Abstract
A frame-shift mutation in the transcript of the ubiquitin-B gene leads to a C-terminally extended ubiquitin, UBB+1. UBB+1 has been considered to inhibit proteasomes, and as such to be the underlying cause for toxic protein buildup correlated with certain neuropathological conditions. We demonstrated that expression of extended ubiquitin variants led to accumulation of heterogeneously-linked polyubiquitin conjugates indicating a pervasive effect on ubiquitin-dependent turnover. 20S proteasomes selectively proteolysed ubiquitin extensions, yet no evidence for inhibition of 26S holoenzymes was found. However, among susceptible targets for inhibition was Ubp6, the primary enzyme responsible for disassembly of lysine-48 linkages at 26S proteasomes. Processing of lysine-48 and lysine-63 linkages by other deubiquitinating enzymes (DUBs) was also inhibited. Disruption of ubiquitin-dependent degradation by extended ubiquitin variants may therefore be attributed to their inhibitory effect on select DUBs, thus shifting research efforts related to protein accumulation in neurodegenerative processes from proteasomes to DUBs.
INTRODUCTION
Ubiquitination is a post-translational modification (PTM) influencing a broad range of regulatory processes in all eukaryotic cells. Although the main function of protein ubiquitination is targeting proteins for selective and rapid degradation within the proteasome, ubiquitination is also found in DNA repair, chromatin dynamics, mRNA export, remodeling multi-subunit complexes, and trafficking of membrane proteins 1,2. Like other PTMs, ubiquitination is reversible; a large family of deubiquitinating enzymes (DUBs) removes ubiquitin (Ub) from its target proteins, thus regulating the affected cellular processes and coordinating their timing 3. Even at proteasomes, DUBs can fine-tune the degradation outcome by removing ubiquitin, remodeling ubiquitin chains, or releasing the substrate 4. Three DUBs are reported to be associated with proteasome complexes: Ubp6/USP14, Rpn11/POH1 (yeast/human nomenclature, Supplementary Results, Supplementary Table 1) and UCHL5 4–6.
Ubiquitin can ligate to an amino group on a substrate, or to one of seven conserved lysines (or the N-terminus) of a previously-attached ubiquitin. Monomeric modifications (monoUb) or polyubiquitin chains (polyUb) of various lengths and branches provide numerous possibilities for differential recognition by ubiquitin receptors. The two most abundant linkages are via Lys48 and Lys63 of ubiquitin 7,8. Lys48-linked polyUb chains are recognized by the 26S proteasome and are the most studied signal for rapid and irreversible protein degradation. Lys63-linked ubiquitin chains are generally thought to signal for nonproteolytic functions such as intracellular localization or complex rearrangements 9. Lys63-linkages were also found to target substrate for proteasomes (primarily in vitro), although this signal for proteolysis is generally considered inefficient 10–12.
Numerous inherited diseases are caused by mutations in elements of the Ubiquitin-Proteasome System (UPS) 1,2. These diseases mainly stem from DNA-encoded malfunctions, yet some can be attributed to mistakes in RNA transcription 13. Altered mRNA is typically removed by nonsense-mediated mRNA decay (NMD) systems 14. Aberrant mRNA that evade this decay mechanism can be translated into misfolded polypeptides that should be recognized by the protein quality control machinery and removed by the UPS. Occasional dinucleotide deletion during transcription of the human ubiquitin-B gene (UBB) results in aberrant mRNA that is stable due to lack of a premature translation termination codon necessary for recognition by NMD 14. This mRNA is translated to a ubiquitin protein elongated at its C-terminus with 19 amino acids (aa), termed UBB+1 (for ‘+1’ reading frame shift) 15, which is unfortunately is a stable protein too.
Ubiquitin sequences and their structural fold are remarkably conserved across species, probably reflecting a selective pressure to maintain the entire molecule for its diverse biological functions. Indeed, a single substitution of glycine by tyrosine at position 76 in UBB+1, results in a ubiquitin molecule that cannot be activated for target modifications, but can serve as a scaffold for ligation on Lys29, Lys48 or Lys63 by an E2 enzyme to produce polyUb chains 16–18. Notably, unlike many polyUb-conjugates, UBB+1 accumulates even in its polyUb-modified form 17. Accumulation of aberrant protein products (including UBB+1) was detected in several tissues 19,20, however in post-mitotic (non-dividing) neuronal cells accumulation is particularly acute with clear medical pathologies 15.
In neurodegenerative disorders such as Alzheimer’s (AD) and Huntington’s diseases, co-appearance of UBB+1 with other polyUb-conjugates and protein aggregates 15,21 likely reflects decreased capacity of the UPS, attributed to a direct inhibition of the 26S proteasome 17,22. In order to better understand the inhibitory properties of UBB+1, we monitored interference of C-terminally extended ubiquitin (Ubext) variants on UPS in yeast cells, a proven model system for UPS research 2, and studied their direct effect on isolated elements of UPS in vitro. Expression of UBB+1 or other Ubext in yeast cells caused massive accumulation of high molecular weight (HMW) ubiquitin conjugates. We did not find any evidence that such Ubext variants inhibited isolated proteasomes, though they did bind tightly to the proteasome-associated DUB, Ubp6. Non-hydrolyzable, Ubext variants (e.g. UbG76V25aa and UBB+1) served as competitive inhibitors of Ubp6 as well as other cytosolic DUBs. These results demonstrate that Ubext variants constitute a potent class of protein-based DUB inhibitors.
RESULTS
Expression of Ubext species perturb the UPS
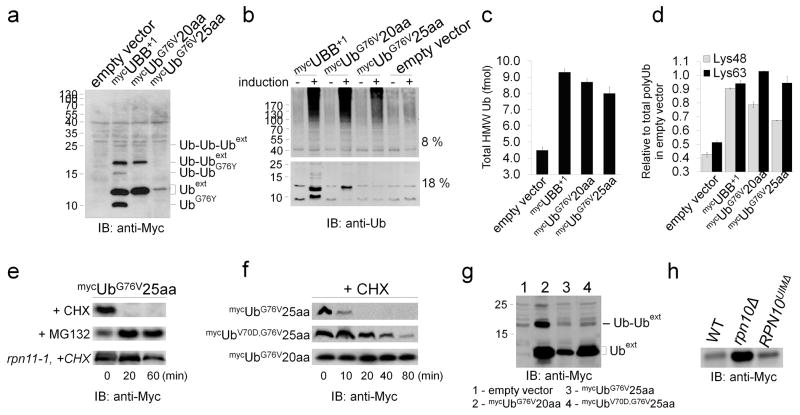
A DNA construct coding for the aberrant UBB+1 protein was expressed in yeast and compared with ubiquitin extended by 20 aa or 25 aa derived from the green fluorescent protein (GFP) sequence (mycUBB+1, mycUbG76V20aa, and mycUbG76V25aa, respectively; Fig. 1a, Supplementary Fig. 1a). As previously reported for mammalian cells 23, the steady-state levels of elongated ubiquitin species in yeast cells differed considerably; ‘+1’ or ‘+20aa’ extensions accumulated to a greater extent than ‘+25aa’ extensions, although they were all stably expressed from identical promoters. Higher molecular weight modifications of the accumulated Ubext variants were also detected (Fig. 1a). These modifications were previously shown to be mono-, di-, or higher order ubiquitination on Lys29, Lys48 and Lys63 of Ubext 16–18 (Supplementary Fig. 1b). Most likely, UbG76V25aa was also ubiquitinated, however, ubiquitinated forms were below detection level due to rapid turnover (see below). UBB+1 was partially processed into a slightly faster-migrating form, which was previously attributed to cleavage at position 76 of UBB+1 (UbG76Y) by the specific ubiquitin C-terminal hydrolase, Yuh1/UCHL3 24. This processed form of UBB+1 also underwent ubiquitination to higher molecular weight species (Fig. 1a).
