purpose of including all patients who underwent cardiac surgery in the study period, those patients whose congenital heart disease diagnosis did not fit the outlined classification scheme were filed under “other”.
Extracardiac Abnormalities
The medical record was reviewed to determine which patients within the study population underwent head and renal ultrasound testing. The results of these tests were collected and classified as either normal or abnormal. Findings on head ultrasound that were classified as abnormal included intraventricular hemorrhages greater than or equal to grade two, agenesis of the corpus callosum, hydrocephalus, Dandy Walker malformation, and agenesis of the vermis. Findings on renal ultrasound that were classified as abnormal included multicystic kidney, single kidney, horseshoe kidney, renal dysplasia and bilateral hydronephrosis.
Cost of Cytogenetic Testing
Cost estimates of each cytogenetic test were obtained and reflect the cost of testing supplies and technologist time to perform the test. They do not reflect the cost of pathologist interpretation. The estimates are current as of May 2014.
Statistical Analysis
Diagnostic yield for each test modality was calculated as the percentage of significant abnormal results obtained divided by the number of tests performed. Chi-square analyses were used to assess for differences in CMA yield between the CHD classes. Each CHD class was analyzed individually and compared with the remainder of the groups. A p-value of 0.05 or less was considered statistically significant. All statistics were performed using IBM® SPSS® Statistics software v. 22.
Results
Patients and Cytogenetic Tests Performed
A total of 535 infants who underwent cardiac surgery were identified within the study period. Demographic data for these patients is summarized in Table 1.
Table 1.
Demographic Characteristics
| n = 535 | |
|---|---|
| Age at surgery, median (range) | 47 days (0 – 358) |
| Sex, n (%) | 310 (58%) males |
| Mortality, n (%) | 29 (5%) |
| CHD Diagnostic Class, n (%) | |
| Conotruncal | 142 (27%) |
| LVOTO | 121 (23%) |
