Abstract
Channid fishes, commonly referred to as “snakeheads”, are currently very important in Asian fishery and aquaculture due to the substantial decline in natural populations because of overexploitation. A large degree of chromosomal variation has been found in this family, mainly through the use of conventional cytogenetic investigations. In this study, we analyzed the karyotype structure and the distribution of 7 repetitive DNA sequences in several Channa species from different Thailand river basins. The aim of this study was to investigate the chromosomal differentiation among species and populations to improve upon the knowledge of its biodiversity and evolutionary history. Rearrangements, such as pericentric inversions, fusions and polyploidization, appear to be important events during the karyotypic evolution of this genus, resulting in the chromosomal diversity observed among the distinct species and even among populations of the same species. In addition, such variability is also increased by the genomic dynamism of repetitive elements, particularly by the differential distribution and accumulation of rDNA sequences on chromosomes. This marked diversity is likely linked to the lifestyle of the snakehead fishes and their population fragmentation, as already identified for other fish species. The karyotypic features highlight the biodiversity of the channid fishes and justify a taxonomic revision of the genus Channa, as well as of the Channidae family as a whole, as some nominal species may actually constitute species complexes.
Introduction
The Channidae family (Actinopterygii, Perciformes) comprises 2 genera (Channa and Parachanna) and 29 recognized species [1]. Although Channa contains 26 exclusively Asian species, the extant 3 species of Parachanna are African. Molecular phylogenetic analysis estimated that both genera diverged between 40–49 Mya, whereas the Channidae family diverged from other Perciformes approximately 54 Mya [2].
Channids are commonly referred to as “snakeheads” due to the presence of large scales on the heads of most species. These fishes are very important in fishery and aquaculture, and over the years, their wild populations have undergone a substantial decline primarily due to overexploitation [3]. Snakeheads are highly evolved air-breathing fishes capable of surviving for months out of the water when buried in moist soil and are able to migrate over land using wriggling movements [4,5]. These traits imply that dispersal and speciation within the family may not necessarily reflect the processes commonly implied for other obligate freshwater fishes [6,7].
Additionally, such behavior also contributes to the large chromosomal variation found in this family. The diploid number varies from 2n = 32 in C. punctata to 2n = 112 in C. gachua [8,9,10], in which Robertsonian rearrangements, pericentric inversions and polyploidy appear to be the main sources of such chromosomal diversity [11,12]. However, cytogenetic studies in this family are still quite scarce. With a few exceptions [12], most studies on the Channidae family used classical cytogenetics data to determine the diploid number and karyotype composition of the species. Therefore, very little is known about other important cytogenetic features, such as the incidence of chromosomal repetitive DNA elements and their evolutionary role in this fish group.
The molecular organization and cytogenetic mapping of repetitive DNA elements, including satellites, multigene families and microsatellite repeats, have been analyzed in a large number of species, demonstrating their enormous potential for expanding the knowledge of the karyotype differentiation in fishes, as reviewed in [13]. In fact, the correlation between repetitive sequences and chromosomal rearrangements has been widely demonstrated [14] because their accumulation in specific genomic areas may induce chromosome breakages, deletions, inversions and amplifications [15,16].
Thus, considering that the high karyotype diversity in Channidae appears to be correlated with speciation events and repetitive DNAs have proven highly effective in showing genomic differentiations, we analyzed the karyotype structure and the distribution of repetitive DNA elements in four Channa species (C. lucius, C. micropeltes, C. striata and C. gachua). We aimed to investigate chromosomal divergence and probable relationships among species and populations to improve upon the knowledge of the biodiversity and evolutionary events of this important fish family.