Figure 1. Outcomes of Ubext species expression in Saccharomyces cerevisiae.
(a) Different Ubext species expressed from identical promoters have different steady-state cellular levels. (b) Expression of Ubext causes accumulation of HMW polyUb-conjugates. (c) Absolute quantification of the ubiquitin by LC-SRM MS/MS in gel slices (>130 kDa) from panel b (Supplementary Table 3). (d) Dissection of polyUb-conjugates by linkage type using signature peptides as internal Ub-AQUA standards. Error bars represent standard deviation of three measurements. In extracts, the majority of polyUb linkages were via Lys48 and Lys63 of Ub. Abundance of both linkage types increased significantly upon expression of Ubext relative to unperturbed cells. (e, f) Turnover of Ubext upon inhibition of synthesis by CHX chase is stabilized by inhibiting proteasome proteolysis using MG132 or mutations known to cause proteasome structural defects such as rpn11-1. (f, g) Estimation of cellular fate of a Ubext mutated in a site recognized by Ub-receptors (V70D). Compared to UbG76V25aa molecules, biological half-life of UbV70D,G76V25aa was significantly extended (f). Ubiquitin modifications of UbV70D,G76V25aa were not detected as well (g). (h) UbG76V25aa levels in a structural proteasome mutant, rpn10Δ, versus a mutant deleted for the UIM of Rpn10 reveals that the UIM is dispensable for biological turnover of this Ubext.
Next, we investigated the impact of Ubext variants on UPS function by estimating the polyUb content (using a protocol for rapid trichloroacetic acid (TCA) protein precipitation 7) after Ubext induction (Fig. 1b). For all three evaluated variants, Ubext expression was correlated with accumulation of high molecular weight (HMW) (>100 kDa) ubiquitin-conjugates (Fig. 1b). These conjugates were not detected by anti-Myc antibodies (Fig. 1a), suggesting that Ubext expression led to accumulation of other ubiquitinated substrates. Quantifying Ubext levels and linkage type in whole-cell extracts by the absolute quantification of ubiquitin (Ub-AQUA) technique 7,25 showed that in Ubext-expressing cells, the total ubiquitin content was enriched in HMW conjugates by more than two-fold and that even expression of a non-accumulating variant (mycUbG76V25aa) resulted in a significant increase in these HMW conjugate levels (Fig. 1b, c, Supplementary Table 3). Further detailed Ub-AQUA analysis clarified that both Lys48 and Lys63 were the main linkage types in HMW polyUb-conjugates, with both linkages accumulating to a similar extent in response to Ubext expression (Fig. 1d, Supplementary Table 3).
Ubiquitination converts Ubext into proteasome substrates
Accumulation of heterogeneous HMW polyUb-conjugates suggested defective clearance of natural proteasome substrates. Ubiquitination of Ubext variants (Fig. 1a, Supplementary Fig. 1a) may indicate that they are targets of the proteasome and therefore could be the source of this interference. Indeed, a viable, yet proteolytically-defective proteasome mutant (rpn11-1; Fig. 1e, Supplementary Fig. 1d) stabilized the low abundant variant (mycUbG76V25aa) by decelerating its turnover, confirming it was a proteasome substrate. Moreover, inhibiting proteasomes using MG132 blocked degradation of Ubext completely and led to accumulation of these targets (+MG132; Fig. 1e, Supplementary Fig. 2a). Consistent with previous reports determining minimal unstructured regions required for efficient proteasomal degradation of Ub-fusions 23,26,27, Ub extended by 25 aa was rapidly turned over as determined by a cycloheximide (CHX)-chase (+CHX; Fig. 1e, f, Supplementary Fig. 2a, b), whereas Ub fused in a similar manner to a shorter tail (20 aa) remained stable (Fig. 1f, Supplementary Fig. 2b). These different turnover rates can explain the observed differences in steady-state levels of proteins under investigation in this study (Fig. 1a, Supplementary Fig. 1a). We propose that Ubext variants are generally able to interact with proteasomes and that the length of their C-terminal extensions determines, to an extent, the outcome of this interaction.
In addition to the length of the C-terminal extension, the Ub domain itself could also influence the association of a Ubext molecule with proteasome complexes. Ubext species may be targeted to proteasomes via their own Ub domain either directly (if a monoUb domain is sufficient) or upon additional Ub modification(s) on their N-terminal Ub domain (if polyUb is a preferred signal for proteasome targeting). Efficient proteasome-dependent degradation of a ubiquitin-fusion degradation (UFD) substrate has been shown to require additional ubiquitination (at positions Lys48 and Lys29) 17,28. When we mutated the canonical hydrophobic element for Ub recognition by ubiquitinating enzymes (e.g., E2-25K) 16, no polyubiquitinated forms of UbV70D,G76V25aa were detected (Fig. 1g). Despite the presence of the 25 aa-long destabilizing element, (which is postulated to be sufficient for degradation of Ub molecules 23,27,29,30), UbV70D,G76V25aa accumulated in cells on par with the shorter variants (UBB+1 or UbG76V20aa; Fig. 1f, g, Supplementary Fig. 2b). The observed experimental biological half-life of UbV70D,G76V25aa increased from approximately 5 min (mycUbG76V25aa) to approximately 40 min (mycUbV70D,G76V25aa) (Fig. 1f, Supplementary Fig. 2b). An additional observation highlighting their UFD nature 31 was that Ubext accumulated in an rpn10Δ strain lacking the proteasome-localized ubiquitin-receptor subunit Rpn10/S5A, but not in a mutant lacking only the ubiquitin-interacting motif of this subunit (RPN10UIMΔ; Fig. 1h, Supplementary Fig. 2c). In contrast to rpn10Δ or rpn11-1 proteasome mutants, either of which lead to structurally defective 26S proteasomes 32,33, deletion of proteasome-associated polyUb receptors or shuttles that do not alter proteasome conformation (e.g., RPN10UIMΔ, Rpn13/hRpn13, Dsk2, and Rad23/HR23A), had no effect on turn-over rates of Ubext (Supplementary Fig. 1c, d).