| Septal | 65 (12%) |
| AVSD | 58 (11%) |
| RVOTO | 34 (5%) |
| Complex | 19 (4%) |
| TAPVR | 12 (2%) |
| Heterotaxy | 5 (1%) |
| Other | 79 (15%) |
LVOTO, Left Ventricular Outflow Tract Obstruction; AVSD, Atrioventricular septal defect; RVOTO, Right Ventricular Outflow Tract Obstruction; TAPVR, Total Anomalous Pulmonary Venous Return.
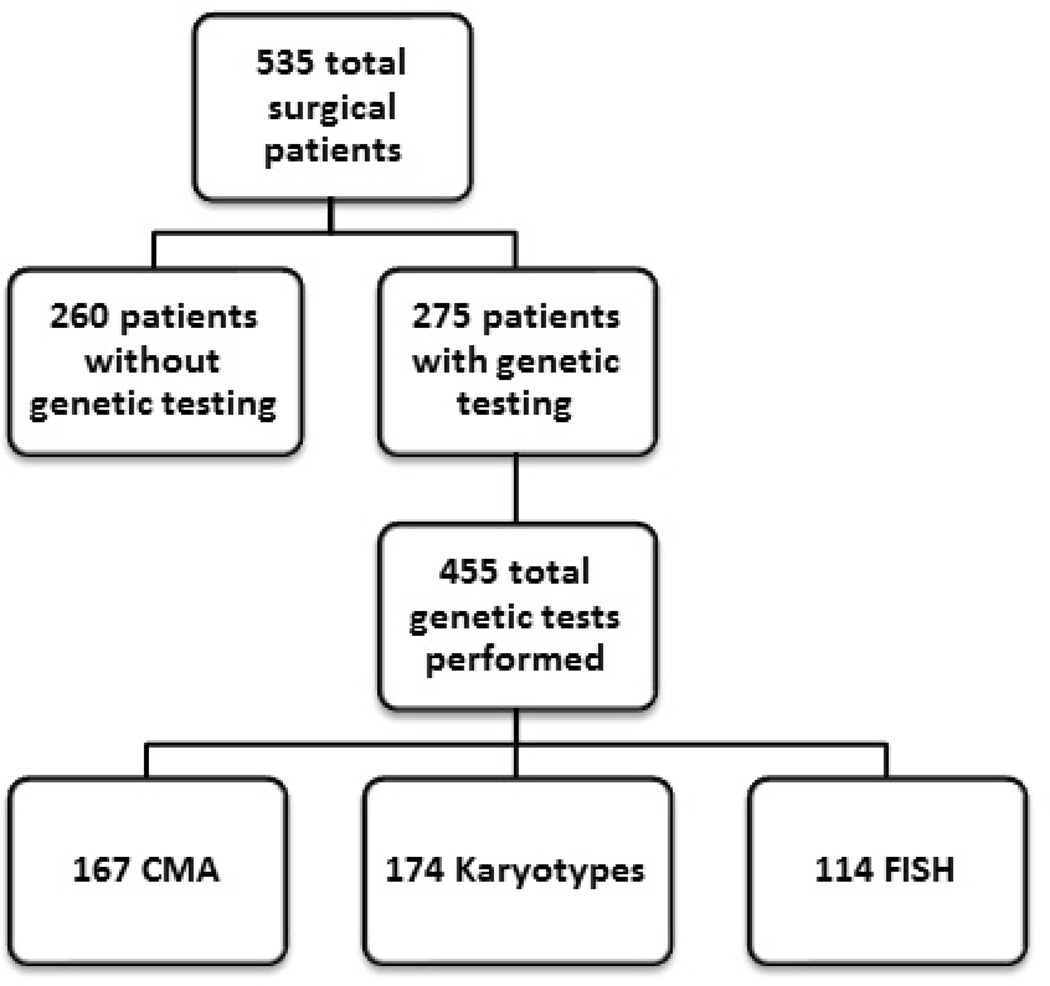
Of these 535 patients, a total of 455 cytogenetic tests were performed in 275 patients. Karyotype was performed most frequently in 33% of infants, followed by CMA in 31% and FISH in 21% (Fig 1). FISH testing performed included probes for 22q11.2 (111/114 tests) and 7q11.23 (3/114 tests). Of those tested, greater than one cytogenetic test was performed in 56% of patients (Table 2). In patients who underwent multiple tests, congruent results (all tests normal or all tests abnormal) were found 92% of the time. Thirty patients underwent both CMA and FISH testing. The increased sensitivity of CMA detected an additional chromosomal abnormality in 10% of these patients (Table 3). As expected, CMA utilization increased over the study period from 2.6 tests performed per month in 2010 to 5.8 per month in 2013 (Table 4).
Figure 1.
Total patient population and cytogenetic testing performed.
Table 2.
Frequency of Cytogenetic Testing
| Number of Cytogenetic Tests | Patients, n (%) (Total n = 535) |
|---|---|
| Single test performed | 121 (23%) |
| Greater than 1 test performed | 154 (29%) |
| Total | 275 (51%) |
Tests included karyotype, FISH, and Chromosome Microarray Analysis (CMA).
Table 3.
Results of Patients Who Underwent Multiple Tests
| Patients with > 1 test | # of Patients | Congruent Test Results (n, %) |
|---|---|---|
| CMA + Karyotype + FISH | 26 | 23 (89%) |
| CMA + Karyotype | 46 | 42 (91%) |
| CMA + FISH | 4 | 4 (100%) |
| Karyotype + FISH | 78 | 73 (94%) |
| Total | 154 | 142 (92%) |
CMA, Chromosome Microarray Analysis; FISH, Florescence In Situ Hybridization. Congruent test results means that the tests performed in that patient were either all normal or all abnormal.
Table 4.
