Abstract
Purpose
Activating germline mutations in CARD11 have recently been linked to a rare genetic disorder associated with congenital B cell lymphocytosis. We describe a patient with a similar clinical phenotype who had a de novo germline G123D CARD11 mutation.
Methods
Whole exome sequencing was performed on DNA from the patient and his biological parents. Laboratory studies examined characteristics of the patient’s B and T lymphocytes. A CARD11 cDNA containing the mutation was transfected into a lymphocyte cell line to gain an understanding of its function. RNA sequencing was performed on samples from the patient and from patients with alternate germline CARD11 mutations and differential gene expression analysis was performed.
Results
The patient had a decade-long history of severe polyclonal B lymphocytosis in the 20,000–90,000 lymphocytes/mm3 range, which was markedly exacerbated by EBV infection and splenectomy at different times. He had a heterozygous germline CARD11 mutation causing a G123D amino acid substitution, which was demonstrated to induce NF-κB activation in unstimulated lymphocytes. In contrast to previous patients with CARD11 mutations, this patient’s B cells exhibited higher expression of several cell cycle progression genes, as well as enhanced proliferation and improved survival following B cell receptor stimulation.
Conclusions
This is the third reported germline and first de novo CARD11 mutation shown to cause congenital B cell lymphocytosis. The mutation was associated with a dramatically greater lymphocytosis than in previously described cases, disproportionate to the level of constitutive NF-κB activation. However, comparative review of the patient’s clinical history, combined with additional genomic and functional analyses, underscore other important variables that may affect pathophysiology or regulate mutant CARD11 function in B cell proliferation and disease. We now refer to these patients as having BENTA disease (B cell Expansion with NF-κB and T cell Anergy).
Keywords: CARD11, B-Cell lymphocytosis, congenital, NF-κB, BENTA
Introduction
Congenital B cell lymphocytosis is a rare clinical syndrome. Recently, two activating germline mutations in CARD11 have been identified as driving disease in patients with what we now refer to as BENTA (B cell Expansion with NF-κB and T cell Anergy) [1]. In these previously described cases, gain-of-function mutations in CARD11 cause constitutive NF-κB activation in lymphocytes leading to a striking naïve B cell expansion but T cell hyporesponsiveness. Similar gain-of-function somatic CARD11 mutations linked to elevated NF-κB activity are relatively common in B cell malignancy, particularly diffuse large B cell lymphoma (DLBCL) [2–6]. Here we report a new patient with BENTA disease who was found by whole exome sequencing to have a heterozygous, germline de novo G123D CARD11 mutation. This mutation is located at the same amino acid residue as one of the previously reported patients, but with a different substitution (G123S, see ref 1). G123D CARD11 has been described as a somatic change in one case of DLBCL [7]. The affected residue lies within the recently described “LATCH” domain of CARD11, which plays a critical role in maintaining CARD11 in a closed, inactive state. An unbiased screen for novel gain-of-function mutations identified a high number of missense mutations in the LATCH domain, including G123D, that could spontaneously activate NF-κB and promote human B cell lymphoma cell survival in vitro [8]. Our discovery of a new BENTA patient harboring a germline G123D mutation offers further insight into how gain-of-function CARD11 mutations perturb lymphocyte development and likely predispose BENTA patients to develop B cell tumors. More importantly, our report highlights several factors that may contribute to exacerbated B cell lymphocytosis in this patient, which improves our understanding of the spectrum of BENTA disease severity.
Case Report
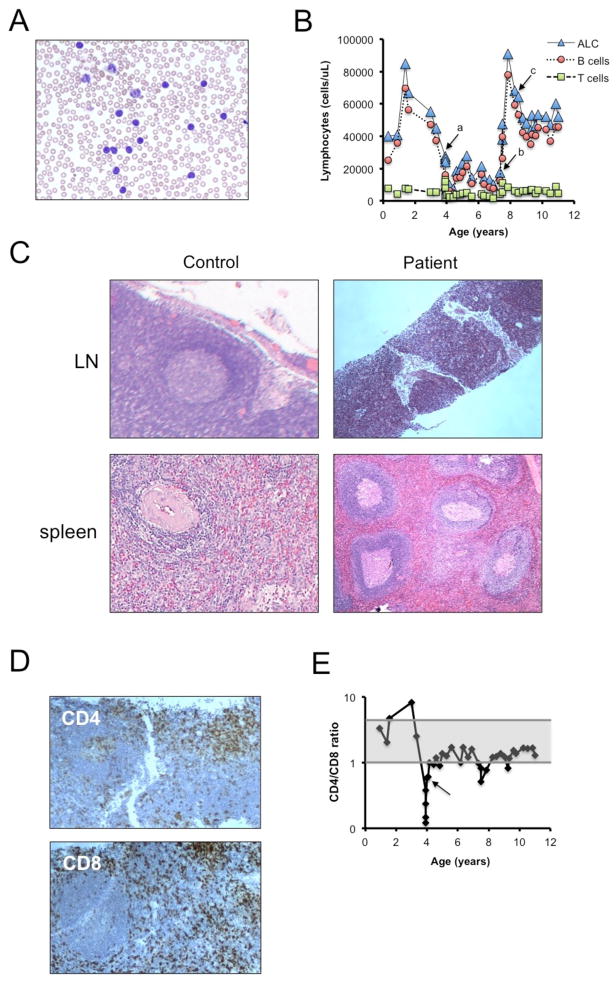
The patient is an 12 year-old boy who presented at 3 months of age with lymphocytosis, splenomegaly, and anemia with a reticulocyte count < 1%. His clinical presentation initially resembled acute lymphoblastic leukemia, but his circulating lymphocytes appeared to be morphologically unremarkable with small resting lymphocytes (Figure 1A). Flow cytometry revealed an excess of mature B lymphocytes that appeared polyclonal with a K:λ ratio of 1.1:1, and normal T cells. His bone marrow aspiration showed normal cellularity with red cell aplasia, without an increase in blasts. Viral testing for cytomegalovirus (CMV), human herpesvirus 6 (HHV6), Epstein-Barr virus (EBV), and parvovirus from bone marrow aspirate were all negative. The reticulocyte count recovered spontaneously. Subsequently, his lymphocyte count continued to range from 50–80×103/mL, composed predominantly of CD19+/CD20+/CD5int/CD27− B cells with normal numbers of CD3+/CD5+/CD7+/TCRαβ+, CD4/CD8 segregated T cells (Figure 1B). Splenomegaly persisted with mild anemia and thrombocytopenia. He had multiple bone marrow biopsies that showed appropriate lineage maturation with marked polyclonal naive B cell lymphocytosis, but was otherwise normocellular except for a slight megakaryocytic hyperplasia. Molecularly he had polyclonal B-cell lymphocytosis by IgH rearrangement, with a constant K:λ ratio of 1:1. Cytogenetic studies of blood and fibroblasts showed 46 XY, inv(2)(p11.2, q13), which is a normal variant; his phenotypically normal father also was found to have the same chromosome 2 inversion.
Figure 1. Clinical history and pathology.
(A) High power (500X) of peripheral blood smear showing circulating lymphocytes. (B) Total circulating lymphocytes (ALC) and B cell and T cell subsets from 3 months of age to present. Arrows mark key events (a = EBV infection; b=splenectomy; c= MTX started). (C) Micrographs of H&E stained sections from lymph node (top, 100X) and laparascopic splenectomy tissue (bottom, 40X) from a normal adult and the patient. (D) Low power (40X) CD4 (top) and CD8 (bottom)-stained lymph node biopsy taken during acute EBV infection. (E) CD4/CD8 ratio of peripheral T cells over time, with an arrow denoting the timing of an acute EBV infection. Shaded area represents normal range (1:1 – 4:1).