Material and Methods
Materials
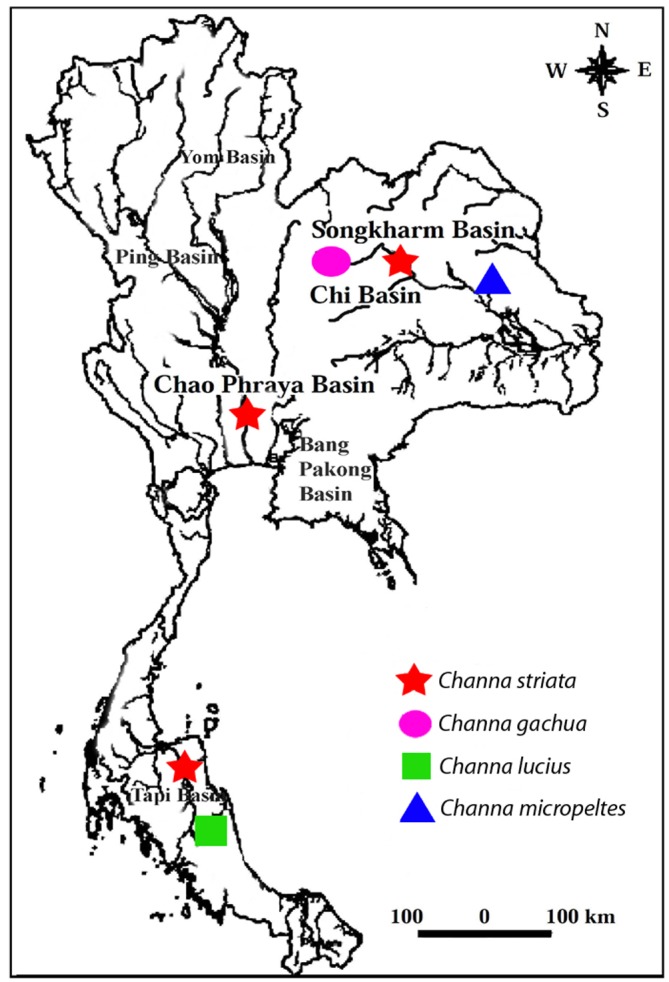
Individuals from both sexes of four Channa species, C. lucius, C. micropeltes, C. striata and C. gachua, were analyzed (Table 1). Fishes were collected from different river basins of Thailand (Fig 1). The specimens were caught using hand-net. After capture, animals were placed in sealed plastic bags containing oxygen and clean water and transported to the research station. The experiments followed ethical protocols and anesthesia with clove oil was administered prior to sacrificing the animals to minimize suffering. The process was approved by the Ethics committee of Khon Kaen University and by the RGJ committee under no. PHD/K0081/2556". Mitotic chromosomes were obtained from cell suspensions of the anterior kidney using the conventional air-drying method [17]. The specimens were deposited in the fish collection of the Cytogenetic Laboratory, Department of Biology Faculty of Science, Khon Kaen University.
Table 1. Collection sites of the analyzed species with the sample sizes.
| Species | N | |
|---|---|---|
| Channa striata | - Chi Basin | (03 ♀; 04 ♂) |
| Channa striata | - Chao Phraya Basin | (06 ♀; 08 ♂) |
| Channa striata | - Tapi Basin | (08 ♀; 07 ♂) |
| Channa micropeltes | - Chi Basin | (12 ♀; 11 ♂) |
| Channa lucius | - Tapi Basin | (08 ♀; 08 ♂) |
| Channa gachua | - Chi Basin | (09 ♀; 07 ♂) |
Fig 1. Collection sites of Channa species analyzed from Thailand.
Chromosome probes and FISH experiments
Two tandemly-arrayed DNA sequences isolated from the genome of an Erythrinidae fish species, Hoplias malabaricus, were used as probes. The first probe contained a 5S rDNA repeat copy and included 120 base pairs (bp) of the 5S rRNA transcribed gene and 200 bp of the non-transcribed spacer (NTS) sequence [18]. The second probe corresponded to a 1,400 bp segment of the 18S rRNA gene obtained via PCR from nuclear DNA [19]. The 5S and 18S rDNA probes were cloned into plasmid vectors and propagated in DH5α Escherichia coli competent cells (Invitrogen, San Diego, CA, USA).
The 5S and 18S rDNA probes were labeled with Spectrum Green-dUTP and Cy5-dUTP, respectively, using nick translation according to the manufacturer’s recommendations (Roche, Mannheim, Germany).
The microsatellites (CA)15, (GA)15, (GC)15, (CGG)10 and (CAA)10 were used as probes and were synthesized according to previous work [20]. These sequences were directly labeled with Cy3 at the 5’ terminus during synthesis by Sigma (St. Louis, MO, USA).
Fluorescence in situ hybridization (FISH) was performed under high stringency conditions on mitotic chromosome spreads [21]. Metaphase chromosome slides were incubated with RNAse (40 μg/ml) for 1.5 h at 37°C. After denaturation of the chromosomal DNA in 70% formamide/2x SSC at 70°C for 4 minutes, the hybridization mixture (2.5 ng/μl probes, 2 μg/μl salmon sperm DNA, 50% deionized formamide, 10% dextran sulphate) was dropped on the slides, and the hybridization was performed overnight at 37°C in a moist chamber containing 2x SSC. The first post-hybridization wash was performed with 2x SSC for 5 min at 65°C, and a final wash was performed at room temperature in 1x SSC for 5 min. Finally, the slides were counterstained with DAPI and mounted in an antifade solution (Vectashield from Vector Laboratories).
Image processing
Approximately 30 metaphase spreads were analyzed to confirm the diploid chromosome number, karyotype structure and FISH results. Images were captured using an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan) with CoolSNAP and the Image Pro Plus 4.1 software (Media Cybernetics, Silver Spring, MD, USA). Chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) or acrocentric (a) according to their arm ratios [22].
Results
Karyotypes
No differences between male and female karyotypes were observed in all species. Due to the small chromosomal size and the resulting difficulty in precisely distinguishing between m, sm and st chromosomes, all were referred to as bi-armed chromosomes, whereas the acrocentric chromosomes were termed mono-armed chromosomes.