Utilization of CMA Over Time
| Year | # CMA | CMA/month |
|---|---|---|
| 2010 | 31 | 2.6 |
| 2011 | 56 | 4.7 |
| 2012 | 57 | 4.8 |
| 2013 | 23 | 5.8 |
CMA, Chromosome Microarray Analysis
Diagnostic Yield of Cytogenetic Testing
Comparing modalities, the diagnostic yield was 10% for karyotype, 12% for FISH and 14% for CMA (Table 5). Chromosomal abnormalities of unknown clinical significance were found 13% of the time when CMA was ordered. Of the clinically significant CMA results, 8/23 (35%) were deletions of chromosome 22q11.2 consistent with DiGeorge Syndrome.
Table 5.
Diagnostic Yield of Cytogenetic Testing
| Frequency (Total n = 455) |
Percent | |
|---|---|---|
| CMA results | ||
| Normal | 122 | 73% |
| Abnormal | 23 | 14% |
| Unknown Clinical Significance | 22 | 13% |
| Total | 167 | |
| Karyotype results | ||
| Normal | 157 | 90% |
| Abnormal | 17 | 10% |
| Total | 174 | |
| FISH results | ||
| Normal | 100 | 88% |
| Abnormal | 14 | 12% |
| Total | 114 |
CMA, Chromosome Microarray Analysis; FISH, Florescence In Situ Hybridization
Cytogenetic Testing Results Stratified by CHD Class
After grouping patients into mutually exclusive CHD classes, diagnostic yield was calculated for each cytogenetic test. The diagnostic yield of karyotype was significantly higher in the septal and atrioventricular septal defect groups (Table 6). The yield of FISH testing was significantly higher in patients with conotruncal defects and lower in patients with left ventricular outflow tract obstruction (LVOTO) (Table 7). CMA was performed most commonly in conotruncal and left ventricular outflow tract obstruction (combined to represent 66% of patients tested). The highest diagnostic yield was found in patients with septal defects (33%, p = 0.01). The diagnostic yields of CMA in the remaining CHD classes were not significantly different from each other (Table 8).
Table 6.
Karyotype Diagnostic Yield by CHD Classification
| CHD Class | Karyotypes Performed |
Abnormal Results |
Diagnostic Yield |
p-value |
|---|---|---|---|---|
| Conotruncal | 61 | 1 | 2% | <0.01 |
| LVOTO | 56 | 2 | 4% | 0.06 |
| RVOTO | 16 | 2 | 13% | 0.70 |
| Septal | 9 | 6 | 67% | <0.01 |
| Complex | 8 | 1 | 13% | 0.79 |
| TAPVR | 4 | 0 | 0% | 0.51 |
| Heterotaxy | 2 | 0 | 0% | 0.64 |
| AVSD | 12 | 5 | 42% | <0.01 |
| Other | 6 | 0 | 0% | 0.41 |
| Total | 174 | 17 | 10% |
LVOTO, Left Ventricular Outflow Tract Obstruction; AVSD, Atrioventricular septal defect; RVOTO, Right Ventricular Outflow Tract Obstruction; TAPVR, Total Anomalous Pulmonary Venous Return. Chi-square analyses were performed comparing the karyotype yield of each CHD class with that of the remaining classes.
Table 7.
FISH Diagnostic Yield by CMA classification
| CHD Class | FISH Performed |
Abnormal Results |
Diagnostic Yield |
p-value |
|---|---|---|---|---|
| Conotruncal | 49 | 11 | 22% | <0.01 |
| LVOTO | 39 | 1 | 3% | 0.02 |
| RVOTO | 7 | 0 | 0% | 0.31 |
| Septal | 3 | 2 | 67% | <0.01 |
| Complex | 5 | 0 | 0% | 0.39 |
| TAPVR | 3 | 0 | 0% | 0.51 |
| Heterotaxy | 1 | 0 | 0% | 0.71 |
| AVSD | 6 | 0 | 0% | 0.35 |
| Other | 1 | 0 | 0% | 0.71 |
| Total | 114 | 14 | 12% |
LVOTO, Left Ventricular Outflow Tract Obstruction; AVSD, Atrioventricular septal defect; RVOTO, Right Ventricular Outflow Tract Obstruction; TAPVR, Total Anomalous Pulmonary Venous Return. Chi-square analyses were performed comparing the FISH yield of each CHD class with that of the remaining classes.
