Abstract
Insect mitochondrial genomes (mtDNA) are usually double helical and circular molecules containing 37 genes that are encoded on both strands. The arrangement of the genes is not constant for all species, and produces distinct gene orders (GOs) that have proven to be diagnostic in defining clades at different taxonomic levels. In general, it is believed that distinct taxa have a very low chance of sharing identically arranged GOs. However, examples of identical, homoplastic local rearrangements occurring in distinct taxa do exist. In this study, we sequenced the complete mtDNAs of the ants Formica fusca and Myrmica scabrinodis (Formicidae, Hymenoptera) and compared their GOs with those of other Insecta. The GO of F. fusca was found to be identical to the GO of Dytrisia (the largest clade of Lepidoptera). This finding is the first documented case of an identical GO shared by distinct groups of Insecta, and it is the oldest known event of GO convergent evolution in animals. Both Hymenoptera and Lepidoptera acquired this GO early in their evolution. Using a phylogenetic approach combined with new bioinformatic tools, the chronological order of the evolutionary events that produced the diversity of the hymenopteran GOs was determined. Additionally, new local homoplastic rearrangements shared by distinct groups of insects were identified. Our study showed that local and global homoplasies affecting the insect GOs are more widespread than previously thought. Homoplastic GOs can still be useful for characterizing the various clades, provided that they are appropriately considered in a phylogenetic and taxonomic context.
Keywords: mitochondrial genomics, gene order analysis, gene order evolution, convergent evolution, Hymenoptera, Lepidoptera
Introduction
The mitochondrial genomes (mtDNA) of insects are usually double helical and circular molecules spanning 15–18 kb (Cameron 2014), though some exceptions exist where the mtDNA is divided into a variable number of mini-circles (e.g., Shao et al. 2009; Cameron et al. 2011; Cameron 2014). Insect mtDNA usually contains 37 genes including 13 protein-encoding genes, 22 tRNAs and the small and large ribosomal RNAs (fig. 1; Cameron 2014). Some mtDNAs have more than 37 genes. This increased number is due to the occurrence of multiple contiguous copies of one or more tRNAs (e.g., Apis florea, Wang et al. 2013; fig. 4).
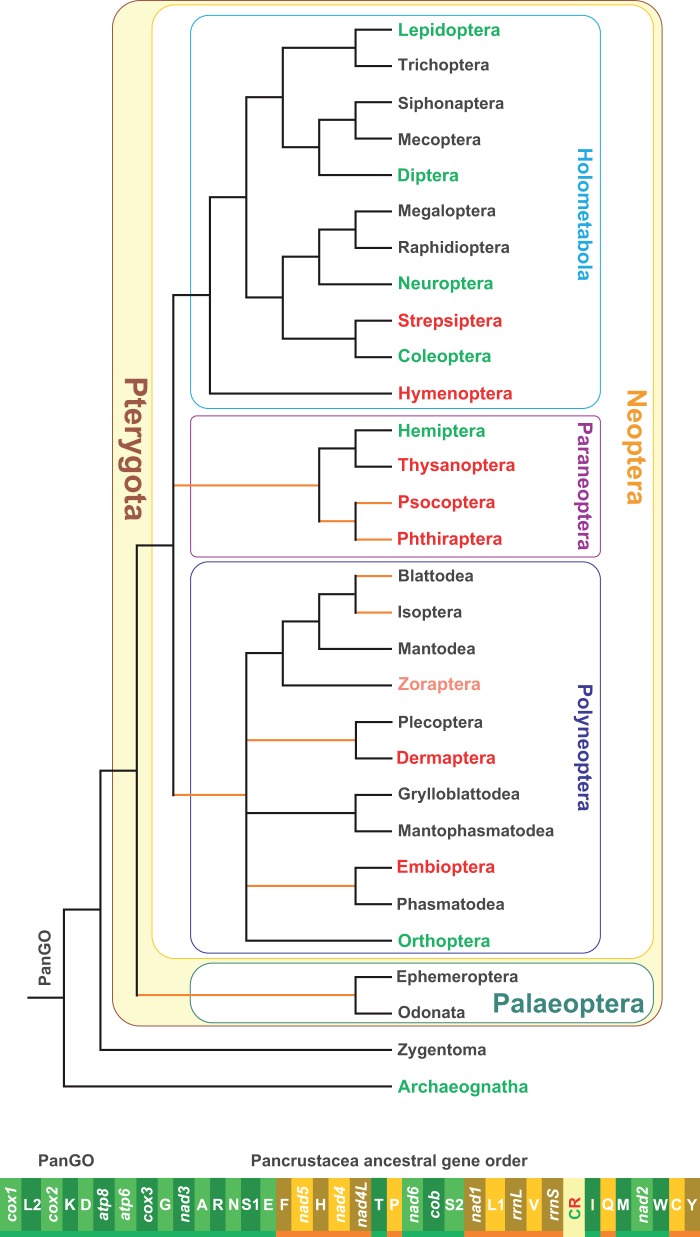
Fig. 1.—
The PanGO and the distribution of GOs in the different orders of Insecta. The PanGO is linearized starting from cox1. The genes encoded on the α-strand (orientation from right to left in fig. 1) are represented in a light/deep green background, whereas those encoded on the β-strand (orientation from left to right in fig. 1) are depicted with a bright yellow/light brown background. The genes on the α-strand are underlined in green, whereas those on the β-strand are underlined in orange. Genes nomenclature: atp6 and atp8: ATP synthase subunits 6 and 8; cob: apocytochrome b; cox1-3: cytochrome c oxidase subunits 1–3; nad1-6 and nad4 L: NADH dehydrogenase subunits 1–6 and 4 L; rrnS and rrnL: small and large subunit ribosomal RNA (rRNA) genes; X: transfer RNA (tRNA) genes, where X is the one-letter abbreviation of the corresponding amino acid, in particular L1 (CTN codon family) L2 (TTR codon family), S1 (AGN codon family) S2 (TCN codon family). The consensus phylogenetic tree depicts the phylogenetic relationships among the 30 existing orders forming the class Insecta. The tree was based principally on the papers of Trautwein et al. (2012) and Peters et al. (2014). The orange branches denote uncertain relationships or possible nonmonophyly of terminal taxa. The orders with black names possess only the PanGO. The pink label for the order Zoraptera denotes that for this taxon, there are no complete mtDNA data currently available. The orders with a green label possess both PanGO as well as alternative GOs. The orders with red label have only GOs that are different from PanGO.