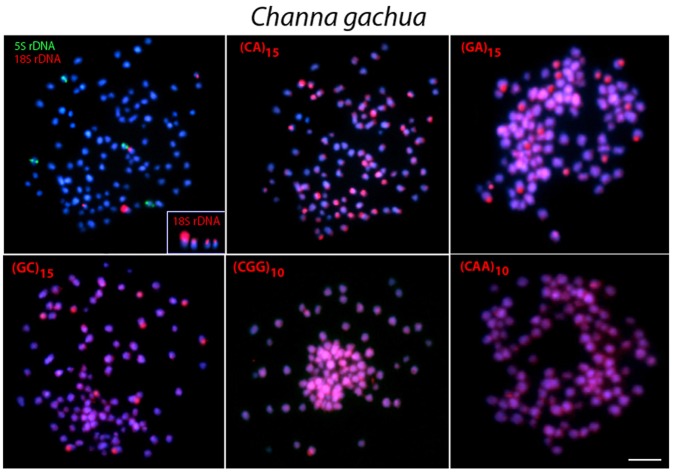
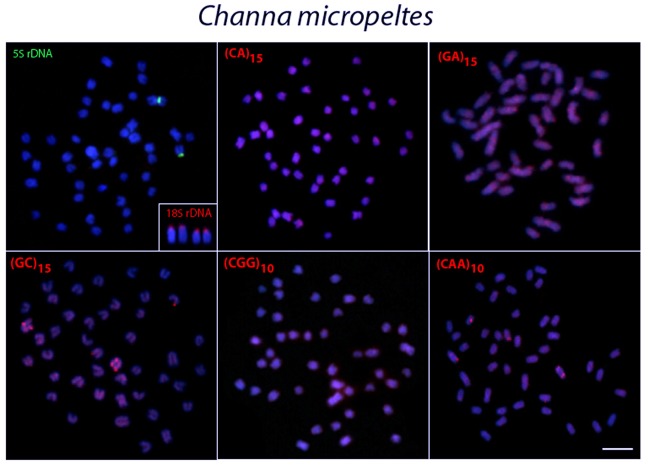
C. micropeltes presented a 2n = 44 (2 bi-armed + 42 mono-armed chromosomes), with the consequent number of chromosome arms (NF) equal to 46. C. lucius presented a 2n = 48 (4 bi-armed + 44 mono-armed chromosomes) with NF = 52, and C. gachua presented a 2n = 104 (8 bi-armed + 96 mono-armed chromosomes) with NF = 112 (Fig 2).
Fig 2. Fish images and karyotypes of Channidae fishes after Giemsa staining.

a = bi-armed (m, sm and st) chromosomes and b = mono-armed (a) chromosomes. Scale bars = 5 μm.
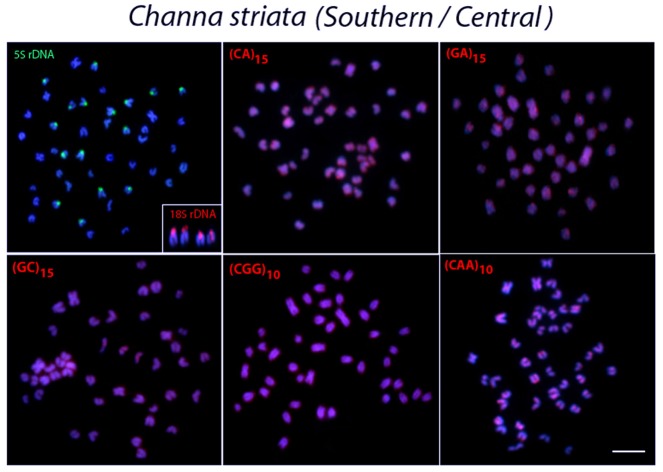
For C. striata, samples of three populations (from southern, central and northeastern Thailand) were analyzed. Both specimens from the southern and central populations presented a 2n = 44 (6 bi-armed + 38 acrocentric chromosomes) with NF = 50. However, all the individuals from the north-northeastern population presented the atypical diploid number of 2n = 43 (7 bi-armed + 36 acrocentric chromosomes) with NF = 50 (Fig 2).
Chromosome mapping of 5S and 18S rDNA sequences
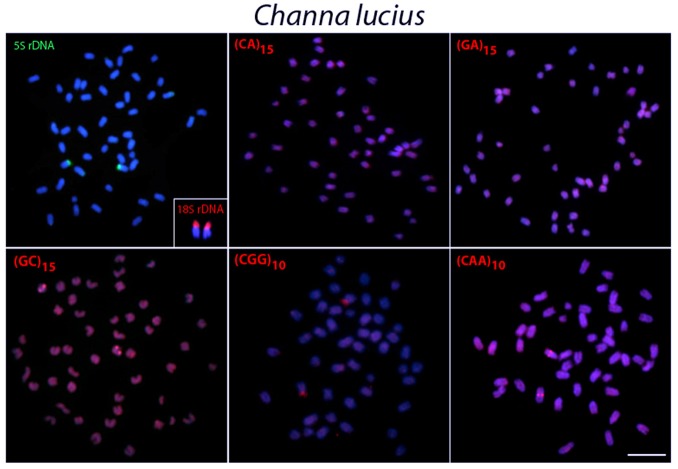
The probe for the 18S rDNA hybridized to the subtelomeric/telomeric region of two chromosome pairs in all species analyzed, except for C. lucius, in which only one pair was labeled with this probe (Figs 3–7, in the detail).