Table 8.
CMA Diagnostic Yield by CHD Classification
| CHD Class | CMAs Performed |
Abnormal Results |
Diagnostic Yield |
p-value |
|---|---|---|---|---|
| Conotruncal | 57 | 6 | 11% | 0.38 |
| LVOTO | 54 | 9 | 17% | 0.45 |
| RVOTO | 16 | 1 | 6% | 0.36 |
| Septal | 12 | 4 | 33% | 0.01 |
| Complex | 8 | 0 | 0% | 0.25 |
| TAPVR | 5 | 0 | 0% | 0.36 |
| Heterotaxy | 4 | 0 | 0% | 0.42 |
| AVSD | 3 | 1 | 33% | 0.32 |
| Other | 8 | 2 | 25% | 0.35 |
| Total | 167 | 23 | 14% |
LVOTO, Left Ventricular Outflow Tract Obstruction; AVSD, Atrioventricular septal defect; RVOTO, Right Ventricular Outflow Tract Obstruction; TAPVR, Total Anomalous Pulmonary Venous Return. Chi-square analyses were performed comparing the CMA yield of each CHD class with that of the remaining classes
Not surprisingly, 5/6 (83%) of the abnormal CMA results in the conotruncal group were deletions consistent with DiGeorge Syndrome. Interestingly, 7/9 (78%) abnormal results in the LVOTO group were infants with hypoplastic left heart syndrome (HLHS) and the overall diagnostic yield for this subgroup was 23% (7/30). Of the two patients in the “other” category who had abnormal CMA, one carried the diagnosis of anomalous left coronary artery from the pulmonary artery (ALCAPA) and the other had dilated cardiomyopathy.
Extracardiac Abnormalities
A total of 345 (72%) patients from the study population underwent screening for extracardiac abnormalities with either a head ultrasound, renal ultrasound or both. Of these, 247 (72%) had at least one genetic test performed.
There was no difference in the frequency of genetic testing performed in patients with abnormal ultrasound results versus those that were normal (70% vs 72%, p = 0.79). Of those who had genetic testing, patients who had abnormal ultrasound results tended to have a higher frequency of genetic abnormalities compared with patients who had normal ultrasound results but this finding was not significant (23% vs 15%, p = 0.28).
Cost of Testing
The current cost estimates for each test at our institution are $450 for karyotype, $429 for FISH, and $670 for CMA. These estimates cover the cost of supplies and technologist time to perform the test. Using the ordering practices and diagnostic yields found in this study, the cost of detecting one clinically significant chromosomal abnormality by karyotype, FISH and CMA was $4,600, $3,500 and $4,800 respectively.
Discussion
This study represents the largest review of cytogenetic testing practices in infants undergoing cardiac surgery in the era of CMA. During the study period, 51% of infants who underwent cardiac surgery had cytogenetic testing performed. Of those tested, 17% had a clinically significant chromosomal abnormality which is concordant with rates found by other studies that reviewed similar patient populations (5, 7). The true prevalence remains unknown as each of these retrospective studies carries an inherent selection bias when universal testing is not performed.
More than one cytogenetic test was performed in 56% of patients tested. Although clinical indications for performing multiple tests may exist (i.e. karyotype to screen for aneuploidy and FISH to screen for DiGeorge Syndrome), duplicate and redundant testing was not infrequent. Specifically, FISH testing for DiGeorge syndrome is often unnecessary in patients who undergo CMA. Although the turnaround time from the collection of the sample to the reporting of results is often considerably faster for FISH (about 4 days versus 7 days), this slightly earlier diagnosis of DiGeorge syndrome infrequently alters counseling, management or surgical decision-making for these infants. In this study, 18% of patients who had CMA also had FISH testing performed. This inefficiency resulted in over $12,000 of unnecessary additional cost. In a similar review by Connor et. al., 41% of patients with CHD had multiple cytogenetic tests performed that were “redundant or overlapping” (7). Limiting this inefficient resource utilization will only become increasingly important in the current healthcare environment and speaks to the need for a standardized, cost-effective approach.