He had febrile episodes requiring hospitalization approximately once per year. At age 4 years he was hospitalized for acute EBV infection including massive adenopathy and increased splenomegaly, which was complicated by immune thrombocytopenic purpura. Although the basic architecture of the patient’s lymph nodes was preserved, an intense B cell infiltrate superimposed upon and obscured both follicular and parafollicular areas (Figure 1C and Supplemental Figure 1). The lymph node biopsy obtained during the EBV infection also showed preserved histological architecture, with a predominant polyclonal B cell population present in both the small follicles and interfollicular zones. A moderate number of T cells (Figure 1D) were present in the interfollicular zone and showed a predominance of CD8 over CD4 T cells as also seen in peripheral blood (Figure 1E). He was treated with intravenous immunoglobulin, rituximab, corticosteroids, and acyclovir with resolution of symptoms, and clearance of plasma EBV by PCR. After the EBV infection his circulating lymphocyte count dropped to the 10 – 20 × 103/μL range, and his CD4:CD8 ratio, which had ranged from 2–8 and which had dropped precipitously to as low as 0.12 during the acute EBV infection, slowly recovered to a range of 0.8–1.7 (Figure 1D). Plasma EBV DNA remained undetectable at age 7.
The patient was diagnosed with otitis media approximately 2–3 times per year and had bilateral tympanostomy tubes placed several times, most recently at age 8 with no further otitis media infections. He had adenoidectomy performed at the same time. He has had occasional episodes of sinusitis and bronchitis.
Despite the lower lymphocyte count after the EBV infection, the patient experienced progressive hypersplenism, and underwent elective laparoscopic splenectomy at age 7. The spleen was markedly enlarged (889 grams) but architecturally unremarkable compared to normal tissue from an older individual (Figure 1C). The patient’s spleen showed follicular hyperplasia and red pulp expansion (Figure 1C) with increased polyclonal B lymphocytes in both follicles and splenic cords. Post-splenectomy his mild anemia and thrombocytopenia resolved, but his lymphocyte count increased to a range of 60–90 × 103/μL, with total WBCs exceeding 100 × 103/μL. To control his lymphocytosis, he was treated with methotrexate 20 mg/m2/week, which decreased his lymphocyte count to the range of 50–60 × 103/μL. After 4 years of treatment, a methotrexate holiday was attempted, and his lymphocyte count did not rise above 80 × 103/μL.
Table 1 summarizes the most recent counts (per μl of blood) and immunophenotyping data collected on this patient. We observed profound lymphocytosis, with elevated numbers of B, T, and NK cells. Although the number of T cells was marginally above normal ranges, they only comprised 10% of total lymphocytes in the blood (normal range = 60–84%). This reduced proportion of T cells directly relates to a striking expansion of B cells (>100-fold over normal range). Indeed we noted a significantly higher overall percentage and absolute number of total B cells compared to other BENTA patients (Table 1). Most of these B cells had an immature or naïve phenotype, with a very low percentage of CD27+ memory B cells. Strikingly, the immature CD10+ B cell pool was almost entirely comprised of CD21+ “T2/T3” transitional B cells, with few CD21lo “T1” cells, similar to previous BENTA patients. K:λ was 1.04, and IgH rearrangement studies showed his B cells were polyclonal. Because of the high WBC, absolute numbers of T cell and NK cell subsets were mildly increased.
Table 1. Flow cytometric analysis of patient PBMC.
Percentages and absolute counts (per microliter of whole blood) are noted for each cell population. Ranges in aged-matched pediatric controls are listed in the right columns [1]; parentheses indicate normal adult ranges when pediatric data were unavailable. Ranges for the 3 previous pediatric BENTA patients (ages 4–8) are included in the middle columns for comparison. New patient was 11 years old at time of phenotyping.
| Marker | Patient | BENTA | Normal Ctrl | |||
|---|---|---|---|---|---|---|
| % | Abs # | % | Abs # | % | Abs # | |
| CD19+ B | 87.4 | 48262 | 53–65 | 4700–8100 | 14–33 | 390–1400 |
| CD20+ IgD+ IgM+ | 84.4 | 46606 | 52–64 | 4650–7950 | 7.4–12.1 | 50–276 |
| CD20+ CD5+ | 54.3 | 29984 | 36–44 | 3150–5500 | (0.7–9.7) | (14–159) |
| CD20+ CD10+ | 35.4 | 19570 | 27–34 | 2250–4100 | (0.1–3.4) | (2–76) |
| CD20+ CD27+ | 1 | 552 | 0.4–1 | 62–76 | 2.9–7.3 | 12–68 |
| CD20+ IgG+ | 0 | 0 | 0.1 | 9–15 | (0.2–2.2) | (4–49) |
| CD19+ IgM− CD38hi | 0 | 11 | 0 | 0–6 | (0–0.1%) | (0–1) |
| CD3+ T | 10 | 5522 | 32–40 | 2250–6250 | 56–75 | 1400–3700 |
| CD4+ T | 4.8 | 2651 | 9–17 | 1200–1400 | 28–47 | 700–2200 |
| CD4+ CD45RA+ | 2.8 | 1546 | 3–17 | 350–1250 | 20–33 | 430–1500 |
| CD4+ CD45RA− | 2 | 1104 | 0–6 | 2–400 | 6–17 | 220–660 |
| CD8+ T | 3.4 | 1877 | 10–28 | 700–4250 | 16–30 | 490–1300 |
| CD8+ CD45RA+ | 1.9 | 1049 | 7–11 | 600–1100 | 16–22 | 380–1100 |
| CD8+ CD45RA− | 1.5 | 829 | 0.1–5 | 5–800 | 3–11 | 90–440 |
| NK cells | 2.3 | 1270 | 4–7 | 250–1100 | 4–17 | 130–720 |
| TCRα/β+ CD4− CD8− | 0.5 | 265 | 4–5 | 250–600 | (0.3–1.3) | (6–23) |
| CD4+ CD25+ Foxp3+ | 0.3 | 177 | 0.3–1 | 40–83 | (1.6–4.3) | (25–89) |
Autoimmunity workup (age 3) was negative for anticardiolipin antibody, antinuclear antibody (ANA), anti-double stranded DNA antibody, antimitochondrial antibody (AMA), anti-SSA/Ro and anti-SSB/La antibodies, and rheumatoid factor. His C3 complement level was normal, but C4 complement level was mildly low at 9 mg/dL (normal range 15–53 mg/dL). He later became ANA positive at (<1:80 to 1:160 (speckled) or 1.7 EU/mL), and transiently positive for anti-SSA at 57.3 EU/mL. RNP, smooth muscle, c-ANCA, p-ANCA, and Sm antibodies were negative with no clinical evidence of autoimmune disease. His immunoglobulin levels have been normal with the exception of a low IgA level (since age 4 years). Despite routine immunizations and additional Pneumovax boosting, he had poor titers to pneumococcus (1/23 serotypes), meningococcus (0/4 serotypes), rubella, measles, and VZV. Poor responses to polysaccharide-conjugate vaccines were previously noted in two BENTA patients [1]. Titers were suboptimal to tetanus and Haemophilus influenzae, but positive to mumps.
Methods
Cells, tissues, and treatments
Patient’s and biological parents’ blood/tissue samples were obtained after written informed consent through protocols established by the Pediatric Oncology Branch and the Lymphocyte Molecular Genetics Unit and approved by the Institutional Review Board (IRB) of the National Institutes of Health (NIH). The parents separately provided consent for a single-patient clinical protocol that was approved by the University of Kentucky IRB. Anonymized buffy coat samples from healthy normal donors were provided by the NIH Blood Bank. Jurkat T cell (E6 and JPM50.6) and BJAB B cell lines were maintained in complete RPMI medium (RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 100 U/ml each of penicillin and streptomycin (Invitrogen)). Peripheral blood mononuclear cells (PBMC) were isolated from whole blood or buffy coats using Ficoll-Hypaque-PLUS (Roche) density gradient centrifugation. Following erythrocyte lysis in ACK buffer (Lonza), pan T cells or naïve B cells were purified by negative selection using magnetic bead sorting kits (EasySep Human T cell Enrichment Kit, EasySep Human Naïve B cell Enrichment Kit, Stem Cell Technologies). CD10+ immature/transitional B cells were subsequently depleted using magnetic beads (CD10+ Positive Selection beads, Stem Cell Technologies). Naïve B cell stimulations were performed in complete RPMI medium using Staphylococcus aureus Cowan I (SAC, Sigma) + 200U/ml rIL-2 (Peprotech), anti-IgM F(ab′)2 (Jackson Immunoresearch, 25 μg/ml) −/+ anti-CD40 (1 μg/ml) or recombinant human BAFF (200 ng/ml) (R&D Systems). For ELISA and plasmablast assays, naïve B cells were stimulated with either anti-IgM F(ab′)2 (Jackson Immunoresearch, 10 μg/ml) or dextran-coupled anti-IgD or anti-IgM (Fina Biosolutions, 100 ng/ml) plus 1 μg/ml anti-CD40 (R&D Systems) and 10 ng recombinant human IL-21 (BioLegend). T cells were stimulated with 2 μg/ml each of anti-CD3ε and anti-CD28 mAbs (BD Biosciences) or MACS iBead particles (1:1 bead:cell ratio) loaded with biotinylated anti-CD2, anti-CD3ε, and anti-CD28 Abs (Miltenyi Biotec) in complete RPMI.