Fig 3. Metaphase plates of Channa lucius mapped with different repeated DNAs.
5S rDNA (green), 18S rDNA (red) and di- and trinucleotide microsatellites (red) as probes. The chromosomes with 18S rDNA sites are showned in enlarged forms in boxes. Scale bars = 5 μm.
Fig 7. Metaphase plates of Channa gachua mapped with different repeated DNAs.
5S rDNA (green), 18S rDNA (red) and di- and trinucleotide microsatellites (red) as probes. The chromosomes with 18S rDNA sites are showned in enlarged forms in boxes. Scale bars = 5 μm.
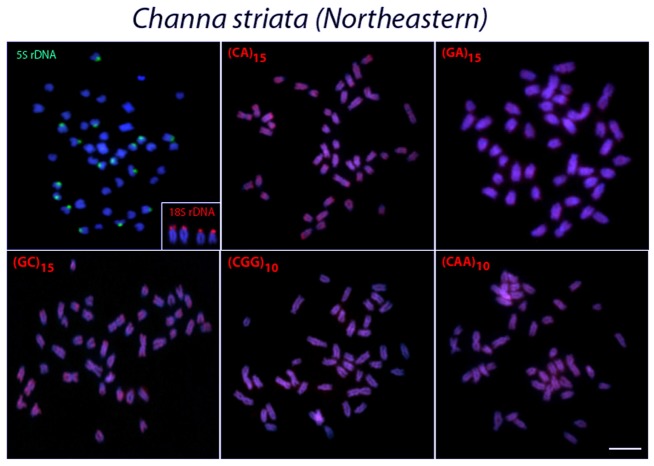
In contrast, 5S rDNA sequences showed a widely distinct distribution among species, although they were always located in the centromeric region of the chromosomes. Thus, 5S rDNA sequences were located in only one chromosome pair in C. micropeltes and C. lucius (Fig 4) but were present in two chromosome pairs in C. gachua (Fig 7). In turn, all 3 populations of C. striata showed a surprising increase in the number of 5S rDNA sites, with 8 chromosomal pairs bearing these sequences (Figs 5 and 6).
Fig 4. Metaphase plates of Channa micropeltes mapped with different repeated DNAs.
5S rDNA (green), 18S rDNA (red) and di- and trinucleotide microsatellites (red) as probes. The chromosomes with 18S rDNA sites are showned in enlarged forms in boxes. Scale bars = 5 μm.
Fig 5. Metaphase plates of Channa striata from southern and central Thailand populations mapped with different repeated DNAs.
5S rDNA (green), 18S rDNA (red) and di- and trinucleotide microsatellites (red) as probes. The chromosomes with 18S rDNA sites are showned in enlarged forms in boxes. Scale bars = 5 μm.
Fig 6. Metaphase plates of Channa striata from northeastern Thailand population mapped with different repeated DNAs.
5S rDNA (green), 18S rDNA (red) and di- and trinucleotide microsatellites (red) as probes. The chromosomes with 18S rDNA sites are showned in enlarged forms in boxes. Scale bars = 5 μm.
Chromosome Mapping of Microsatellite Sequences
In C. micropeltes, C. lucius and in the three populations of C. striata, most microsatellites were abundantly distributed in all chromosomes with the exception of (CG)15, (CGG)10 and (CAA)10, which showed stronger hybridization signals on two chromosomes in C. lucius, and (GC)15 and (CAA)10, which also displayed several more conspicuous sites in C. micropeltes (Figs 3–6). However, in C. gachua, the three dinucleotide microsatellites, (CA)15, (GA)15 and (GC)15, were highly accumulated in several chromosomes (Fig 7).
Discussion
Chromosomal evolution among Channa species
Morphological diversification among most Perciformes fishes is not accompanied by a conspicuous karyotype diversification [23]. However, this scenario does not appear to be the case for the Channidae family, in which a remarkable chromosomal diversity is observed.
The Thailand region contains seven recognized species of the genus Channa, with cytogenetic data known for six of them [24]. The evolutionary relationships for Asian snakeheads fishes and their probable divergence times within the Channidae family were proposed by one study [2] based on phylogenetic analyses. In addition, a putative evolutionary pathway based on the karyotypes of some Channa species highlighted a parallel with Adamson’s data [25]. Among these species, C. lucius occupies the most basal position and is closely related to C. diplogramme and C. micropeltes. In fact, these species are characterized by a region of gular scales that is also present in the African sister group Parachanna but absent in the majority of other Asian species [26]. Accordingly, C. lucius possesses 2n = 48 chromosomes, which was proposed as the basal number for Perciformes [27]. The decrease in the diploid number in the other Channa species analyzed (with the exception of C. gachua), with concomitant changes in the number of the bi-armed chromosomes, suggests that fusions and pericentric inversions were the main chromosomal rearrangements related to the karyotypic evolution of this genus. In fact, such rearrangements have been frequently detected in several species of Perciformes and are considered to be effective tools to characterize derived karyotypes [28]. Indeed, it is likely that major changes in karyotype structure may lead to a reorganization of co-adapted gene complexes; for this reason, these changes are an important evolutionary source [29,30].