Ubext are potential substrates of 20S CP
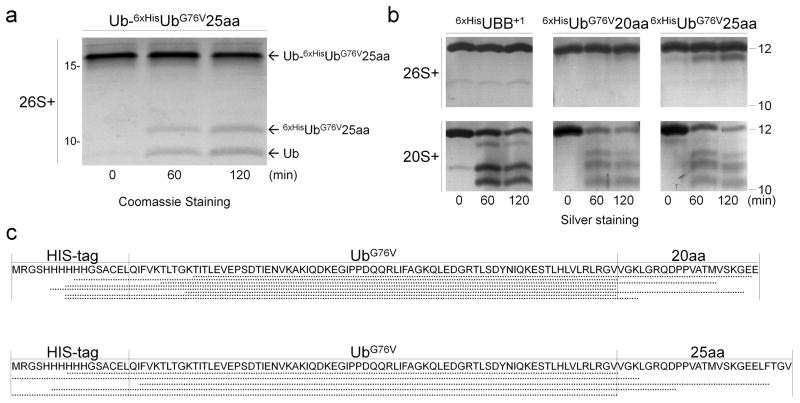
To better understand the impact of Ubext species on UPS function, we investigated the proteasome action on Ub-Ubext (ubiquitinated Ubext was the main modified form of Ubext identified in whole cell extract, Fig. 1), which we created by ubiquitinating recombinant UbG76V25aa with the E2-25K enzyme in vitro. Purified 26S proteasomes processed Ub-UbG76V25aa into its two domains (Ub and UbG76V25aa; Fig. 2a), indicating that the primary action of 26S proteasomes was deubiquitination of the isopeptide bond. Deubiquitination by 26S proteasomes could explain the observed excess of unmodified Ubext expressed in cells (Fig. 1a, g, Supplementary 1a). In vitro, all three tested Ubext variants were relatively insensitive to proteolysis by 26S proteasomes (Fig. 2b, Supplementary Fig. 3a). Even UbG76V25aa, extended by 25 amino acids (a tail that was sufficient to render it a substrate in vivo), was only mildly trimmed but remained stable. Proteolysis by 26S proteasomes occurs within a subcomplex termed the 20S proteasome core particle (CP). To recognize, anchor and process ubiquitin-conjugates, a 19S regulatory particle (RP) attaches to the 20S CP together forming the 26S proteasome holoenzyme. Surprisingly, 20S CP, which lack the 19S RP subunits that interact with ubiquitin (such as receptors, or DUBs), were able to trim Ubext non-processively regardless of the C-terminal extension length (Fig. 2b; Supplementary Fig. 3a). The reaction generated heterogeneous products. Some degradation of longer substrates (‘+20/25 aa’) was even discernible (Fig. 2b). The striking differences in catalytic activities of 20S CP vs. 26S proteasomes cautions that great care should be taken to completely separate these two species in order to guarantee unambiguous results.
Figure 2. Ubext molecules are processed by isolated 20S core particles.
(a) Purified 26S proteasomes deubiquitinate Lys48-linked extended di-Ub (Ub-6xHisUbG76V25aa), releasing the distal Ub from the proximal Ubext. (b) Purified 6xHisUbext are subjected to process by 20S (bottom) but not by 26S (top). (c) Schematic representation of products of Ubext digestion by 20S, as identified by MS (Supplementary Table 4).
Next, we analyzed the products of this degradation to better understand the interaction between the 20S proteasome subcomplex and its substrates and the mechanism of the subsequent degradation reaction. Ubext reaction products were separated by high-performance liquid chromatography (HPLC) and analyzed by mass spectrometry (MS). The molecular mass of each product in the sample was translated to a peptide sequence using the FindPept algorithm (http://web.expasy.org/findpept/) 34 (Supplementary Table 4). The resulting product repertoire implied that the 20S CP trimmed the unstructured stretches at both termini of Ubext, releasing the tightly packed globular ubiquitin domain as a stable product (Fig. 2c). Without 19S RP, even extensions shorter than 25 aa were removed by 20S CP, indicating that they were accessible to the proteolytic β catalytic sites at the center of the 20S catalytic core. Nevertheless, despite the presence of multiple putative cleavage sites inside the Ub sequence as predicted by the PAProC algorithm (http://www.paproc.de/expl1.html) 35 (Supplementary Fig. 3b), only few products generated by ‘nibbling’ at the globular Ub domain were detected following extensive digestion with 20S CP. To summarize, the primary action of 26S proteasomes was deubiquitination of ubiquitinated-Ubext, whereas 20S CP processed loosely folded extensions.
No evidence for direct inhibition of proteasomes by Ubext
Unmodified Ubext species, which are not susceptible to proteasome-dependent degradation, may accumulate and consequently block proteasomes competitively 36. We investigated the interaction between the 26S proteasome complex and Ubext by Far-Western analysis during which purified 26S proteasome complexes were resolved on SDS-PAGE and immobilized on a nitrocellulose membrane for a protein-protein association assay (Fig. 3a). During proteins transfer to a nitrocellulose membrane a portion of immobilized proteins undergoes refolding which facilitates interactions with analyte proteins. Membranes were exposed to either UbG76V25aa, Ub-GFP, or free ubiquitin and washed extensively. By blotting with anti-Ub antibodies, we identified two proteasome subunits as being stably associated with Ubext (Rpn1 and Ubp6; Fig. 3a). This observation was confirmed by using purified recombinant Rpn1 and Ubp6 subunits (Fig. 3a). Both Rpn1 and Ubp6 have been documented to associate with ubiquitin-fusion domains 37,38. Therefore it was not surprising that Ub-GFP was a bit more promiscuous, associating with these two proteasome subunits and several more (Fig. 3a). As expected, neither of the single Ub-domain-containing targets, Ub-GFP and Ubext, stably interacted with the polyUb-receptor, Rpn10 (Fig. 3a). This result was consistent with the ubiquitin-interacting motif (UIM) of Rpn10 being dispensable for turnover of Ubext in vivo (Fig. 1h). Notably, unlike free Ub which was labile, Ub-containing proteins bound proteasome subunits, suggesting the existence of additional structural elements in Ubext that differentiate their fate from free Ub.
Figure 3. In vitro, Ubext molecules associate with proteasomes without inhibiting them.
(a) Identification of proteasome components interacting with Ubext- by Far-Western technique. Purified 26S proteasome and selected recombinant proteasome subunits were resolved by SDS-PAGE and immobilized on a nitrocellulose membrane. Relative positioning of each potential ligand is shown by Ponceau staining (left). Due to the epitope tag, a slight retardation in migration of recombinant proteins (6xHisRpn1, 6xHisRpn10, and 6xHisUbp6) relative to endogenous proteasome subunits is observed. Identical membranes were incubated with purified recombinant 6xHisUbG76V25aa, 6xHisUb-GFP or 6xHisUb as noted and washed. Residual analyte associated to immobilized proteins that succeeded to refold during immobilization on the membrane was detected by anti-Ub staining. Asterisk (*) points to possible association of Rpn13 with the Ub-GFP fusion. (b) Ub-independent proteolysis of β-casein by either 20S or 26S purified proteasomes was followed in the absence (top) or presence of 5-fold molar excess of 6xHisUBB+1 to substrate (bottom). (c) Processing of 6xHisUb-GFP by purified 26S proteasomes was followed in the absence (top left) or presence of Ubext (6xHisUBB+1, 6xHisUbG76V25aa or Ub-6xHisUbG76V25aa). (b, c) The residual amounts of unprocessed substrates at each time point are indicated above the gel images as [uM].
Despite our initial hypothesis, Ubext variants did not competitively inhibit the hydrolytic activities of 26S proteasomes. Presence of excess UBB+1 had no effect on Ub-independent proteolysis of β-casein by either purified 26S or 20S proteasomes (Fig. 3b). In the reciprocal direction, casein did not interfere with processing of Ubext by 20S as it was the preferred substrate (~30 min. half-life of casein vs. ~120 min for Ubext; Fig 2b, 3b). Proteasome-dependent processing of the model substrate, Ub-GFP, was also unaffected by excess of UBB+1 or of UbG76V25aa (Fig. 3c). Even in the presence of Ub-Ubext (demonstrated above to be a direct substrate for proteasomes; Fig. 2a), deubiquitination of Ub-GFP was observed (Fig. 3c) suggesting that the two substrates did not compete with each other effectively.