Proliferation/survival assays and ELISAs
Purified naïve B cells or T cells were plated at 1×105 cells/well in triplicate (200 μl/well) in 96-well flat-bottom microtiter plates and stimulated as described above. Four days later, wells were pulsed with 1 μCi of tritiated (3H-) thymidine (Perkin Elmer) and incubated for 24 hrs prior to collection on a mash harvester. Incorporation of 3H-thymidine was quantified as counts per million. Alternatively, naïve B cells were labeled with PBS containing 0.5 μM carboxyfluorescin succinyl ester (CFSE, Invitrogen), washed 2x in complete RPMI, and stimulated as above for 5 days prior to flow cytometric analysis. In separate experiments, cell supernatants were harvested in duplicate on day 6 post-stimulation with the reagents described above. IgM and IgG levels were measured using Ready-Set-Go ELISA kits (eBioscience) according to the manufacturer’s instructions. For B cell survival assays, cell viability of resting or activated B cells was measured over 5 days using a flow cytometer, using forward/side scatter parameters and propidium iodide exclusion to gate on viable cells.
Antibody staining and flow cytometry
Before or after stimulation, 0.5–1×106 cells were washed in cold FACS buffer (1x PBS, 1% FBS, 0.1% sodium azide) and stained for 30 min on ice with appropriate antibodies. B cells were stained with 5 μg of FITC anti-CD86, APC anti-CD25, and PE anti-CD83. T cells were stained with 5 μg of FITC anti-CD2, PE anti-CD69, and APC anti-CD25. For plasmablast generation assays, naïve B cells were stimulated as described above, harvested on day 6 and stained with PE anti-IgD and FITC anti-CD38. BJAB B cell transfectants were stained with PE anti-CD83 24 hrs post-transfection. All antibodies were obtained from BD Biosciences. Cells were subsequently washed in cold FACS buffer and acquired on a C6 Accuri Flow Cytometer (BD Biosciences). GFP reporter expression in transfected JPM50.6 cells was measured in similar fashion following 1 wash in cold FACS buffer. Cell cycle analysis of naïve B cells using propidium iodide was performed as previously described [9]. CFSE dilution was measured on day 5 post-stimulation. Data analysis was performed using C6Flow (BD Biosciences) or FlowJo software (TreeStar).
Immunofluorescence analysis
Purified primary B or T cells were adhered to poly-L-lysine coated slides and fixed for 30 min with 2% paraformaldehyde (Electron Microscopy Sciences), then permeabilized for 5 min in 0.1% Triton X-100/PBS. After blocking for 30 min in 1% FCS/PBS, cell spots were incubated overnight with the following Abs: anti-CARD11 (Abcam), anti-BCL10, anti-MALT1, anti-p65, anti-RelB (Santa Cruz Biotech), anti-phospho-IKKα/β (Cell Signaling Technology). Cell spots were washed 3x in PBS, then incubated with goat anti-rabbit or anti-mouse secondary Abs conjugated to AlexaFluor 488, 568, or 647 (Invitrogen) plus a 1:10000 dilution of Hoechst 33342 (30 min per Ab). Following 6x PBS washes, coverslips were attached using Fluoromount (Southern Biotech). Fluorescent images were acquired on a Zeiss 710 confocal laser scanning microscope using a 63x oil immersion objective, and analyzed using Zeiss ZEN software. Jurkat or BJAB cells transfected with Venus-CARD11 were prepared and analyzed in similar fashion; CARD11-FLAG was visualized after staining with an AlexaFluor 647-conjugated anti-FLAG Ab (Cell Signaling Technology).
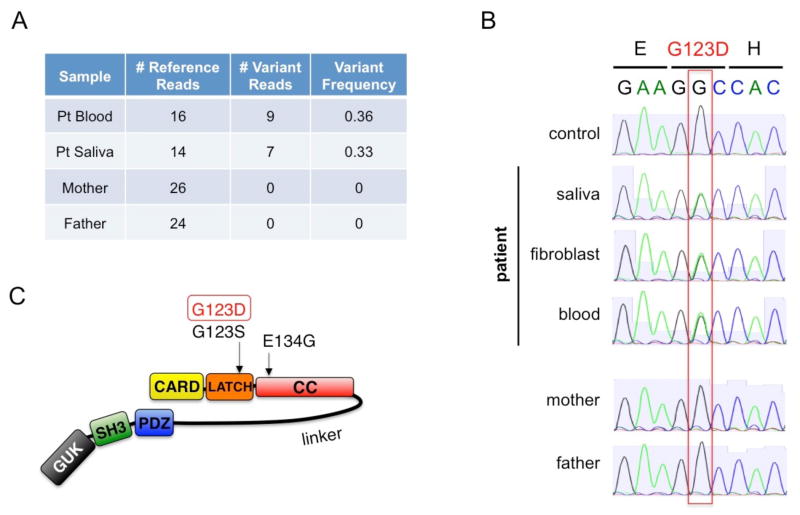
Exome Sequencing
Exome capture was performed on DNA extracted from PBMC and saliva using the SureSelectXT exon kit (Agilent). Paired-end 50/35bp sequencing was performed using the SOLiD™ platform (Life Technologies), yielding 164–214 million reads per sample. Sequences were mapped to the human reference genome (build 37) using the BFAST algorithm with default parameters, with a resulting average median coverage of 30x in the exome capture region. Technical duplicates were removed using Picard, and the GATK algorithm was used to call variants. Because both parents were phenotypically normal with no lymphocytosis, we tested for de novo mutations in the patient using the following filters to minimize false positive calls: coverage ≥ 20 at variant position in all samples (both parents, blood and saliva DNA from patient), variant allele frequency =0 in both parents and ≥ 0.3 in both blood and saliva samples. Application of these filters resulted in only 1 remaining candidate mutation, a C→T change on chromosome 7 position 2984162 causing a G123D amino acid change in CARD11. The presence or absence of this mutation in DNA samples from the patient, mother, and father was confirmed by Sanger sequencing (USU BIC Genomics Facility) after PCR amplification of CARD11 exon 5 using the following primers: CARD11-Ex4F (5′-CTACACACTCTCATAGCCCCATC-3′); CARD11-Ex4R (5′-TGTGCCTGCACCTGCTTTATG-3′). The mutation was described according to Human Genome Variation Society recommendations, using GenBank Reference Sequences NC_000007.13 (DNA), NM_032415.4 (mRNA), and NP_115791.3 (protein) based on NCBI Annotation Release 104. Coding DNA variations are described with the A of the ATG translation initiation codon designated as nucleotide +1. Note that previous studies referred to a human CARD11 sequence that omitted the first 7 N-terminal amino acid residues based upon NP_115791.2 [1, 3].
CARD11 transfections
An expression construct containing WT CARD11 and a C-terminal 3xFLAG tag driven by a hybrid EF-1/HTLV-1 promoter (pUNO-CARD11) was purchased from Invivogen. Venus-CARD11 fusion constructs were generated as previously described [1]. Site-directed mutagenesis to obtain G123D CARD11 was performed using specific primers for linear amplification, followed by DpnI digestion of methylated DNA. Mutations were verified by Sanger sequencing (USU BIC Genomics Facility). JPM50.6/E6 Jurkat cells or BJAB cells (5×106/transfection) were transfected with 5–10 μg DNA using a BTX Electroporator (BTX Harvard Apparatus, 260V, 950mF), then incubated in complete RPMI medium for 24–48 hrs prior to analysis.