Karyotype diversification processes and morphological patterns are often indicators of the lifestyle of a species [31,32]. Although Channidae species have a wide geographical distribution, their low vagility facilitates the maintenance of chromosomal rearrangements in small populations and, consequently, an increase in their chromosomal diversity. This same scenario is also found with the Neotropical fish Hoplias malabaricus (Erythrinidae), which represents a “species complex” with a wide geographic distribution and a low vagility. This fish is outstanding in its karyotypic diversity, with well-defined population differences in diploid numbers and chromosomal morphology and distinct sex chromosome systems [33,34].
The in situ investigation of repetitive DNA elements provided useful characteristics for comparative genomics at the chromosomal level, offering new insights into the karyotype evolution of Channa species. The number and distribution of ribosomal genes were not conserved among species, although the distribution of 18S rDNA sequences was less variable than that of 5S rDNA. All species presented four 18S rDNA sites, except C. lucius, in which only two incidences of this type of sequence were found (Figs 2–7). In contrast, the distribution pattern of the 5S rDNA sites showed a large variation. Thus, whereas 2 or 4 sites were mapped on the chromosomes of C. lucius, C. micropeltes and C. gachua, a total of 16 sites were observed in all C. striata populations analyzed (Figs 2–7). In this sense, C. striata is thought to be the most differentiated species, considering the 5S rDNA distribution in the karyotype. Coincidentally, these data support the chromosomal phylogeny proposed by one report [25], where C. striata was located in a different evolutionary path than that shared by C. lucius, C. micropeltes and C. gachua.
Hypervariability in number and location of rDNA loci was previously described for several fish groups [35–38]. In the red wolf fish, Erythrinus erythrinus, it was demonstrated that chromosomal rearrangements and genomic modifications due to rDNA spreading were significant events during the evolutionary history of this fish [35,36]. Additionally, the synteny of 5S rDNA genes and a retrotransposable element also indicated that chromosomes may have undergone rearrangements mediated by retrotransposon activity during evolution [35]. However, at the moment, we have no information on the possible role of transposable elements in increasing the number of 5S rDNA sites in C. striata, which will be the goal of further investigations. In fact, the particular incidence of such elements in the genome of this species will be a useful tool to strengthen this hypothesis.
Microsatellites, which are abundant repeated sequences that are present in all eukaryotic genomes studied thus far, are found either between the coding regions of structural genes or between other repetitive sequences [39]. In fish species such as H. malabaricus, Triportheus trifurcatus, Oplegnathus fasciatus and Leporinus elongatus, microsatellite repeats show a conspicuous accumulation in the telomeric regions of chromosomes [40–43]. However, only a discrete banding pattern was observed for some microsatellites in the Channa species analyzed, with a small accumulation of (GC)15, and (CAA)10 in specific chromosomal pairs in C. micropeltes and C. lucius, with this later also presenting also a (CGG)10 accumulation (Figs 3 and 4). However, C. gachua represented an exception to this general scenario, since a strong accumulation of some repeats was presented in several chromosomes (Fig 7). As this species has a polyploid origin [25], this differential accumulation of repetitive sequences may be due to its particular evolutionary history, as discussed below.
Chromosomal variability among C. striata populations
A comparative population analysis showed that C. striata differs in chromosome number and karyotype structure. Thus, although populations from southern and central Thailand presented 2n = 44 chromosomes, all the individuals from the northeastern population presented an atypical 2n = 43 chromosomes (Fig 1) and were distinguished by the presence of one particularly large m chromosome. Accordingly, some previous studies have also demonstrated that C. striata from Thailand contains a conspicuous karyotypic variation, with diploid numbers ranging from 2n = 40 to 2n = 44 [24,25].
In the multi-locus molecular phylogeny proposed by [2], the same C. striata populations were sampled; whereas all other Channa species formed a single monophyletic group, the divergence among C. striata populations was the highest. At the intra-specific level, C. striata populations were separated into three distinct and highly divergent clades. In accordance with the cytogenetic data, populations from central and southern Thailand were placed in a single group, whereas populations from northeastern Thailand comprised a divergent group [2].
Previous analysis of C. striata from the same northeastern Thailand region showed a karyotype structure of 2n = 42 chromosomes, with 8 bi-armed + 34 mono-armed chromosomes, i.e., with a reduction in the diploid number and the addition of two large m chromosomes compared with individuals from central and southern populations, which is likely due to centric fusion rearrangements [24]. In view of these findings, it is possible that C. striata with 2n = 43 chromosomes may correspond to a hybrid form from parental specimens with 2n = 42 x 2n = 44 chromosomes or, alternatively, to specimens heterozygotic for the centric fusion that gave origin to the 2n = 42 karyotype.