Ubext display ability to interfere with certain DUBs
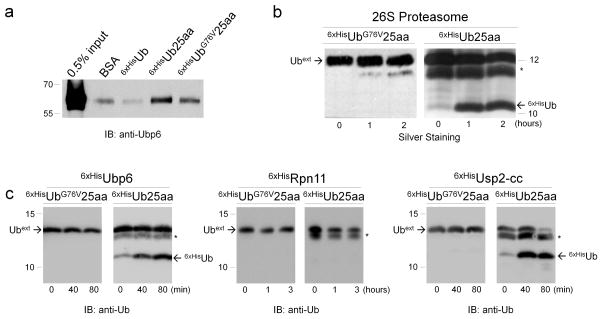
Although they did not perturb the function of 26S proteasomes, Ubext species of different lengths, did interact with a proteasome subunit Ubp6 (Fig. 3a, 4a, Supplementary Fig. 4a, b). Ubext differed from free Ub (which did not bind Ubp6 effectively; Fig. 3a, 4a, Supplementary Fig. 4b), by a C-terminal extension and a substitution of glycine with valine at position 76. These changes should make Ubext uncleavable by most DUBs 5. Reversing position 76 to glycine (Ub25aa) resulted in even tighter binding to Ubp6 in a pull-down experiment (Fig. 4a, Supplementary Fig. 4b), and conversion of Ubext into a substrate of the 26S proteasome (Ub25aa was processed by 26S proteasomes to yield free and stable Ub; Fig. 4b). The ability of the 26S proteasome to deubiquitinate Ub25aa is likely attributed to the proteasome-associated DUB, Ubp6, which was also capable as a free enzyme to deubiquitinate Ub25aa (Fig. 4c). Ubp6 also deubiquitinated di-Ub (Supplementary Fig. 4c) in a similar manner to that by intact proteasome (Fig 3). In contrast, another proteasome DUB that belongs to the MPN+ metalloprotease family, Rpn11, was inactive on these substrates (Fig. 4c), whereas a cytosolic DUB (the catalytic core of Usp2; Usp2-cc), also acted on Ub25aa (Fig. 4c), indicating that Ubext variants could be general substrates for cysteine-based DUBs, depending on the identity of amino acid at position 76. Enzymatic activity of isolated Rpn11 independent of the proteasome complex was confirmed (Supplementary Fig. 5a).
Figure 4. Ubext molecules are targets of DUBs.
(a) Pull-down of recombinant proteasome-associated DUB, Ubp6, with Ubext columns. Specified proteins were covalently immobilized on an activated CH-sepharose column, incubated with purified recombinant 6xHisUbp6, and residual eluted analyte was visualized with anti-Ubp6 antibodies. (b) Identity of residue 76 of Ubext changes the outcome of the reaction with 26S proteasome. 6xHisUbG76V25aa or 6xHisUb25aa were incubated with 26S proteasomes (same as in Fig. 2b), and reaction mixtures were evaluated. An asterisk (*) marks an E. coli protein that persistently co-purified with 6xHisUb25aa. (c) Susceptibility of Ubext to isolated DUBs. Unlike extended WT Ub UbG76V25aa was not susceptible to deubiquitinating activity of purified recombinant 6xHisUbp6, 6xHisRpn11 and 6xHisUsp2-cc.
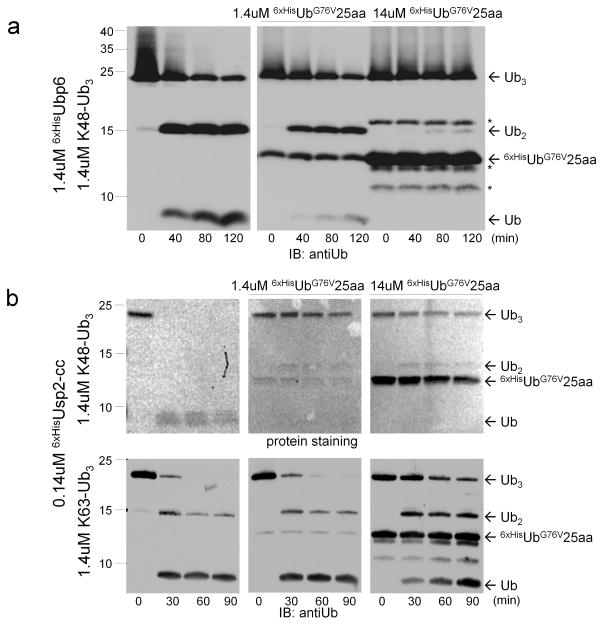
By binding DUBs, uncleavable Ubext could function as competitive DUB inhibitors. To test this hypothesis, we studied the effect of UbG76V25aa on deubiquitination of Lys48-linked polyUb chains by Ubp6. Ubp6 efficiently disassembled Lys48-linked polyUb chains representing the typical targeting signal for most proteasome substrates (Fig. 5a). This DUB activity was completely abolished by UbG76V25aa (Fig. 5a). The inhibitory effect of Ubext was not limited to proteasome-associated DUBs from yeast; Usp2, a fast-acting cytosolic DUB from mammals was also inhibited by Ubext (Fig. 5b). Both Lys48- and Lys63-linked activities of Usp2 were effectively inhibited by Ubext (Fig. 5b). This result provides a possible mechanistic link between expression of extended Ub mutants and the accumulation of polyUb-conjugates with diverse linkage types in cells (Fig. 1). At the same time, conclusions should be taken on a case-by-case basis as UbG76V25aa did not interfere with OTUB1 or Rpn11 (Supplementary Fig. 5).
Figure 5. Extended Ub mutants are protein-based DUB inhibitors.
(a) Deubiquitination of Lys48- or Lys63-linked Ub3 chains by 6xHisUbp6 (a) or 6xHisUsp2-cc (b) was carried out in the absence (left) or presence of 1× (middle) or 10× (right) of 6xHisUbG76V25aa. Disassembly of chains into monoUb was monitored over 2 hrs, as indicated. An asterisk (*) marks an E. coli protein that persistently co-purified with 6xHisUbG76V25aa.
DISCUSSION
The current work characterizes the molecular basis for perturbation of the UPS by UBB+1, an abnormal ubiquitin protein prevalent in neurodegenerative tissues 15,21 as well as in a few non-neuronal pathologies 19,20. Naturally, all Ub is synthesized as fusions to either additional copies of ubiquitin or to ribosomal subunits; yet, these fusion proteins do not inhibit cellular protein turnover. As we demonstrated herein, UBB+1 is characterized by a single site mutation at residue 76 of Ub that generates a fusion protein that is not susceptible to DUB action. As a result Ubext cannot be activated by ubiquitination enzymes but can be ubiquitinated at different positions. Due to its C-terminal extension, Ubext mimics Ub-conjugates sufficiently to bind some DUBs tighter than free Ub. Binding leads to processing of the Ub-conjugate if the amino acid at position 76 is the naturally occurring glycine and not valine. Thus, in contrast to other Ub-conjugates, uncleavable Ubext species occupy the active site of DUBs causing an inhibitory effect which can end up altering the general cellular ubiquitin landscape. Inefficient UPS may impact the capability of cells to dilute or process aggregates. This may be one reason why UBB+1 is a pathology commonly associated neurodegeneration. Even though we found that Ubext species inhibit certain DUBs directly, we observed no substantial changes in the proteolytic activity of proteasome complexes in the presence of a molar excess of UBB+1 or any other stable Ubext variants, suggesting that Ubext molecules do not efficiently compete with other substrates for direct access to the proteolytic chamber of the proteasome. In fact, Ubext variants were actually substrates of proteasomes, particularly of 20S core particles. This leaves open the possibility for indirect inhibition of the proteasome as an explanation of interference with intracellular proteolysis measured in correlation with accumulating Ubext 17,36, apparently regardless of extension length (Fig. 1).