Transcriptome profiling by RNA-seq
Sequencing libraries were prepared from 100 ng total RNA input using the TruSeq RNA Sample Prep Kit v2 (Illumina) with barcoded adapters. Library integrity and size distribution was determined using an Experion DNA 1k Analysis LabChip (Bio-Rad) before measuring library concentration using the Illumina/Universal Library Quantification Kit (KAPA) before clustering on a cBot (Illumina). Sequencing was performed on a Genome Analyzer IIx (Illumina) at single-read 100bp length with 22–40 million reads passing filter per sample after demuxing. FASTQ data output from CASAVA was preprocessed and filtered using FastQC and FASTQ Groomer before generation of FPKM expression values by TopHat2 and Cufflinks on a Galaxy open platform. Differential transcript expression analysis was performed using Cuffdiff and Comparative Marker Selection. Sample specific differential expression candidates were identified from the group comparison subset with expression values greatest or least by 2 standard deviations of the mean values.
Quantitative PCR analysis
cDNA was generated from 100 ng total RNA input using the iScript Advanced cDNA Synthesis Kit (Bio-Rad). qPCR was performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) using SsoFast EvaGreen Supermix (Bio-Rad). Primers span an exon-exon boundary and single amplicons of predicted size were confirmed by gel-electrophoresis and melt-curve analysis. Raw Ct values were calculated as an average of four technical replicates. Comparative expression values were calculated after normalizing with values from the housekeeping gene HPRT. Primers used were: CCNC forward = 5′ - GGC AAT GGT TTG CTG AGC TT – 3′; CCNC reverse = 5′ - GAG GTG GTT TTG GTT TTG GCA – 3′; CDK6 forward = 5′ - TGA GGC ACC TGG AGA CCT T – 3′; CDK6 reverse = 5′ - GTC TGT TCG TGA CAC TGT GC – 3′; EIF2AK2 forward = 5′ - TGA AGG TAG ATC AAA GAA GGA AGC A – 3′; EIF2AK2 reverse = 5′ - ATT CCC CAT GGA TAA TCC TTC TGA – 3′; HPRT forward = 5′ - GCA GAC TTT GCT TTC CTT GG – 3′; HPRT reverse = 5′ - TAT CCA ACA CTT CGT GGG GT – 3′; N4BP2 forward = 5′ - GGA GTT GCT CGC ATC AAA CC – 3′; N4BP2 reverse = 5′ - GCA TGA CTT TCA AGC ACC CTG – 3′; ORC2 forward = 5′ - AGG GCC ATT CTG CTT CAG AC – 3′; ORC2 reverse = 5′ - AGA CTA CGA GGT GCT GTT GAC – 3′.
KREC assay
Peripheral Blood Mononuclear cells (PBMCs) were stained and sorted as previously described to obtain CD10+CD21+CD27− immature/transitional and CD10−CD21+CD27− naïve cells [1]. The replication history of sorted fractions was evaluated using a PCR-based assay for measuring κ-deletion recombination excision circles (KREC). The ratio of KREC joints (signal joint) to the Jκ-Cκ recombination genomic joints (coding joint) for each subset was determined as previously described [10][10][11].
Results
Proliferation and functional responses in subject’s B cells
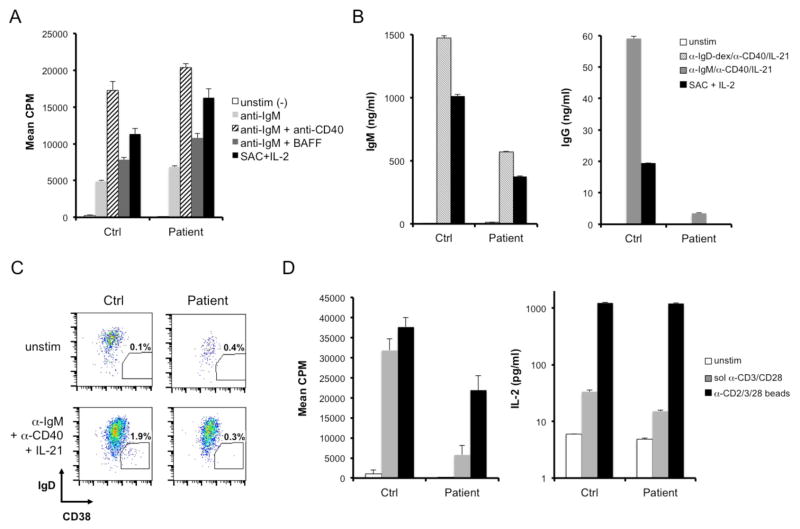
We first evaluated the proliferative capacity of the patient’s B cells in response to various activating stimuli. Proliferation was evaluated by 3H-thymidine incorporation after 4 days in culture. Naïve B cells from the patient exhibited slightly enhanced proliferation in response to B cell receptor crosslinking with anti-IgM, −/+ anti-CD40 or recombinant BAFF, relative to normal controls (Figure 2A). Patient B cells also proliferated robustly in response to heat-killed Staphylococcus aureus Cowan I (SAC) plus IL-2, a potent polyclonal B cell mitogen. Neither control nor patient B cells exhibited appreciable 3H-thymidine incorporation in the absence of stimulation, suggesting that the patient’s resting B cells were not actively proliferating. B cells also failed to proliferate in response to PHA, BAFF, IL-1, IL-2, IL-4, IL-5, or IL-6 alone (data not shown). In response to activating stimuli, patient B cells also showed comparable blastogenesis and upregulation of NF-κB-dependent cell surface markers (CD25, CD86, CD83) relative to controls (Supplemental Figure 2). Despite normal signs of naïve B cell activation, patient B cells secreted ~30% less IgM when stimulated with polyclonal dextran-coupled anti-IgD, a T cell-independent type 2-like stimulus known to induce robust IgM production from human B cells [11] (Figure 2B). Moreover, IgG production was severely impaired from the patients’ B cells in response to bivalent anti-IgM plus anti-CD40 and IL-21, which can induce class switching in vitro in human B cells [12] (Figure 2B). IgM and IgG production from patient B cells was also decreased after stimulation with heat-killed Staphylococcus aureus Cowan (SAC) plus IL-2. Defective immunoglobulin secretion and class-switching in patient B cells was consistent with poor generation of IgD−CD38hi plasmablasts in vitro in response to these stimuli (Figure 2C). These data indicate an intrinsic defect in B cell differentiation for this patient beyond the naïve, mature stage, which may help to explain his poor titers to vaccines and the dearth of circulating class-switched and memory B cells noted in Table 1.
Figure 2. B and T cell activation in a suspected BENTA patient.
(A) Proliferation of naïve, CD10− B cells purified from the patient and a normal control donor was measured by 3H-thymidine incorporation in response to various B cell activating stimuli for 4 days. The polyclonal mitogen SAC + IL-2 served as a positive control. Patient responses were significantly higher than control for all stimuli (p < 0.02) except anti-IgM + anti-CD40. (B) Secreted IgM and IgG levels were measured by ELISA in cell supernatants following 6 days of activation with various B cell activating stimuli. (C) Plasmablast generation from control and patient naïve B cells was quantified by flow cytometry following 6 days in culture left unstimulated (top row) or activated with anti-IgM, anti-CD40, and IL-21 (bottom row). Gates denote the % of cells that differentiated into IgD−CD38hi plasmablasts. (D) Proliferation of purified T cells was measured as in (A) following 4 day stimulation with soluble anti-CD3ε + anti-CD28 mAbs (gray) or bead-coupled anti-CD2/CD3/CD28 Abs (black) (left panel). IL-2 secretion was measured by ELISA in cell supernatants following 3 days of stimulation as described above (right panel). For (A–D), data show mean ± standard deviation of triplicate wells.