When comparing CHD classes, we only found a significant difference in CMA yield in patients with septal defects. This finding should be interpreted with caution given the small number of patients tested in this group and the fact that our sample did not undergo universal testing. Additionally, 25% of these patients had extracardiac abnormalities which may have precipitated genetic testing. However, previous studies have shown that differences exist amongst CHD classes with regard to prevalence of chromosomal abnormalities and recurrence risk in families (10, 18). Furthermore, it seems that differences exist within classes of CHD for specific defects. For example, D-TGA has consistently been shown to not be associated with significant genetic abnormalities nor show patterns of increased risk for recurrence (5, 18–20). In this study, no patients with D-TGA were found to have clinically significant chromosomal abnormalities (18 had CMA testing and all were normal). Any algorithm developed that standardizes testing practices in infants with CHD should be strongly influenced by the type of CHD present.
We found the cost of detecting one clinically significant chromosomal abnormality by CMA during the study period was approximately $4,800. This is likely an underestimate given that these figures do not include the cost of the interpretation of a pathologist which is significantly more demanding for CMA. Seven of the 23 abnormal CMA results were deletions consistent with DiGeorge syndrome and all of them would have been detected by FISH. Given that FISH is a less expensive test and the results are reported in a more timely fashion, the cost-effectiveness of CMA as a first-line universal screening test in patients with conotruncal lesions is further called into question.
Alternatively, CMA may be an appropriate first-line cytogenetic test for select patient populations. In this study, it had a particularly high yield for detecting abnormalities in infants with hypoplastic left heart syndrome. Importantly, the majority of these abnormalities were unlikely to be detected by karyotype or FISH. This finding has not been well described and warrants further study.
Unfortunately, data regarding differential ordering practices of individual physicians was not available for analysis as part of this study but we suspect a high degree of variability. At our institution, the cardiac intensivist plays the predominant role in ordering genetic testing of infants in the perioperative period. The decision of whether to order any testing and which test to order is often made at the discretion of the physician. Consultation from a geneticist is obtained if there is evidence of syndromic CHD (dysmorphic features, extracardiac anomalies etc.) or in the setting of an abnormal cytogenetic test result.
In the current study, CMA found chromosomal abnormalities of unknown clinical significance in 13% of patients tested. As our understanding of the genetic contribution to CHD improves, a proportion of those results may one day be re-classified as clinically significant abnormalities. However, the effect of a genetic abnormality of “unknown clinical significance” may in fact be very significant on the psyche of our patients and their families. Zyblewski et. al. found that chromosomal abnormalities rather than severity of heart disease resulted in a 14-fold increased likelihood of parents choosing termination for a fetus with prenatally diagnosed CHD (21). Clearly, any genetic diagnosis may carry significant burdens to the family and only consented testing should be considered.
Limitations
This study is limited by its retrospective design. Not all patients underwent genetic testing and therefore it is unknown whether the prevalence of chromosomal abnormalities found in this study represents an underestimate or overestimate. Additionally, the retrospective design limits our ability to evaluate the clinician’s reasoning behind ordering cytogenetic testing. The amount of genetic testing performed in this population may also be underestimated by this study given that infants may have had testing performed at another institution prior to or after their operation. The interpretation of differences between CHD classes was limited by infrequent testing in some groups.