Consistent with other findings 27,39, in vitro analysis of Ubext degradation products by the proteasome demonstrated that the proteasome requires a minimal length of unstructured stretches or loosely folded terminal extensions to allow the entry and passage of a compact domain through the 19S complex. In 26S proteasomes, polyUb chains anchor the substrate to dedicated receptors, orienting the substrate such that ATPases can start unfolding and threading the substrate into the 20S proteolytic chamber (Fig. 6a, b). We observed that a requirement for a minimal extension length was less stringent for the unregulated 20S CP, which lacks the proteasomal Ub receptors, and as such is likely to engage substrates based on unstructured regions or hydrophobic protrusions. The absence of a 19S regulatory subcomplex reduces the distance between Ubext and the proteolytic β subunits in the 20S core. Without coordination from substrate-anchoring subunits in regulatory complexes hydrolytic processing by the 20S proteasome has undefined directionality. This property likely enables 20S CP to trim Ubext species from both N- and C-termini, astonishingly, allowing even shorter extensions to convert proteins into substrates for proteolysis by free 20S CP. However, without help from 19S RP ATPases, tightly folded globular ubiquitin molecule cannot pass through the narrow ‘gate’ formed by the α-subunits and enter the proteolytic core (Fig. 6c, d), leading to our observation that the majority of the Ub domain escaped degradation by the 20S proteasome after the extensions had been cleaved from Ubext. Given that 26S holoenzymes appear to be the primary proteasome specie in living cells 40,41, unmodified Ubext is expected to be reasonably stable, yet upon ubiquitination some may be turned over and removed by the 26S proteasomes. Unregulated 20S CP is expected to have only a minor contribution to treatment of short extensions (as shown in Fig. 2), yet may be useful in removing damaged or misfolded proteins following mild denaturing conditions such as heat shock or oxidative stress. Therefore, a shift in proteasome population favoring 20S CP is a logical prediction to occur post-stress.
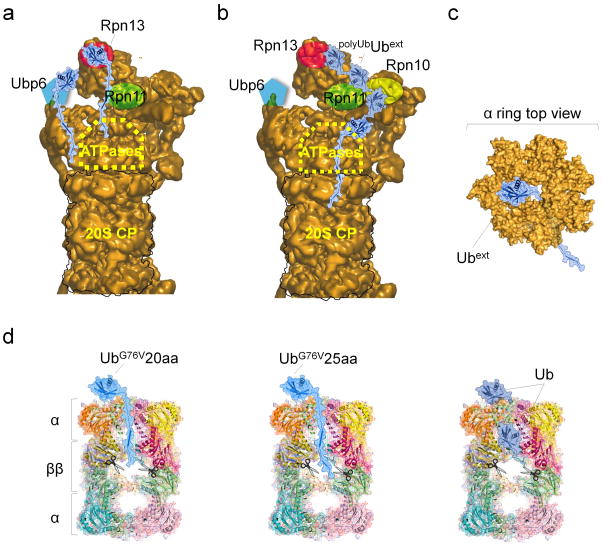
Figure 6. A model of Ubext-proteasome engagement: implications for efficient proteolysis by proteasomes.
Modeling putative engagement sites of Ubext summarizes potential elements (in both proteasomes and substrates) required for efficient proteasome proteolysis. Models of extended Ub molecules were created by modifying coordinates of WT Ub (PDB-1UBQ). These species were manually docked onto potential binding sites on 26S based on an available 3D model (EMD-2165) (a, b). Putative locations of Rpn13 (published as a monoUb-receptor) or of Ubp6 (experimentally determined in Fig. 3a, 5a as a Ubext receptor) distance these substrates from the catalytic protease active sites (a). After Ubext additional ubiquitination, cooperation with a polyUb-binding subunit, Rpn10, may bring the unstructured tail closer (b). Threading the unstructured extension through an open channel centered in 20S α-ring (PDB-1VSY), suggests that processing of Ubext by the protease catalytic sites (scissors) may be initiated by 20S without complete unfolding of the globular Ub domain (c, d).
We identified Ubp6 as the primary receptor for Ubext proteins at the proteasome, and demonstrated that this subunit could even bind ubiquitin-fusion proteins independently. None of the other tested Ub-receptors (Rpn10UIM, Rpn13, Rad23, Dsk2) were essential for recognition or for anchoring Ubext to proteasomes. Occupation of the Ubp6 active site has wider ramifications as previous studies demonstrated that this association initiates a series of downstream events such as ATPase activity, unfolding of the substrate, and polypeptide hydrolysis of the conjugated substrate 42. To be efficiently degraded by 26S proteasomes, ubiquitin should be (poly)ubiquitinated and should also have a loosely folded stretch, as in Ubext (Fig. 6b). However, the current study demonstrated that interaction of short Ub-modifications (e.g., Ub-UbG76V25aa) with proteasomes resulted in their rapid disassembly rather than degradation of the conjugate. This difference may stem from the ability of longer polyUb chains to bind multiple receptors (Fig. 6b) leading to ample time for the ATPases in the Base subcomplex of the 19S RP to latch onto a loosely folded region of the conjugate substrate, unravel the polypeptide, and transfer it to the 20S core for proteolysis. Inhibiting deubiquitination activity of proteasome associated DUBS, particularly Ubp6/USP14, is of intensive therapeutic efforts in combating cancer and other diseases 43–45. In contrast to highly specific USP14 inhibitors, some small molecules are promiscuous enough to target a subset of proteasome-associated DUBs, UCHL5/UCH37 and Ubp6/USP14, yet exhibit only minimal effect on others 43. The cellular response to “inhibitors of 19S regulatory particle deubiquitinating activity” was accumulation of polyubiquitin conjugates and interference with UPS-dependent protein turnover in a manner that was distinct from direct inhibition of 20S CP proteasome proteolytic sites 44. Such an outcome is intriguingly reminiscent of Ubext in the current study, thereby adding Ubext to the arsenal of experimental tools employed in research of UPS in etiology of human disease.
Resistance of 26S holoenzymes to the inhibitory influence of Ubext variants suggests that DUB(s) other than Ubp6 in the proteasome holoenzyme are unaffected by this class of inhibitors. This resistant DUB is most likely Rpn11, which requires for its enzymatic activity simultaneous recognition of multiple Ub moieties in a single chain 46,47. The potency of Rpn11 as a proteasome-associated DUB for polyUb disassembly may explain why proteasome mechanisms appeared largely unaffected by Ubext species. The general inhibitory effect of Ubext proteins on UPS probably lies with cysteine-based DUBs of the USP/Ubp family. Independent of the extension length, Ubext variants led to pervasive accumulation of heterogeneously-linked polyUb conjugates suggesting an interference of Ubext with ubiquitin signaling beyond proteasome-dependent turnover. Interference of Ubext with deubiquitination affects multiple Ub-dependent pathways, possibly clarifying the underlying link between UBB+1 and protein aggregation that contributes to neurodegenerative diseases 48,49. An inducible protein-based broad-specificity DUB inhibitor could be a powerful tool to perturb the ubiquitin landscape for studying the role of DUBs in cellular maintenance and response to changing conditions.
ONLINE METHODS
Antibodies
Anti-Ub (Dako), anti-Myc and anti-GFP (Santa Cruz Biotechnology), anti-Ubp6 (a generous gift from Dan Finley).
E. coli Plasmids and Yeast Strains
Detailed in the Supplementary Results (Supplementary Tables 5 and 6).
Construction of plasmids
For expression in yeast, mycUBB+1, mycUbG76V20aa and mycUbG76V20aa were amplified by PCR from plasmids provided by N. Dantuma and were cloned using Sac1 and Kpn1 into GAL4 or ADH promoter-carrying vectors, pYES2 (Invitrogen) and YEplac181 respectively. mycUb20, mycUb25, mycUbV70D,G76V25aa constructs were created by side-directed mutagenesis of plasmids described in preceding sentence.