In contrast to B cells, T cells purified from the patient demonstrated relatively poor proliferation in response to anti-CD3/anti-CD28 stimulation, relative to normal control T cells (Figure 2D). Although potent stimulation with anti-CD2/CD3/CD28-coated beads induced more patient T cell proliferation, 3H-thymidine incorporation was still significantly below control T cells. Parallel experiments tracking cell division via dilution of carboxyfluorescein succinimidyl ester (CFSE) demonstrated congruent results for both CD4+ and CD8+ T cells (data not shown). Patient T cells also secreted ~50% less IL-2 in response to soluble anti-CD3/anti-CD28 stimulation, although IL-2 production was comparable to control T cells in response to anti-CD2/CD3/CD28-coupled beads (Figure 2D). Mild T cell hyporesponsiveness, coupled to overt B cell expansion in vivo with impaired Ig production and memory/plasma cell formation, was consistent with BENTA disease in this patient.
Discovery of germline de novo CARD11 mutation
Based on the striking patient phenotype and lack of other affected family members, we hypothesized that the variant of interest was likely to be a de novo mutation. After informed consent, DNA was extracted from samples taken from patient’s blood and saliva as well as from blood taken from the unaffected biological parents. Whole exome sequencing was performed for these 4 samples. Bioinformatics analysis was performed to identify candidate mutations that fit this model and to filter likely false positive variant calls. The analysis yielded only 1 candidate variant, a de novo heterozygous CARD11 mutation in exon 5 position 368G→A, G123D (Figure 3A). Sanger dideoxy DNA sequencing performed on samples from patient’s blood, saliva, and fibroblasts, as well as parents’ blood, confirmed the mutation as de novo germline in the patient (Figure 3B). This mutation is at the same amino acid position as one of the previously described BENTA cases but with different residue change (G123S) [1], and the exact mutation was reported as a somatic change in one case of DLBCL [7]. G123D is located within the LATCH domain (residues 112–130), in contrast to most other somatic CARD11 mutations that are located just downstream in the coiled-coil CC domain, [8] (Figure 3C).
Figure 3. De novo, heterozygous missense CARD11 mutation (G123D) detected in new BENTA patient.
(A) Summary of exome sequencing data at chr7:2984162. A C→T substitution in exon 5 of CARD11 was detected in reads obtained from DNA libraries derived from patient blood and saliva, with no mutation evident in parents’ samples. (B) Sanger dideoxy-DNA sequencing confirmation of the G123D missense mutation in genomic DNA derived from patient samples (blood, saliva, fibroblasts), but not mother and father. The reference sequence is shown in reverse complement orientation (G->A substitution), reflecting the direction of protein translation. (C) Schematic diagram of the CARD11 protein. The G123D mutation in the putative LATCH domain is denoted, relative to other BENTA-associated mutations.
CARD11 G123D mutant spontaneously aggregates and drives constitutive NF-κB activation
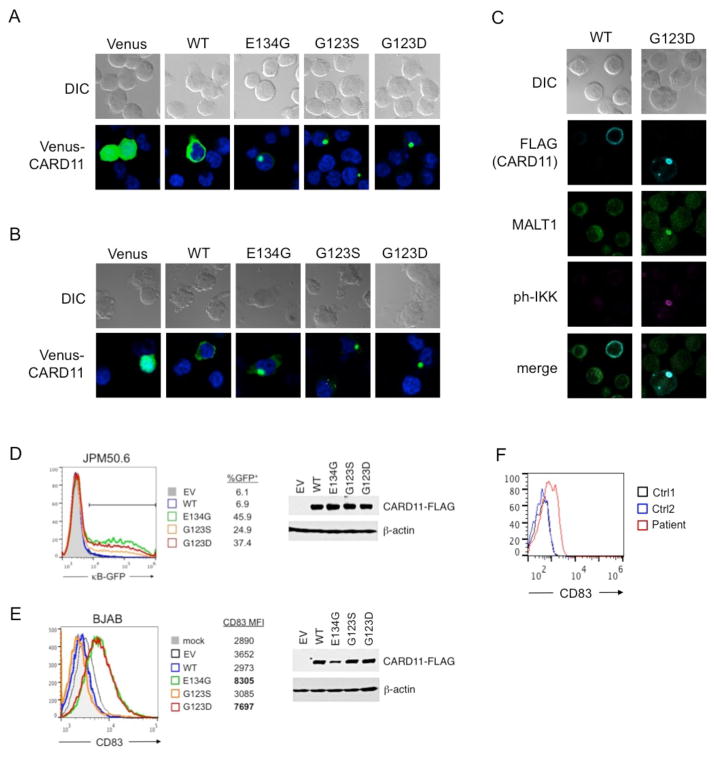
In resting lymphocytes, WT CARD11 is maintained in a closed, inactive state until antigen receptor engagement triggers phosphorylation of serines in the inhibitory linker domain (ID). An ensuing conformational change following phosphorylation allows CARD11 to multimerize and recruit numerous molecules required for IκB kinase (IKK) activation. Gain-of-function mutations in either the CC or LATCH are thought to induce CARD11 signaling by disrupting intramolecular contacts with the inhibitory linker domain, inducing spontaneous CARD11 aggregation without requiring antigen receptor signaling [1, 8]. Similar to other BENTA-associated mutants, G123D CARD11 formed large, often single cytoplasmic aggregates when expressed as a Venus fusion protein in both Jurkat T cells (Figure 4A) and BJAB B cells (Figure 4B). By contrast, WT CARD11 remained evenly distributed throughout the cytoplasm. Microscopy analysis using FLAG-tagged CARD11 constructs revealed substantial colocalization with MALT1 and active, phosphorylated IKK within clusters of G123D CARD11 (Figure 4C), suggesting that mutant CARD11 aggregates are sites of active IKK signaling.
Figure 4. Spontaneous aggregation and constitutive signaling of G123D CARD11 in B and T cell lines.
(A) Ectopic expression of WT, G123D, and other BENTA-associated CARD11 mutants (E134G, G123S) as N-terminal Venus fusion proteins in transfected Jurkat T cells. Single, punctate spots of Venus-CARD11 mutants indicate aggregation, relative to cytoplasmic distribution of WT. Empty vector (Venus only) is shown for comparison; data are representative of at least 3 experiments. (B) Venus-CARD11 proteins were transfected and visualized as in (A) in BJAB B cells. Data are representative of at least 3 experiments. (C) Jurkat T cells transfected with WT or G123D CARD11-FLAG (blue) were analyzed by confocal microscopy using AlexaFluor647-anti-FLAG plus anti-MALT1 (green) and anti-phospho-IKKα/β Abs (red). MALT1 and phospho-IKK were detected using AlexaFluor488 and 594-conjugated secondary Abs, respectively. Similar results were obtained for transfected BJAB B cells. Data are representative of two separate experiments. (D) CARD11-deficient Jurkat cells (JPM50.6) harboring an NF-κB responsive GFP reporter cassette were transfected with empty vector (EV), WT or various mutant CARD11-FLAG constructs. At 48 hours post transfection, the percentage of GFP+ cells (indicating active NF-κB) was analyzed by flow cytometry. CARD11-FLAG expression was confirmed by immunoblotting of cell lysates 48 hours post-transfection; actin serves as a loading control. Data are representative of 3 separate experiments. (E) CD83 expression was measured on BJAB B cells 24 hours post-transfection with CARD11-FLAG constructs as in (D). Mean fluorescence intensity (MFI) of CD83 is listed for each construct, and CARD11-FLAG expression in cell lysates was confirmed by immunoblotting. Data are representative of 3 separate experiments. (F) CD83 expression on unstimulated, naïve B cells isolated from the patient and two independent controls. Data are representative of two independent blood draws.