One remarkable feature present in all populations analyzed was the high number of 5S rDNA sites. Several adaptive conditions with significant evolutionary relevance may be associated with the rapid dispersion of specific sequences over a short time period [44]. For example, in the sister species Coregonus albula and C. fontanae, the spreading of rDNA sites affected the recombination rates in both genomes and led to a rapid genomic divergence, resulting in a faster ecological speciation event [37]. C. striata is currently the most common snakehead fish found in southern Asia and is largely exploited in the fish market due to its large natural distribution that includes many freshwater habitats. Thus, as well as in the Coregonus species, a rapid genome differentiation could help intensify divergences among distinct populations of C. striata. In fact, the sum of the available data for C. striata shows a clear chromosomal differentiation in this important fish group, which may correspond to a possible species complex.
The polyploidy status of C. gachua
The chromosomal analysis indicates that C. gachua corresponds to a polyploid species within the genus Channa with 4n = 104 chromosomes. However, this is not the only known case of polyploidy in this genus, as C. stewartii represents an autotetraploid species [11]. Although polyploidy represents an uncommon trait in higher vertebrates, it occurs independently and often repeatedly in many fish species [45]. The advantage of polyploidy in fishes is not only an increase in their potential for adaptability to the environment but also an enhancement of their reproductive efficiency [45]. In fact, C. gachua can live in higher mountain areas with fluctuating climates and can tolerate very stagnant, poorly oxygenated, turbid and even foul water, besides being the dominant species in the habitat [10,46].
A high degree of karyotype diversity has been reported for C. gachua, with the diploid number ranging from 78 to 112 chromosomes [10]. One of these populations, with 2n = 52 chromosomes, may likely be linked to the origin of the current analyzed polyploid individuals. Thus, as considered for C. striata, the karyotypic data for C. gachua corroborate the view that distinct populations of this species cannot be considered an evolutionary unit and likely resemble an assemblage of species with unresolved taxonomy.
The repetitive DNA fraction of the genome plays an important role during polyploidization and post-polyploidization changes [47,48]. After polyploidy, an eventual diploidization process may occur that leads to the loss of a subset of genes and the accumulation of repetitive DNA elements in many genomic areas. Because these sequences evolve faster than unique sequences and genes, they tend to colonize these new “ghost towns” and rapidly accumulate on the chromosomes [37,49]. Additionally, a parallel can also be traced to the evolution of the sex chromosomes, in which the “gene deserts” found in the sex-specific chromosomes (Y or W) serve as ideal niches for the long-term survival of these elements due to the weaker selection in these regions [50,51]. Similar events could also explain the strong accumulation of repeated DNA elements that was found in several chromosomes of C. gachua compared with the other diploid Channa species analyzed.
However, a repetitive DNA complex can also mediate ectopic chromosomal exchanges when homologous and (or) non-homologous chromosome recombination moves sequences within and between genomes. Furthermore, insertions of repeated DNA elements may create a new crossing-over ‘hot spot’ that promotes homologous or non-homologous chromosome rearrangements. The latter include spontaneous translocations, inversions and deletions and are potential mechanisms for rapid genome reorganization during speciation and stabilization of polyploids [14]. Therefore, whether the accumulation of repetitive DNA elements is the cause or the consequence of the cytogenetic events responsible for the emergence of polyploidy in C. gachua still requires further investigation.
Conclusions
Channidae fishes present a remarkable evolutionary dynamism. In the genus Channa, this attribute is mediated by different chromosomal rearrangements, such as pericentric inversions, fusions and polyploidization, resulting in a high karyotypic variation among species and populations. Additionally, such variability is also reinforced by the dynamism of repetitive elements in the genome, especially by the differential distribution and accumulation of rDNA sequences among chromosomes. Although not yet completely understood, this marked diversity is likely linked to the lifestyle of these fishes and to population fragmentation, as already identified for other fish species. In turn, these karyotypic features justify a taxonomic revision of the genus Channa, and of the Channidae family as a whole, in light of the observation that some nominal species may actually constitute species complexes.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was financially supported by the Genetics and Environmental Toxicology Research Group (Khon Kaen University) and by the Brazilian agency CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nelson JS. Fishes of the World. 4rd ed New York: John Wiley & Sons; 2006. [Google Scholar]