For E. coli expression plasmids, open reading frames (ORFs) of UBB+1, UbG76V20aa, UbG76V20aa, Ub20, Ub25 were amplified by PCR from the corresponding yeast vectors described above, and ORFs of Rpn10, Rpn11, and Ubp6 were amplified from S. cerevisiae genomic DNA. All PCR products were cloned into pQE30 (QIAGEN) using Sac1 and Pst1.
Protein overexpression in yeast
Plasmids for expression of proteins in yeast were transformed according to the lithium acetate/polyethylene glycol protocol. For the steady-state levels of protein analysis, yeast strains overexpressing different Ub species under control of the constitutive ADH promoter were grown on selective dextrose-containing SD-LEU medium for 6 hours (ODA600=1). Strains overexpressing proteins under control of GAL4 promoter were grown on selective dextrose-containing SD-URA medium to ODA600=1, after which the medium was changed to galactose-containing SGal-URA for 20 hrs. Cells were harvested at indicated times and lysed by vigorous vortexing with glass beads. Extracted proteins were TCA precipitated, dissolved in 2× Laemmli loading buffer and boiled. Whole cell extract was resolved through 8–18% Tris-Glycine SDS-PAGE, and proteins transferred to a nitrocellulose membrane for immunoblotting.
Expression and purification of recombinant proteins
Vectors carrying the gene for 6xHis-tagged proteins were electro-transformed into bacteria strain M15 or Bl21(DE3) for recombinant protein expression. The cells were grown in LB medium supplied with 75 μg/mL ampicillin (and 25 μg/mL kanamycin for M15 strain) at 37°C to ODA600=0.6–0.8, then IPTG was added to a final concentration of 1mM to induce protein overexpression. The cells were then grown over night at 16°C and harvested using TBS buffer (50 mM Tris pH 7.4, 150 mM NaCl). The pellet was resuspended in a 7-fold volume of lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM Imidazole) supplemented with protease inhibitors. The cells were squeezed twice through a French Press (Thermo Scientific) and the obtained lysate was clarified by centrifugation at 30,590×g for 20 min at 4°C. The supernatant was loaded on 5 mL HisTrap FF column (GE Healthcare) which was previously equilibrated with the lysis buffer. Weakly-bound non-specific proteins were washed out by 10% elute buffer (50 mM Tris pH 8.0, 300 mM NaCl, 400 mM imidazole) and then 6xHis-tagged proteins were eluted by 70% elute buffer using ÄKTA FPLC. The purity of fractions was analyzed by SDS-PAGE, selected fractions were combined and dialyzed against PBS buffer (10 mM Phosphate buffer, 2.7 mM KCl, 140 mM NaCl), and proteins were stored at −80°C.
Purification of recombinant proteins on gel filtration column
Selected fractions after HisTrap FF affinity purification were subsequently resolved on a gel filtration column to obtain a higher level of purity. Superdex S75 column (GE Healthcare) was equilibrated with PBS buffer and 1-mL sample was loaded on the column. Fractions were collected at 0.4 mL/min flow rate. Presence of protein and its purity was verified by resolving fractions with SDS-PAGE followed by Coomassie staining.
Purification of the 26S and 20S proteasomes
Proteasomes were purified from wild type yeast (MY58; Supplementary Table 5) lysates by anion-exchange and gel filtration chromatography as described previously 50. Briefly, yeast cells were grown for three days in YPD medium. After harvesting, the cell pellet was resuspended in a 2-fold volume of buffer A (25 mM Tris pH 7.4, 10 mM MgCl2, 10% glycerol, 4 mM ATP, 1 mM DTT) and lysed in a French press. Extract was clarified by centrifugation at 30,590×g for 20 min at 4°C and filtered through cheesecloth. The supernatant was fractionated on a 100 mL DEAE-Affigel Blue column (Bio-Rad), followed by anion exchange chromatography and gel filtration chromatography. Anion exchange was performed by an XK26 column packed with 50 mL of Resource Q resin (GE Healthcare). Proteins were resolved on a 500 mL gradient of 100 to 500 mM NaCl at 4 mL/min. 20 mL fractions were collected and screened for the ability to hydrolyze Suc-LLVY-AMC (Millipore).
Fractions containing the peak of activity, eluting between 270 to 330 mM NaCl, were pooled, desalted, concentrated to 0.5 mL by use of Vivaspin concentrators with a molecular weight cut-off (MWCO) of 30 kDa, and further resolved by gel filtration using Sephacryl S-300 column (GE Healthcare). Samples were run isocratically in buffer A at a flow rate of 1 mL/min and 2 mL fractions were collected and screened for peptidase activity. Fractions from the broad peak of peptidase activity were frozen at −80°C until needed.
The 20S proteasome was purified by depleting proteasome holoenzyme (26S) samples of ATP and separating the core particle from the regulatory particle on an 8 mL ceramic hydroxyapetite column (GE Healthcare), using a 10–400 mM potassium phosphate gradient at pH 7.4. 8 mL fractions were collected and screened for the ability to hydrolyze Suc-LLVY-AMC after activation by 0.01% SDS. Fractions containing the peak of activity were pooled, concentrated to 1 mL by use a Vivaspin concentrator with a MWCO of 30 kDa, dialyzed into buffer A and stored at −80°C.
Peptidase assay for proteasome activity
Proteasome samples (10 μl) were added to 40 μl of 0.1 mM Suc-LLVY-AMC in buffer A. After 15 min incubation at 30°C, the reaction was stopped by adding 1 mL of 1% SDS. The fluorescence of released AMC was measured using a Synergy 2 fluorometer (BioTek) (Ex. 380 nm; Em. 440 nm).
PolyUb chain synthesis
The components (enzymes and substrates; Supplementary Table 6) for synthesis of K48- and K63-linked tri-Ub and of the Ub-UbG76V25aa K48-linked dimer were expressed in BL21 Rosetta E. Coli. The ubiquitin monomers (UbK48R/K63R and UbD77) for chain synthesis were expressed without an affinity tag and purified with perchloric acid precipitation followed by cation exchange on a 5 mL HiTrap SP column (GE Healthcare) as published 51. E2 conjugating enzymes, E2-25K and Ubc13 were expressed as GST fusions and purified with a 10 mL GST-Trap column (GE Healthcare). MMS26xHis a component necessary for Ubc13 in K63 chain synthesis was purified according to published protocol 52 and 6xHisE1 was purified as described 53. Purification of the yeast Ub C-terminal hydrolase (YUH1) was based on published protocol 54. YUH1 was expressed without an affinity tag, and after lysis and centrifugation the supernatant was loaded onto a 10-mL anion column (HiTrap QF) in 50 mM Tris, pH 7.6. Then, YUH1 was eluted with 5 CV steps of 15%, 30%, 50%, and 100% anion elution buffer (50 mM Tris, 1 M NaCl, pH 7.6). The 30% anion elution fractions containing the 25 kDa YUH1 were pooled, exchanged into PBS buffer pH 7.4 and further purified with a Superdex 75 60/120 column (GE Healthcare) with a flow rate of 0.4 mL/min.
K48- and K63-linked tri-Ub were created sequentially from two separate ubiquitin chain reactions. Each ubiquitin chain reaction was performed in a 2-mL volume containing, 20 mg of each ubiquitin monomer, 5 mM TCEP, 15 mM ATP, 15 mM MgCl2, 10 μM (GSTE2-25K or GSTUbc13/MMS26xHis), 500 nM E1, ATP regenerating system (20 mM creatine phosphate, 1.2 U/mL inorganic yeast pyrophosphate, and 1.2 U/mL creatine phosphokinase), and 50 mM Tris pH 8.0. Reactions were incubated in a 30°C water bath for 20 hrs. First, UbK48R/K63R and UbD77 were reacted with either GSTE2-25K or GSTUbc13/MMS26xHis to form K48-Ub2 and K63-Ub2, respectively. Di-Ub were purified using cation and size exclusion steps and the proximal D77 block was removed using YUH1 according to published protocol 12. Then, the di-Ub with a free G76 was allowed to react with UbD77 under the same conditions, producing tri-Ub. tri-Ub was purified from resulting product mix by size exclusion chromatography. The K48-Ub3 and K63-Ub3 were dialyzed agaisnt the desired buffer and stored at −20°C until needed.