Through a quantitative signaling screen, the G123D CARD11 mutation was recently identified to drive NF-κB-dependent transcriptional activity in overexpression experiments [8]. To confirm this finding and compare to other mutants, we expressed the wild-type (WT) and mutant CARD11 constructs in CARD11-deficient JPM50.6 cells (a derivative of Jurkat T cells) containing an NF-κB-driven GFP reporter gene. In contrast to WT CARD11 or empty vector alone, G123D CARD11 induced GFP expression in JPM50.6 T cells without requiring T cell receptor stimulation (Figure 4D). Specific NF-κB activity associated with G123D CARD11 expression was comparable to other BENTA-linked mutants, although G123S CARD11 was consistently weaker in this assay, despite equal expression of all mutants tested. Constitutive NF-κB activation was also observed in BJAB B cells transfected with G123D CARD11, as assessed by upregulation of the NF-κB-dependent marker CD83 (Figure 4E). Surprisingly, G123S failed to induce CD83 despite comparable expression, suggesting that selected NF-κB target genes may be differentially affected by various gain-of-function missense mutations, even at the same codon. Indeed, this pattern was congruent with steady-state CD83 expression measured on naïve B cells, which was elevated in this patient (Figure 4F) and BENTA patients harboring the E134G mutation, but not G123S [1]. Taken together, these data strongly suggest G123D CARD11 spontaneously signals and promotes NF-κB activation in lymphocytes independently of antigen receptor stimulation.
CARD11 aggregation and NF-κB activity is increased in patient lymphocytes
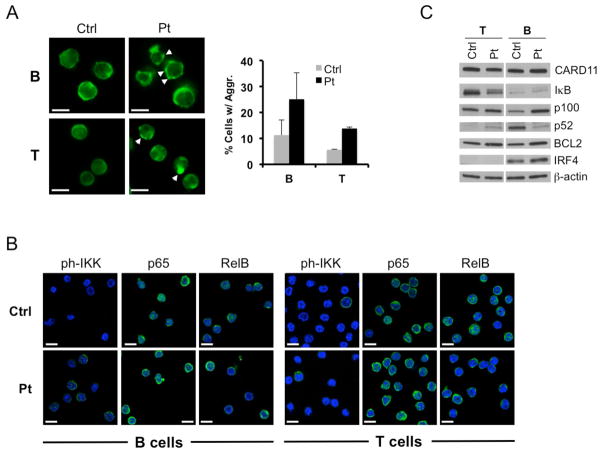
We next examined several indicators of NF-κB activity in resting B and T cells derived from our patient and healthy donors. Using confocal microscopy, we detected a higher proportion of the patients’ lymphocytes with punctate aggregates of CARD11 relative to control, with more found in B cells than T cells (Figure 5A). In some cases, particularly for patient T cells, we observed large single clusters of CARD11 resembling those in our transient transfections (Figure 5A, bottom right). B cells from patient and control contained comparable levels of IκB expression (Figure 5C) as well as nuclear localization of RelA (p65) or RelB (Figure 5B). It was also difficult to discern any difference in phosphorylated IKKα/β levels between patient and control B cells (Figure 5B). However, immunoblotting of cell lysates revealed higher levels of p100, BCL2, and IRF4 - all direct gene targets induced by canonical NF-κB signaling (Figure 5C). Similar to other BENTA patients, p52 levels were decreased in this patient’s B cells, implying that cleavage of p100 resulting from non-canonical NF-κB signaling may be less prevalent. In contrast, phosphorylated IKKα/β in patient T cells as well as nuclear localization of RelA (p65) or RelB were similar relative to control (Figure 5B). However, IκB levels were decreased and patient T cells displayed increases in BCL2 and p100 expression, with slightly more p52 detected (Figure 5C). Taken together, these data imply that in spite of its gain-of-function potential, G123D CARD11 is likely not signaling constitutively in the majority of the patients’ peripheral lymphocytes, contributing to only a mild increase in NF-κB activation. This may reflect the dynamic nature of NF-κB signaling, which may only be “on” transiently in a fraction of primary BENTA patient lymphocytes at any given time.
Figure 5. Evidence of marginally elevated CARD11-induced NF-κB signaling in patient lymphocytes.
(A) Confocal microscopy analysis of CARD11 expression (green) in purified B and T cells from the patient (Pt) and a normal donor (Ctrl). The percentage of cells with visible CARD11 aggregates (white arrowheads) was quantified by scoring ~200 cells from multiple fields. Scoring data represent mean ± standard deviation from two blinded scorers; p<0.03 (B cells), p<0.01 (T cells). Scalebars = 5 μm. (B) Confocal microscopy imaging of phospho-IKK, p65 (RelA) and RelB in control and patient lymphocytes. Scalebars = 5 μm. (C) Immunoblotting of total lysates from purified T cells and naïve B cells prepared from the patient (Pt) and a normal donor (Ctrl). Detected proteins are listed at right; β-actin serves as a loading control. Data are representative of two separate blood draws.
Concomitant hyperproliferation and decreased apoptosis sensitivity in patient B cells following activation
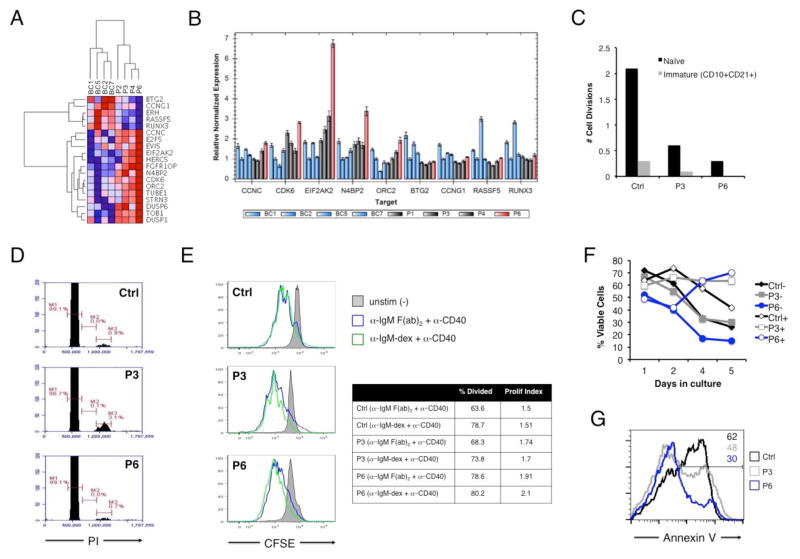
The distinguishing phenotypic feature in this BENTA patient is more severe B cell lymphocytosis (Table 1). However, this profound B cell accumulation correlates neither with the potency of the G123D mutation to drive NF-κB (Figure 4D–E), nor with the frequency of resting B cells showing relatively increased NF-κB activity (Figure 5). To search for broader gene expression differences in our patient, we performed RNA-Seq analysis on naïve B cells purified from four BENTA patients (including this patient, herein referred to as P6) versus four normal donor B cell controls (BC1, BC2, BC5, and BC7). Hierarchical cluster analysis revealed that the transcriptome profiles of all four BENTA patients clustered separately from three of four controls, with P4 (G123S) and P6 (G123D) bearing closer resemblance relative to siblings P2 and P3 (E134G) (Supplemental Figure 3). We then identified specific genes that were differentially expressed in P6 B cells (−/+ 2 standard deviations) compared to mean expression values from both patients and controls. Subsequent gene ontology analysis of this subset revealed a significant signature of 19 cell cycle-related genes that distinguished P6 B cells from controls and other BENTA patients (Figure 6A). Multiple oncogenes associated with cell cycle progression (e.g. CCNC, EVI5, CDK6) were elevated in BENTA patient cells, with highest expression levels observed in P6 [13–16], whereas several transcriptional repressors and/or tumor suppressor genes (e.g. CCNG1, RASSF5) appeared to be downregulated (Figure 6A) [17–19]. Genes associated with cell cycle progression and NF-κB-dependent transcription, including some co-opted for EBV replication and lymphomagenesis (CDK6, N4BP2, ORC2, EIF2AK2), were validated by qPCR as significantly overexpressed in P6 B cells compared to both controls as well as other BENTA patients (Figure 6B) [20–23]. Genes in the tumor suppressor/transcriptional repressor category were expressed at lower levels in all BENTA patients compared to controls and expression levels between P6 and other patients were not significantly different by qPCR analysis (Figure 6B).
Figure 6. Comparative analysis of enhanced cell cycle progression and survival of patient B cells upon stimulation.