Conclusions
Upon review of our current practice for ordering cytogenetic testing in infants undergoing cardiac surgery, it is evident that redundant testing is performed resulting in suboptimal resource utilization. Chromosome microarray analysis detected abnormalities of unknown clinical significance quite frequently and we caution its use as a first-line, universal cytogenetic screening test in this population. This test may have more utility in select patient populations such as HLHS where the yield appears high for detecting abnormalities of clinical significance that are incapable of being identified by karyotype or FISH. Our findings advocate for the development of a screening algorithm that risk-stratifies based on classification of CHD, cost-effectiveness and clinical suspicion which would provide a practical, data-driven approach to genetic testing in this population. In order to develop such an algorithm, the optimal study design would prospectively screen all infants born with congenital heart disease with karyotype, FISH and CMA and be large enough to determine true differences between classes of CHD. Such a study seems difficult and costly for a single center to pursue but may be feasible with a multi-centered, collaborative approach.
Table 9.
Extracardiac Abnormalities
| Abnormal | Normal | p-value | |
|---|---|---|---|
| Ultrasound Results | 50 | 295 | |
| Genetic Testing Performed | 35 (70%) | 212 (72%) | 0.79 |
| Abnormal Genetic Test Results | 8 (23%) | 33 (15%) | 0.28 |
Ultrasound results includes all head and renal ultrasounds performed within the study population. Genetic testing performed included all karyotypes, FISH testing, and CMA performed within the study population and whether any of these tests were abnormal.
Acknowledgements
This work was supported in part by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under Award Number T32 HL007710. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest Statement: The authors have no financial, personal or industrial conflicts of interest to disclose.
Disclosures:
All authors have approved the manuscript in its final form and have full knowledge of its submission for publication.
Author Contributions
Dr. Scheurer contributed in study design, data collection and analysis, interpretation of data as well as drafting of manuscript and approval of final version.
Drs. Kavarana and Chowdhury contributed in drafting of manuscript including approval of its final form.
References
- 1.Bjornard K, Riehle-Colarusso T, Gilboa SM, Correa A. Patterns in the prevalence of congenital heart defects, metropolitan Atlanta, 1978 to 2005. Birth defects research Part A, Clinical and molecular teratology. 2013;97(2):87–94. doi: 10.1002/bdra.23111. [DOI] [PubMed] [Google Scholar]
- 2.Ferencz C, Rubin JD, McCarter RJ, Brenner JI, Neill CA, Perry LW, et al. Congenital heart disease: prevalence at livebirth. The Baltimore-Washington Infant Study. American journal of epidemiology. 1985;121(1):31–36. doi: 10.1093/oxfordjournals.aje.a113979. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39(12):1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 4.Nora JJ, Nora AH. Familial risk of congenital heart defect. American journal of medical genetics. 1988;29(1):231–233. doi: 10.1002/ajmg.1320290134. [DOI] [PubMed] [Google Scholar]
- 5.Baker K, Sanchez-de-Toledo J, Munoz R, Orr R, Kiray S, Shiderly D, et al. Critical congenital heart disease--utility of routine screening for chromosomal and other extracardiac malformations. Congenital heart disease. 2012;7(2):145–150. doi: 10.1111/j.1747-0803.2011.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breckpot J, Thienpont B, Peeters H, de Ravel T, Singer A, Rayyan M, et al. Array comparative genomic hybridization as a diagnostic tool for syndromic heart defects. The Journal of pediatrics. 2010;156(5):810–817. 7 e1–7 e4. doi: 10.1016/j.jpeds.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 7.Connor JA, Hinton RB, Miller EM, Sund KL, Ruschman JG, Ware SM. Genetic testing practices in infants with congenital heart disease. Congenital heart disease. 2014;9(2):158–167. doi: 10.1111/chd.12112. [DOI] [PubMed] [Google Scholar]