The Ub-UbG76V25aa K48-linked dimer was created in a single ubiquitin chain reaction with UbK48R/K63R and 6xHisUbG76V25aa monomers in the presence of GSTE2-25K under the same conditions described above for diUb. Following completion, the reaction was diluted into 10 mL of His-buffer A (20 mM sodium phosphate, 500 mM NaCl, pH 7,4) and slowly hand injected on to a 1 mL HisTrap column (GE healthcare). An extra 10 mL of His-buffer A was injected to washout any remaining proteins and the column was eluted with 10mL of His-buffer B (20 mM sodium phosphate, 500 mM NaCl, 500 mM Imidazole, pH 7.4). Eluted proteins were concentrated and dialyzed against PBS buffer pH 7.4 using a centrifugal unit. Finally, the concentrated elution was loaded onto a Superdex 75 60/120 column (GE Healthcare) in PBS buffer pH 7.4 and separated with a flow rate of 0.4 mL/min. Pure Ub-UbG76V25aa appeared as a separate peak and was confirmed using 15% SDS-PAGE.
CHX-chase assay
The strains bearing plasmids for Ubext expression under ADHp control were grown on SD-LEU selective media overnight at 30°C, except of rpn11-1, which was grown at 25°C. Cells were diluted 100-fold into fresh media and allowed to grow until ODA600=1. Different cultures were normalized by OD and supplemented with 250 μg/ml CHX (Sigma-Aldrich). Equal cell numbers were taken at the indicated time points after CHX treatment. Cells were pelleted and proteins were extracted by vigorous shaking with glass beads. TCA-precipitated proteins were dissolved in 1× Laemmli loading buffer, boiled for 5 min and loaded on 16.5% Tris-Tricine SDS-PAGE. After the proteins were resolved, they were transferred to a nitrocellulose membrane and immunoblotted.
Proteasome inhibition by MG132
To estimate degradation rates of proteasome substrates, wild type strain expressing mycUbG76V25aa under ADH promoter was grown to ODA600=1 as mentioned above. MG132 (EMD Millipore) was added to a final concentration of 100 μM. Equal amounts of cells equilibrated by OD measurements were taken at the indicated time points and pelleted. TCA-precipitated proteins were dissolved in 1× Laemmli loading buffer, boiled for 5 min and resolved by 16.5% Tris-Tricine SDS-PAGE followed by immunoblotting.
Far-Western analysis
For far-Western procedure, recombinant proteins (0.8 μg) or purified whole proteasome 26S complex (5 μg) were resolved on 12% Tris-Glycine SDS PAGE and electro-transferred to a nitrocellulose membrane under wet conditions (1X Tris-Glycine buffer, 20% methanol) 55. After blocking with low-fat 5% milk solution in TBST buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.05% Tween-20), the membranes were incubated with 20 μg of analyte recombinant proteins for 2 hrs in TBST buffer. Afterwards, the analyte proteins were washed and immunoblotted for analyte proteins.
In-vitro processing assay
All proteasome reactions (degradation as well as deubiquitination) were performed in buffer A (25 mM Tris pH 7.4, 10 mM Mg2Cl, 10% glycerol, 1 mM ATP and 1 mM DTT. The reaction mixes were kept at 30°C and at the indicated time points, the reactions were stopped by taking aliquots and boiling with 5× Laemmli loading buffer. Reaction products were separated on SDS-PAGE and either stained with non-specific protein dyes (Coomassie or Silver staining) or immunoblotted.
For analyzing 6xHisUBB+1 inhibition ability on β-casein degradation, 1.8 μM β-casein (Sigma-Aldrich) was incubated with 0.018 μM 26S/20S proteasome and 9.2 μM 6xHisUBB+1. As a control, the same reaction without 6xHisUBB+1 was tested. The deubiquitination of 6xHisUb-GFP was performed by mixing 0.25 μM 6xHisUb-GFP with 10-fold less 26S proteasome complex. To inhibit the reaction, 2.5 μM of potential inhibitor was added (6xHisUBB+1, 6xHisUbG76V25aa or Ub-6xHisUbG76V25aa), where indicated. Processing of elongated ubiquitin species was examined when they were incubated with 26S or 20S proteasome complexes at 50 fold molar excess of the substrates. Ability of recombinant DUB proteins to remove the elongation from Ubext was tested by mixing the extended substrate with DUB enzymes at 1:1 molar ratio. The reactions to analyze DUB inhibition contained 1.4 μM triUb-linkages (Lys48 or Lys63), 1.4 μM 6xHisUbp6 (or 0.14 μM Usp2-cc), and 1.4 μM or 14 μM 6xHisUbG76V25aa. The reaction without 6xHisUbG76V25aa served as control to demonstrate the native DUB activity of the enzymes.
HPLC and MS monitoring of Ubext degradation process
The degradation reaction of recombinant elongated ubiquitin species by 20S proteasome was performed as described before. At noted time points, the reaction products were separated on a Thermo instrument (Spectra System p4000) using an analytical column (Jupiter 5 micron, C18/C4, 300Å, 150 × 4.6 mm) and a flow rate of 1.2 mL/min. The linear gradient used to elute the bound peptides was integrated from load buffer (water with 0.1% (v/v) trifluoroacetic acid (TFA)) and elute buffer (acetonitrile with 0.1% TFA). Manually-collected peptides corresponding to HPLC-detected peaks were analyzed by MS carried out using LCQ Fleet Ion Trap (Thermo Scientific). Masses identified in MS were converted to peptide sequences using the FindPept algorithm (http://web.expasy.org/findpept/) (Supplementary Table 4).
Pull downs
Fifty μg of each of the indicated recombinant Ubext species were immobilized on activated CH-sepharose beads (Sigma-Aldrich). Non-occupied active groups were blocked with 0.1 M Tris base buffer and bound proteins were exposed to interaction with 60 μg analyte protein (6xHisUbp6) for 2 hrs. After numerous wash procedures, 6xHisUbp6 was eluted by buffer containing 8 M Urea. Eluted fractions were concentrated by TCA protein precipitation and separated by 12% SDS-PAGE followed by immunobloting with anti-Ubp6 antibody.
Ub-AQUA analysis
Yeast cell extract preparation for AQUA analysis
Yeast strain carrying Ubext expression plasmids with GAL4 promoter elements were grown for overnight at SD-URA medium. The cells were diluted into fresh medium and grown additional 4 hours to ODA600=1, then density normalized cells were pelleted and resuspended with SGAL-URA. The cells were harvested after 20 hrs. Samples were prepared by TCA lysis and precipitation and separated on 4–12% SDS PAGE.
Sample preparation for AQUA analysis
Gel regions above 130 kDa were excised, and prepared for Ub-AQUA by liquid-chromatography selected reaction monitoring (LC-SRM) in a TSQuantum Ultra (ThermoElectron) as published 56,57.