(A) Heat map of 19 differentially expressed cell cycle-related genes in naïve B cells from the patient (P6) relative to other BENTA patients and B cell controls. Color intensity represents fragments per kilobase per million (FPKM) values derived from paired-end RNA-Seq analysis. (B) Quantitative PCR validation of the expression of selected genes in controls versus BENTA patients, derived from the cell cycle signature outlined in (A). (C) KREC analysis of in vivo replication history (displayed as # of cell divisions) for sorted immature and mature naïve B cells from P6, P3, and a normal control. (D) Cell cycle profiling of naïve B cells from control, P3, and P6. Markers represent the percentage of cells in G1 (M1), S (M2), and G2/M (M3) phases of the cell cycle. Data in (C) and (D) are representative of two experiments. (E) CFSE dilution analysis of naïve B cells left unstimulated (gray filled histogram) or activated with anti-IgM F(ab)2 plus anti-CD40 (blue) or dextran-conjugated anti-IgM plus anti-CD40 (green) for 5 days. The percentage of divided cells and the proliferative index (total # of divisions/total # of dividing cells) are displayed at right for activated samples. Data are representative of three separate experiments. (F) Cell survival assays for naïve B cells purified from a normal control (black), P3 (gray), or P6 (blue) −/+ stimulation with anti-IgM F(ab)2 plus anti-CD40. Cell viability was measured over 5 days in culture. Data are representative of two separate experiments. (G) Quantitation of apoptotic cells following BCR stimulation. The percentage of Annexin V+ apoptotic cells (inset numbers) within the actively dividing population (based on CFSE dilution) from (F) was determined on day 5 by flow cytometry.
Based on these data, we next asked whether P6 B cells were cycling more through homeostatic proliferation. We first estimated the number of cell divisions B cells underwent after exiting the bone marrow using PCR-based quantification of kappa-deletion recombination excision circles (KRECs). KRECs are generated during the last stages of gene rearrangement at the immunoglobulin kappa light chain locus, and diluted 2-fold with each cell division [24]. KREC analysis indicated that circulating naïve and immature/transitional B cells isolated from P3 and P6 had actually divided fewer times in vivo relative to a normal donor (Figure 6C). Likewise, cell cycle profiling of naïve peripheral B cells using propidium iodide showed no increase in cells in the S or G2/M phases for P6 relative to P3 or control (Figure 6D). Thus these data suggest that profound B cell lymphocytosis in BENTA patients, including P6, is not explained by heightened homeostatic turnover. In contrast, a slightly greater proportion of P6 naïve B cells divided more vigorously than control or P3 B cells following stimulation with bivalent and dextran-conjugated anti-IgM, suggesting P6 B cells can undergo more robust proliferation after B cell receptor (BCR) engagement (Figure 6E). Strikingly, the proportion of viable, activated P6 B cells actually increased over time, despite lower relative viability on P6 cells on day 1 (Figure 6F). Indeed, the proportion of Annexin V+ cells within the actively dividing population was substantially lower for P6 B cells (30%) compared to control (62%) (Figure 6G), suggesting P6 B cells were far less sensitive to apoptosis triggered by BCR activation. P3 B cells also maintained better viability over time relative to control B cells, which declined over time. Hence enhanced accumulation of activated P6 B cells in vitro may be explained by both greater proliferation upon exogenous stimulation and decreased cell death sensitivity following BCR stimulation. In line with our results, transduction of several gain-of-function CARD11 mutations into murine B cells was recently shown to protect those cells from antigen-induced cell death [25]. In comparison to previous BENTA cases reported [1], these new findings suggest that severe B cell lymphocytosis in this patient may be partly explained by mildly elevated proliferation following stimulation, and significantly better cell survival in a BCR-dependent manner.
Discussion
Here we report our discovery of a patient with a de novo germline mutation in CARD11 (G123D), associated with profound B cell lymphocytosis. The mutation was discovered through whole exome sequencing, where we detected the presence of a heterozygous mutation in the patient that was absent in both biological parents. As the previous 4 reported patients with germline CARD11 mutations included a father-pair of children and an adopted child [1], this is the first case that is a proven de novo mutation. We conclude this patient belongs to a group of patients with mutations of this gene and similar clinical features, which we refer to as BENTA disease. Differentiating features of BENTA disease include splenomegaly, specific expansion of both naïve and transitional T2/T3 B cells in the blood with no overt evidence of autoimmunity, poor humoral responses to selected vaccines, and mild T cell hyporesponsiveness. CARD11 expression is largely restricted to lymphoid cells, explaining why the involvement of other organs has not been observed to date in BENTA patients.
The distinguishing feature of this patient is a remarkably severe B cell lymphocytosis, with 5–10x more B cells in the blood than other BENTA patients described to date. Similar to previous BENTA patients, excessive B cell accumulation in this patient is likely not explained by greater homeostatic proliferation, further implying that enhanced output of T2/T3-like immature B cells from the bone marrow is a major driver of B cell accumulation in this syndrome [1]. By contrast, transcriptome profiling and cellular assays suggested P6 B cells may be specifically “primed” for enhanced cell cycle progression and protection from apoptosis triggered by BCR ligation. As more BENTA patients are identified, it will be interesting to study whether expression levels of specific cell cycle or apoptosis-related genes may predict the extent of B cell accumulation in any given patient. Jeelall et al. recently demonstrated that similar gain-of-function CARD11 mutants (including G123S) can switch antigen-induced B cell deletion into hyperproliferation of murine B cells [25]. Our data implies G123D may work in a similar fashion for P6 B cells, although other gene modifications may play a role as well. Indeed, the potency of this mutant may manifest more clearly with BCR crosslinking, dependent on activation of NF-κB or other signaling pathways (e.g. JNK) tied to CARD11 function.
In addition to the differential functional consequences of the G123D CARD11 mutation demonstrated, important details in the clinical history of this patient likely contribute to the severe B cell lymphocytosis observed. Similar to the first BENTA patient described [1], B cell lymphocytosis was dramatically exacerbated following splenectomy (Figure 1B), suggesting that removal of this important reservoir for B cell maturation contributes to a perceived spike in circulating transitional and mature B cells. Furthermore, the only other BENTA patient infected with EBV (P4) also showed comparatively higher B cell counts. EBV is a well-known driver of B cell blastogenesis and proliferation, and a contributing factor to persistent polyclonal B cell lymphocytosis (PPBL), a phenotypically similar disorder to BENTA [26]. Thus EBV could represent a relevant prognostic indicator of lymphocytosis severity in BENTA patients.
The CARD11 mutation found in our patient lies at the same codon as another unrelated patient, but with a different amino acid change (G123D versus G123S). Both missense mutations have been tied to separate cases of DLBCL [7, 3], and our data confirms both are capable of stimulating NF-κB activation in B and T cells. A recent screen for gain-of-function CARD11 mutations outside of the CC domain identified G123D as a strong NF-κB inducer and illuminated the LATCH domain (residues 112–130) as an important negative regulator of CARD11 signaling [8]. The potency of G123D suggested by this study may explain in part why B cell numbers are substantially increased in this patient relative to other BENTA subjects. However, our JPM50.6 reporter assay indicated G123D induces comparable levels of NF-κB activity relative to E134G, slightly above the activity of G123S. More surprisingly, we found that the NF-κB-dependent gene CD83 is induced by G123D (and E134G) but not G123S, suggesting different gain-of-function mutations (even at the same codon) may signal differently in BENTA patient lymphocytes. Further structure-function and gene expression studies are required to determine how different activating CARD11 mutations potentially control distinct subsets of NF-κB-regulated genes that influence the presentation and severity of BENTA phenotypes. Indeed, our previous clinical workup implies that T cell immunodeficiency is more severe in BENTA patient 4 (G123S mutation), manifesting in chronic infections with EBV, molluscum contagiosum and BK virus ([1] and data not shown).