- 2. Adamson EAS, Hurwood DA, Mather PB. A reappraisal of the evolution of Asian snakehead fishes (Pisces, Channidae) using molecular data from multiple genes and fossil calibration. Mol Phylogenet and Evol. 2010;56: 707–717. 10.1016/j.ympev.2010.03.027 [DOI] [PubMed] [Google Scholar]
- 3. Lim KKP, Ng PKL. A Guide to the Freshwater Fishes of Singapore. Singapore: Singapore Science Centre; 1990. [Google Scholar]
- 4. Berra TM. Freshwater Fish Distribution. Chicago: University Of Chicago Press; 2007. [Google Scholar]
- 5. Sayer MDJ. Adaptations of amphibious fish for surviving out of water. Fish Fish. 2005;6: 186–211. [Google Scholar]
- 6. Near TJ, Benard MF Rapid allopatric speciation in Logperch Darters (Percidae:Percina). Evolution. 2004;58: 2798–2808. [DOI] [PubMed] [Google Scholar]
- 7. Rainboth WJ. The Taxonomy, Systematics, and Zoogeography of Hypsibarbus, A New Genus of Large Barbs (Pisces, Cyprinidae) from the Rivers of Southeastern Asia. Berkeley: University of California Press; 1996. [Google Scholar]
- 8. Donsakul T. Chromosome study on five species of channid fishes (Channa, family Channidae). from Thailand Fac Sc. Dept Biol: Wichian Magtoon 29th Kasetsart Univ. Ann. Conf; Bangkok (Thailand); 1991. [Google Scholar]
- 9. Naorem S, Bhagirath T. Chromosomal differentiations in the evolution of channid fishes—molecular genetic perspective. Caryologia. 2006;59(3): 235–240. [Google Scholar]
- 10. Kumar R, Kushwahaa B, Nagpurea NS, Beherab BK, Lakrac WS. Karyological and molecular diversity in three freshwater species of the genus Channa (Teleostei, Perciformes) from India. Caryologia. 2013;66 (2)109–119. [Google Scholar]
- 11. Rishi KK, Haobam MS. A chromosomal study on four species of snake heads (Ophiocephalidae: pisces) with comments on their karyotypic evolution. Caryologia. 1990;43: 163–167. [Google Scholar]
- 12. Singh M, Barman AS. Chromosome breakages associated with 45S ribosomal DNA sequences in spotted snakehead fish Channa punctatus . Mol Biol Rep. 2013;40: 723–729. 10.1007/s11033-012-2112-z [DOI] [PubMed] [Google Scholar]
- 13. Cioffi MB, Bertollo LAC. Chromosomal Distribution and Evolution of Repetitive DNAs in Fish In: Garrido R, editor. Repetitive DNAs Genome Dynamics. Basel: Karger; 2012. pp. 197–221. 10.1159/000337950 [DOI] [PubMed] [Google Scholar]
- 14. Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120: 351–357. 10.1159/000121084 [DOI] [PubMed] [Google Scholar]
- 15. Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster . Bioessays. 1994;16:269–75. [DOI] [PubMed] [Google Scholar]
- 16. Dimitri P, Arca B, Berghella L, Mei E. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster . Proc Natl Acad Sci. 1997;94: 8052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bertollo LAC, Takahashi CS, Moreira-Filho O. Cytotaxonomic considerations on Hoplias lacerdae (Pisces Erythrinidae). Braz J Genet. 1978;2: 103–120. [Google Scholar]
- 18. Martins C, Ferreira IA, Oliveira C, Foresti F, Galetti PM Jr. A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica. 2006; 127: 133–141. [DOI] [PubMed] [Google Scholar]
- 19. Cioffi MB, Martins C, Centofante L, Jacobina U, Bertollo LAC. Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet Genome Res. 2009;125: 132–141. 10.1159/000227838 [DOI] [PubMed] [Google Scholar]
- 20. Kubat Z, Hobza R, Vyskot B, Kejnovsky E. Microsatellite accumulation in the Y chromosome of Silene Latifolia . Genome. 2008;51: 350–356. 10.1139/G08-024 [DOI] [PubMed] [Google Scholar]
- 21. Pinkel D, Straume T, Gray J. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83: 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52: 201–220. [Google Scholar]
- 23. Molina WF, Maia-Lima FA, Affonso PRAM. Divergence between karyotypical pattern and speciation events in Serranidae fish (Perciformes). Caryologia. 2002; 55: 299–305. [Google Scholar]
- 24. Supiwong W, Jearranaiprepame P, Tanomtong A. A New Report of Karyotype in the Chevron Snakehead Fish, Channa striata (Channidae, Pisces) from Northeast Thailand. Cytologia. 2009;74(3): 317–322 [Google Scholar]
- 25. Tanomtong A, Supiwong W, Jearranaiprepame P, Khakhong S, Kongpironchuen C, Getlekha N. A New Natural Autotetraploid and Chromosomal Characteristics of Dwarf Snakehead Fish, Channa gachua (Perciformes, Channidae) in Thailand. Cytologia. 2014;79 (1): 15–27. [Google Scholar]
- 26. Musikasinthorn P, Taki Y. Channa siamensis (Gunther, 1861), a junior synonym of Channa lucius (Cuvier in Cuvier and Valenciennes, 1831). Ichthyol. Res. 2001;48: 319–324. [Google Scholar]
- 27. Brum MJI, Galetti PM Jr.. Teleostei ground plan karyotype. J. Comput. Biol. 1997;2: 91–102. 9228610 [Google Scholar]