LC-SRM analysis of Ub-AQUA peptides
The yeast versions of Ub-AQUA peptides were monitored by LC-SRM in a TSQuantum Ultra (ThermoElectron) according to the transitions presented in Supplementary Table 2. For each SRM, monoisotopic m/z values for parent and fragment ions were derived from GPMAW (Lighthouse Data). Parent ion charge state, optimal fragment ion, collision energy and instrument resolution were all empirically determined. Digested samples containing peptide standards were injected using a Famos autosampler (LCPackings) and loaded onto a reversed phase column (Denali C18, 1mm × 15 mm, Vydac) at a flow rate of 60μl per minute buffer A (5% (acetonitrile) ACN, 0.4% acetic acid, 0.005% (heptafluorobutyric acid) HFBA). Peptides were eluted using an Agilent 1100 HPLC across a 19 min linear gradient from 15% to 25 % buffer B (95% ACN, 0.4% acetic acid, 0.005% HFBA). Individual SRM transitions were monitored across a 1.2 m/z window for 0.1 sec. Analytically, the run was divided into segments such that 8 SRM transitions (4 peptide pairs) were monitored during a given duty cycle.
Data analysis
For each peptide, the areas under the curve were determined for the native (trypsin digested) and synthetic (isotope labeled) SRM transitions. The product of this ratio with the known abundance of each synthetic peptide was calculated to determine the abundance of each peptide standard. For the Lys63 polyUb peptide, two distinct SRM transitions were monitored, with the reported values reflecting the average of these two measurements. The total amount of ubiquitin in a sample was averaged from independent measurements using peptides surrounding the Lys63 and Lys48 loci and the N-terminus.
Supplementary Material
Acknowledgments
This research was funded by NIH GM095755 to DF and MHG and a USA-Israel Binational Science Foundation Grant to DF and MHG 2009487.
Footnotes
AUTHOR CONTRIBUTIONS
DK, MHG, NR, MAN, and DF designed experiments. DK carried out in vivo and in vitro proteasomal experiments. SPG and DSK aided in aqua analysis. MAN and DZ synthesized all Ub conjugates. AB and PS collected and analyzed processed Ub fragments. DK, MAN, and MGH prepared the manuscript.
Financial interests
The authors declare no competing financial interests
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–60. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 4.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of Ubiquitin Chains by Proteasome-associated Deubiquitinating Enzymes. Molecular & Cellular Proteomics. 2011;10:5. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–38. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 6.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–82. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziv I, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics. 2011;10:M111 009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dammer EB, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–65. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun LJ, Chen ZJ. The novel functions of ubiquitination in signaling. Current Opinion in Cell Biology. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. Embo J. 2009;28:359–71. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? Embo Journal. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21:727–40. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newmann AJ. Pre-mRNA splicing. Current Opinion in Genetics & Development. 1994;4:298–304. doi: 10.1016/s0959-437x(05)80057-7. [DOI] [PubMed] [Google Scholar]
- 14.Maquat LE. Nonsense-mediated mRNA decay. Curr Biol. 2002;12:R196–7. doi: 10.1016/s0960-9822(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 15.van Leeuwen FW, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279 doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 16.Ko S, et al. Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity. J Biol Chem. 2010;285:36070–80. doi: 10.1074/jbc.M110.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsten K, et al. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J Cell Biol. 2002;157:417–27. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Tijn P, et al. Mutant ubiquitin decreases amyloid beta plaque formation in a transgenic mouse model of Alzheimer’s disease. Neurochem Int. 2012;61:739–48. doi: 10.1016/j.neuint.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Wu SS, et al. Coexpression and accumulation of ubiquitin +1 and ZZ proteins in livers of children with alpha(1)-antitrypsin deficiency. Pediatric and Developmental Pathology. 2002;5:293–298. doi: 10.1007/s10024-001-0202-3. [DOI] [PubMed] [Google Scholar]
- 20.Fratta P, et al. Mutant ubiquitin UBB+1 is accumulated in sporadic inclusion-body myositis muscle fibers. Neurology. 2004;63:1114–1117. doi: 10.1212/01.wnl.0000138574.56908.5d. [DOI] [PubMed] [Google Scholar]
- 21.de Pril R, et al. Accumulation of aberrant ubiquitin induces aggregate formation and cell death in polyglutamine diseases. Hum Mol Genet. 2004;13:1803–13. doi: 10.1093/hmg/ddh188. [DOI] [PubMed] [Google Scholar]
- 22.Lam YA, et al. Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhoef LG, et al. Minimal length requirement for proteasomal degradation of ubiquitin-dependent substrates. FASEB J. 2009;23:123–33. doi: 10.1096/fj.08-115055. [DOI] [PubMed] [Google Scholar]
- 24.Dennissen FJA, et al. Mutant ubiquitin (UBB+1) associated with neurodegenerative disorders is hydrolyzed by ubiquitin C-terminal hydrolase L3 (UCH-L3) Febs Letters. 2011;585:2568–2574. doi: 10.1016/j.febslet.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Matiuhin Y, et al. Extraproteasomal Rpn10 Restricts Access of the Polyubiquitin-Binding Protein Dsk2 to Proteasome. Molecular Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Shabek N, Iwai K, Ciechanover A. Ubiquitin is degraded by the ubiquitin system as a monomer and as part of its conjugated target. Biochem Biophys Res Commun. 2007;363:425–31. doi: 10.1016/j.bbrc.2007.08.185. [DOI] [PubMed] [Google Scholar]
- 28.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–43. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 29.Shabek N, Ciechanover A. Degradation of ubiquitin The fate of the cellular reaper. Cell Cycle. 2010;9:523–30. doi: 10.4161/cc.9.3.11152. [DOI] [PubMed] [Google Scholar]
- 30.Shabek N, Herman-Bachinsky Y, Ciechanover A. Ubiquitin degradation with its substrate, or as a monomer in a ubiquitination-independent mode, provides clues to proteasome regulation. Proc Natl Acad Sci U S A. 2009;106:11907–12. doi: 10.1073/pnas.0905746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu HY, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo Journal. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 33.Rinaldi T, et al. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gattiker A, Bienvenut WV, Bairoch A, Gasteiger E. FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics. 2002;2:1435–44. doi: 10.1002/1615-9861(200210)2:10<1435::AID-PROT1435>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Nussbaum AK, Kuttler C, Hadeler KP, Rammensee HG, Schild H. PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics. 2001;53:87–94. doi: 10.1007/s002510100300. [DOI] [PubMed] [Google Scholar]
- 36.van Tijn P, et al. Dose-dependent inhibition of proteasome activity by a mutant ubiquitin associated with neurodegenerative disease. J Cell Sci. 2007;120:1615–23. doi: 10.1242/jcs.03438. [DOI] [PubMed] [Google Scholar]
- 37.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 38.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–71. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–31. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–4. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 41.Metcalfe MJ, Huang Q, Figueiredo-Pereira ME. Coordination between proteasome impairment and caspase activation leading to TAU pathology: neuroprotection by cAMP. Cell Death Dis. 2012;3:e326. doi: 10.1038/cddis.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288:7781–90. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Arcy P, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–40. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 45.Nag DK, Finley D. A small-molecule inhibitor of deubiquitinating enzyme USP14 inhibits Dengue virus replication. Virus Res. 2012;165:103–6. doi: 10.1016/j.virusres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 47.Sato Y, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–U19. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 48.Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2009;32:150–9. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Wooten MW, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–9. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 50.Glickman M, Coux O. Purification and characterization of proteasomes from Saccharomyces cerevisiae. Curr Protoc Protein Sci. 2001;Chapter 21(Unit 21):5. doi: 10.1002/0471140864.ps2105s24. [DOI] [PubMed] [Google Scholar]
- 51.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–14. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–43. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 53.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–24. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–87. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. Journal of Biological Chemistry. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 56.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–73. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 57.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.