Despite the ability of G123D CARD11 to induce NF-κB, our results suggest that only a fraction of patient lymphocytes are actively signaling through NF-κB at any given time. NF-κB induction is known to be both transient and oscillatory, reflecting multiple layers of feedback regulation in place to suppress its oncogenic potential [27]. In fact, constant NF-κB activity associated with B cell tumors is often coupled to deleterious mutations in critical tumor suppressor genes like A20 that normally function to turn off NF-κB [28]. Thus it is likely that B cell lymphoma/leukemia development in BENTA patients is dependent on additional mutations that disable feedback regulation or enhance the pro-proliferative and survival functions tasked to active NF-κB. Moreover, levels of NF-κB signaling observed in primary patient cells are also likely influenced by the nature of the CARD11 mutation itself, the maturation state of affected lymphocytes, and/or the clinical management of each patient. For example, substantial B cell lymphocytosis in the patient’s bone marrow suggests elevated output of immature B cells is partly responsible for B cell lymphocytosis, especially since these cells appear to replicate less upon egress (Figure 6C). It is possible that an initial burst of NF-κB activity, coupled to the first dramatic upregulation of CARD11 expression noted after the pre-B cell stage, is the major driver for B cell proliferation in the bone marrow. In contrast, negative feedback mechanisms may dominate in mature, resting B cells to modulate NF-κB activation in the presence of sustained expression of mutant CARD11. Although somatic mutations in CARD11 are frequently found in DLBCL, additional genomic alterations are clearly required for full-blown B cell transformation in BENTA, as noted in the first patient described that developed B-CLL in adulthood [1].
In conclusion, the discovery and characterization of a new patient with a unique de-novo germline CARD11 mutation advances our understanding of BENTA, a newly recognized, congenital B cell lymphoproliferative disorder with features of immunodeficiency. This report identifies important molecular, cellular, and clinical correlates that help elucidate phenotypic differences between patients with germline CARD11 mutations, shedding new light on the role of CARD11 in B and T cell biology. In addition to BENTA, the recent description of two patients with loss-of-function CARD11 mutations and combined immunodeficiency further highlights the critical role of CARD11 in lymphocyte function and development [29, 30]. Further studies are warranted to elucidate how CARD11 dysregulation can manifest in impaired lymphocyte function as well as selective B cell expansion and eventual transformation. In addition, we posit that profound B cell accumulation in BENTA makes these patients a natural laboratory for studying how additional genomic changes coupled with CARD11 dysregulation may culminate in B cell transformation.
Supplementary Material
Acknowledgments
We thank the patient and their family for participating in this study. We also thank Dr Koneti Rao for clinical consultation and Ms. Shakuntala Guprasad for flow cytometry support. This work was supported in part by the Intramural Research Program of the National Institutes of Health, the National Institute for Allergy and Infectious Diseases, and the National Cancer Institute. The authors also acknowledge support from the Concern Foundation and an intramural grant from Uniformed Services University of the Health Sciences (to A.L. Snow), and from DanceBlue, a student-run effort at the University of Kentucky to support pediatric oncology clinical care and research.
Footnotes
The authors declare they have no conflict of interest.
References
- 1.Snow AL, Xiao W, Stinson JR, Lu W, Chaigne-Delalande B, Zheng L, et al. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J Exp Med. 2012;209(12):2247–61. doi: 10.1084/jem.20120831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. nature08638 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–9. doi: 10.1126/science.1153629. 1153629 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. cshperspect.a000109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamason RL, McCully RR, Lew SM, Pomerantz JL. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry. 2010;49(38):8240–50. doi: 10.1021/bi101052d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–21. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan W, Schaffer TB, Pomerantz JL. A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival. Mol Cell Biol. 2013;33(2):429–43. doi: 10.1128/MCB.00850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snow AL, Lambert SL, Natkunam Y, Esquivel CO, Krams SM, Martinez OM. EBV can protect latently infected B cell lymphomas from death receptor-induced apoptosis. J Immunol. 2006;177(5):3283–93. doi: 10.4049/jimmunol.177.5.3283. [DOI] [PubMed] [Google Scholar]
- 10.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205(8):1797–805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunswick M, Finkelman FD, Highet PF, Inman JK, Dintzis HM, Mond JJ. Picogram quantities of anti-Ig antibodies coupled to dextran induce B cell proliferation. Journal of immunology. 1988;140(10):3364–72. [PubMed] [Google Scholar]
- 12.Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, et al. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175(12):7867–79. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 13.Baron BW, Anastasi J, Bies J, Reddy PL, Joseph L, Thirman MJ, et al. GFI1B, EVI5, MYB--additional genes that cooperate with the human BCL6 gene to promote the development of lymphomas. Blood cells, molecules & diseases. 2014;52(1):68–75. doi: 10.1016/j.bcmd.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Law ME, Theis JD, Gamez JD, Caron LB, Vrana JA, et al. Clinicopathologic features of CDK6 translocation-associated B-cell lymphoproliferative disorders. The American journal of surgical pathology. 2009;33(5):720–9. doi: 10.1097/PAS.0b013e3181934244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishida T, Kobayashi N, Tojo T, Ishida S, Yamamoto T, Inoue J. CD40 signaling-mediated induction of Bcl-XL, Cdk4, and Cdk6. Implication of their cooperation in selective B cell growth. J Immunol. 1995;155(12):5527–35. [PubMed] [Google Scholar]
- 16.Liu ZJ, Ueda T, Miyazaki T, Tanaka N, Mine S, Tanaka Y, et al. A critical role for cyclin C in promotion of the hematopoietic cell cycle by cooperation with c-Myc. Mol Cell Biol. 1998;18(6):3445–54. doi: 10.1128/mcb.18.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horne MC, Donaldson KL, Goolsby GL, Tran D, Mulheisen M, Hell JW, et al. Cyclin G2 is up-regulated during growth inhibition and B cell antigen receptor-mediated cell cycle arrest. The Journal of biological chemistry. 1997;272(19):12650–61. doi: 10.1074/jbc.272.19.12650. [DOI] [PubMed] [Google Scholar]
- 18.Horne MC, Goolsby GL, Donaldson KL, Tran D, Neubauer M, Wahl AF. Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression. The Journal of biological chemistry. 1996;271(11):6050–61. doi: 10.1074/jbc.271.11.6050. [DOI] [PubMed] [Google Scholar]
- 19.Katagiri K, Ueda Y, Tomiyama T, Yasuda K, Toda Y, Ikehara S, et al. Deficiency of Rap1-binding protein RAPL causes lymphoproliferative disorders through mislocalization of p27kip1. Immunity. 2011;34(1):24–38. doi: 10.1016/j.immuni.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Lin SS, Lee DC, Law AH, Fang JW, Chua DT, Lau AS. A role for protein kinase PKR in the mediation of Epstein-Barr virus latent membrane protein-1-induced IL-6 and IL-10 expression. Cytokine. 2010;50(2):210–9. doi: 10.1016/j.cyto.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, et al. Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS pathogens. 2011;7(2):e1001275. doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe N, Wachi S, Fujita T. Identification and characterization of BCL-3-binding protein: implications for transcription and DNA repair or recombination. The Journal of biological chemistry. 2003;278(28):26102–10. doi: 10.1074/jbc.M303518200. [DOI] [PubMed] [Google Scholar]
- 23.Zheng MZ, Qin HD, Yu XJ, Zhang RH, Chen LZ, Feng QS, et al. Haplotype of gene Nedd4 binding protein 2 associated with sporadic nasopharyngeal carcinoma in the Southern Chinese population. Journal of translational medicine. 2007;5:36. doi: 10.1186/1479-5876-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204(3):645–55. doi: 10.1084/jem.20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeelall YS, Wang JQ, Law HD, Domaschenz H, Fung HK, Kallies A, et al. Human lymphoma mutations reveal CARD11 as the switch between self-antigen-induced B cell death or proliferation and autoantibody production. J Exp Med. 2012;209(11):1907–17. doi: 10.1084/jem.20112744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larcher C, McQuain C, Berger C, Mitterer M, Quesenberry PJ, Huemer HP, et al. Epstein-Barr virus-associated persistent polyclonal B-cell lymphocytosis with a distinct 69-base pair deletion in the LMP1 oncogene. Annals of hematology. 1997;74(1):23–8. doi: 10.1007/s002770050250. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298(5596):1241–5. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 28.Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114(12):2467–75. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 29.Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. The Journal of allergy and clinical immunology. 2013;131(2):477–85. e1. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 30.Greil J, Rausch T, Giese T, Bandapalli OR, Daniel V, Bekeredjian-Ding I, et al. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. The Journal of allergy and clinical immunology. 2013;131(5):1376–83. e3. doi: 10.1016/j.jaci.2013.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.