- 28. Molina WF. Chromosomal changes and stasis in marine fish groups In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG. editors. Fish Cytogenetics. Science Publishers: Enfield; 2007. pp. 69–110. [Google Scholar]
- 29. Butlin RK. Recombination and speciation. Mol Ecol. 2005;14: 2621–2635. [DOI] [PubMed] [Google Scholar]
- 30. Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25: 660–669. 10.1016/j.tree.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 31. Karr JR, James FC. Eco-morphological configurations and convergent evolution of species and communities In: Cody ML, Diamond JM. editors. Ecology and Evolution of Communities. Cambridge: Harvard University Press; 1975. pp. 258–291. [Google Scholar]
- 32. Wainwright P C, Reilly SM (1994) Ecological morphology: integrative organism biology. Chicago, The University of Chicago Press, 367p [Google Scholar]
- 33. Bertollo LAC, Born GG, Dergam JA, Fenocchio AS, Moreira-Filho O. A biodiversity approach in the Neotropical Erythrinidae fish, Hoplias malabaricus. Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chromosome Res. 2000; 8:603–613. [DOI] [PubMed] [Google Scholar]
- 34. Bertollo LAC. Chromosome evolution in the Neotropical Erythrinidae fish family: an overview, In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG, editors. Fish cytogenetics. NH, USA: Enfield; 2007. pp. 195–211. [Google Scholar]
- 35. Cioffi MB, Martins C, Bertollo LAC. Chromosomal spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol Biol. 2010;10: 217 10.1186/1471-2148-10-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martins NF, Bertollo LAC, Troz WP, Feldberg E, Valentin FCS, Cioffi MB. Differentiation and evolutionary relationships in Erythrinus erythrinus (Characiformes, Erythrinidae). Comparative chromosome mapping of repetitive sequences. Rev Fish Biol Fisher. 2013;23: 261–269. [Google Scholar]
- 37. Symonová R, Majtánová Z, Sember A, Staaks GBO, Bohlen J, Freyhof J, et al. Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress—activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol Biol. 2013; 13: 42 10.1186/1471-2148-13-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lima-Filho PA, Bertollo LAC, Cioffi MB, Costa GWWF, Molina WF. Karyotype divergence and spreading of 5S rDNA sequences between genomes of two species: darter and emerald gobies (Ctenogobius, Gobiidae)”. Cytogenet Genome Res. 2014;142: 197–203. 10.1159/000360492 [DOI] [PubMed] [Google Scholar]
- 39. Tautz D, Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984; 25 12(10): 4127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cioffi MB, Kejnovsky E, Bertollo LAC. The chromosomal distribution of microsatellite repeats in the wolf fish genome Hoplias malabaricus, focusing on the sex chromosomes. Cytogenet and Genome Res. 2011;132: 289–296. [DOI] [PubMed] [Google Scholar]
- 41. Dongdong X, Bao L, Bertollo LAC, Cioffi MB. Chromosomal mapping of microsatellite repeats in the rock bream fish Oplegnathus fasciatus, with emphasis of their distribution in the neo-Y chromosome. Mol Cytogenet. 2013;6:12 10.1186/1755-8166-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poltronieri P, Marquioni V, Bertollo LAC, Keynovský E, Molina WF, Liehr T, et al. Comparative chromosomal mapping of microsatellites in Leporinus species (Characiformes, Anostomidae): Unequal accumulation on the W chromosomes. Cytogenetic and Genome Res. 2014;142: 40–50. 10.1159/000355908 [DOI] [PubMed] [Google Scholar]
- 43. Yano CF, Poltronieri J, Bertollo LAC, Artoni RF, Liehr T, Cioffi MB. Chromosomal Mapping of Repetitive DNAs in Triportheus trifurcatus (Characidae, Characiformes): Insights into the Differentiation of the Z and W Chromosomes. PLoS ONE. 2014; 9(3): e90946 10.1371/journal.pone.0090946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. David P, Chen NWG, Pedrosa-Harand A, Thareau V, Sévignac M, et al. A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol. 2009;151: 1048–1065. 10.1104/pp.109.142109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev. Fish Biol Fish. 2003;13: 237–246. [Google Scholar]
- 46. Rahman AKA. Freshwaterfishes of Bangladesh. Department of Zoology, University of Dhaka, Bangladesh: Zoological Society of Bangladesh; 1989. [Google Scholar]
- 47. Feldman M, Levy AA. Allopolyploidy: a shaping force in the evolution of wheat genomes. Cytogenet and Genome Res. 2005;109: 250–258. [DOI] [PubMed] [Google Scholar]
- 48. Ma XF, Gustafson JP. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet and Genome Res. 2005;109: 236–249. [DOI] [PubMed] [Google Scholar]
- 49. Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Ann Bot (Lond). 1989; 64: 315–324 [Google Scholar]
- 50. Kejnovsky E, Hobza R, Kubat Z, Cermak T, Vyskot B. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity. 2009; 102:533–541. 10.1038/hdy.2009.17 [DOI] [PubMed] [Google Scholar]
- 51. Matsunaga S Junk DNA promotes sex chromosome evolution. Heredity. 2009; 102: 525–526 10.1038/hdy.2009.36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.