- 8.Ferencz C, Neill CA, Boughman JA, Rubin JD, Brenner JI, Perry LW. Congenital cardiovascular malformations associated with chromosome abnormalities: an epidemiologic study. The Journal of pediatrics. 1989;114(1):79–86. doi: 10.1016/s0022-3476(89)80605-5. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez JH, Shirali GS, Atz AM, Taylor SN, Forbus GA, Zyblewski SC, et al. Universal screening for extracardiac abnormalities in neonates with congenital heart disease. Pediatric cardiology. 2009;30(3):269–273. doi: 10.1007/s00246-008-9331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman RJ, Rasmussen SA, Botto LD, Riehle-Colarusso T, Martin CL, Cragan JD, et al. The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatric cardiology. 2011;32(8):1147–1157. doi: 10.1007/s00246-011-0034-5. [DOI] [PubMed] [Google Scholar]
- 11.Andrews RE, Simpson JM, Sharland GK, Sullivan ID, Yates RW. Outcome after preterm delivery of infants antenatally diagnosed with congenital heart disease. The Journal of pediatrics. 2006;148(2):213–216. doi: 10.1016/j.jpeds.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 12.McDonald R, Dodgen A, Goyal S, Gossett JM, Shinkawa T, Uppu SC, et al. Impact of 22q11.2 deletion on the postoperative course of children after cardiac surgery. Pediatric cardiology. 2013;34(2):341–347. doi: 10.1007/s00246-012-0454-x. [DOI] [PubMed] [Google Scholar]
- 13.O'Byrne ML, Yang W, Mercer-Rosa L, Parnell AS, Oster ME, Levenbrown Y, et al. 22q11.2 Deletion syndrome is associated with increased perioperative events and more complicated postoperative course in infants undergoing infant operative correction of truncus arteriosus communis or interrupted aortic arch. The Journal of thoracic and cardiovascular surgery. 2014 doi: 10.1016/j.jtcvs.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simsic JM, Coleman K, Maher KO, Cuadrado A, Kirshbom PM. Do neonates with genetic abnormalities have an increased morbidity and mortality following cardiac surgery? Congenital heart disease. 2009;4(3):160–165. doi: 10.1111/j.1747-0803.2009.00281.x. [DOI] [PubMed] [Google Scholar]
- 15.Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 16.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, Hauser NS, et al. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatric research. 2008;64(4):358–363. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 17.Botto LD, Lin AE, Riehle-Colarusso T, Malik S, Correa A National Birth Defects Prevention S. Seeking causes: Classifying and evaluating congenital heart defects in etiologic studies. Birth defects research Part A, Clinical and molecular teratology. 2007;79(10):714–727. doi: 10.1002/bdra.20403. [DOI] [PubMed] [Google Scholar]
- 18.Peyvandi S, Ingall E, Woyciechowski S, Garbarini J, Mitchell LE, Goldmuntz E. Risk of congenital heart disease in relatives of probands with conotruncal cardiac defects: An evaluation of 1,620 families. American journal of medical genetics Part A. 2014 doi: 10.1002/ajmg.a.36500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burn J, Brennan P, Little J, Holloway S, Coffey R, Somerville J, et al. Recurrence risks in offspring of adults with major heart defects: results from first cohort of British collaborative study. Lancet. 1998;351(9099):311–316. doi: 10.1016/s0140-6736(97)06486-6. [DOI] [PubMed] [Google Scholar]
- 20.Song MS, Hu A, Dyamenahalli U, Chitayat D, Winsor EJ, Ryan G, et al. Extracardiac lesions and chromosomal abnormalities associated with major fetal heart defects: comparison of intrauterine, postnatal and postmortem diagnoses. Ultrasound in obstetrics & gynecology: the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2009;33(5):552–559. doi: 10.1002/uog.6309. [DOI] [PubMed] [Google Scholar]
- 21.Zyblewski SC, Hill EG, Shirali G, Atz A, Forbus G, Gonzalez J, et al. Chromosomal anomalies influence parental treatment decisions in relation to prenatally diagnosed congenital heart disease. Pediatric cardiology. 2009;30(8):1105–1111. doi: 10.1007/s00246-009-9514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]