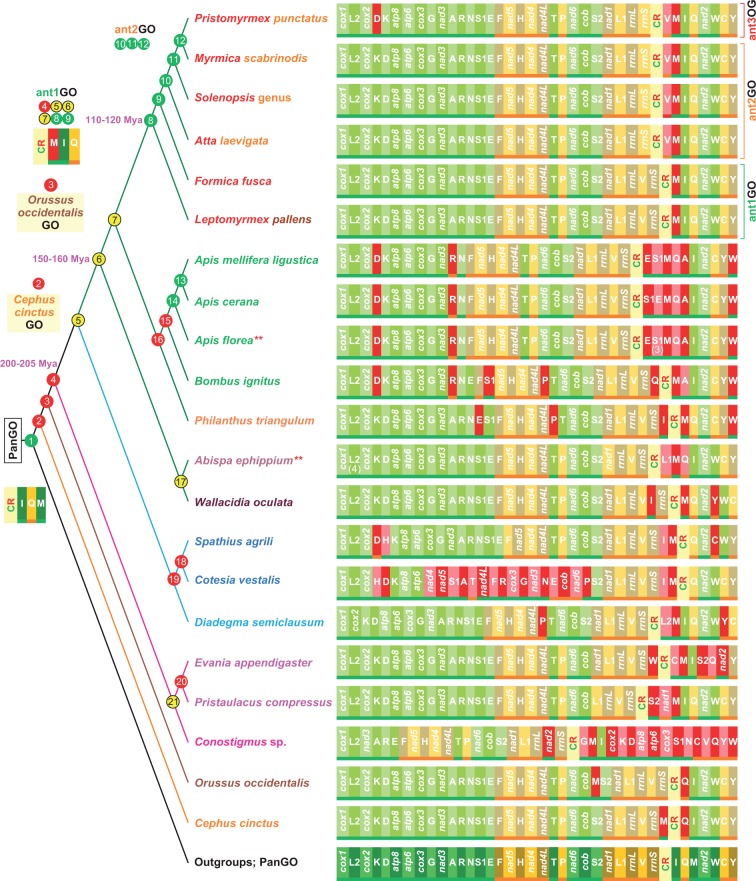
Fig. 4.—
The evolution of GOs in Hymenoptera. Nodes are colored according the output of the TreeREx program (Bernt et al. 2008). Green node, consistent node. Yellow node, 1-consistent node; red node, fallback node. The different colors reflect the level of uncertainty that characterizes the reconstruction of the GO. The red nodes exhibit the highest level of uncertainty with respect to alternative GOs, whereas yellow and green nodes are more reliable in their reconstructions. The GO reconstructions are provided solely for nodes that are pivotal to this article. For a detailed reconstruction for all nodes, included the TreeREx scores see the supplementary figure S3, Supplementary Material online. *, multiple copies of S1 (3) considered just once in the TreeREx analysis; **, multiple copies of L2 considered just once in TreeREx analysis. The dating times are expressed in millions of years and were obtained from Grimaldi and Engel (2005). The genomic and genetic nomenclature, as well as the color scheme, are the same as in figure 1. The genes that changed their position relative to PanGO are shown with a pink/red background.
The insect mtDNA genes are encoded on both strands of DNA (referred to herein as the α- and β-strands). Genes can overlap, be adjacent or separated by intergenic spacers composed of a variable number of nucleotides. The major intergenic spacer that is always present is the Control Region (CR). The CR is variable in length (hundreds to thousands of base pairs) and contains the origin of replication for the mtDNA (Saito et al. 2005). It encodes the elements necessary to maintain the mtDNA; thus, it is important to take into account its position in the mtDNA sequence. Insect mtDNAs exhibit different gene orders (GOs). With respect to a reference GO, a gene can be transposed (i.e., moved to a different placement on the same strand), inverted (i.e., moved to the opposite strand but occupying the same placement in the mtDNA), or both inverted and transposed (a combination of the first two events). It is not completely understood how genes move, although various models have been proposed (Cameron 2014). An intramitochondrial recombination process is invoked to model gene inversion (Dowton and Campbel 2001). Simple transposition can be explained by a duplication and random loss (drl) model (Moritz et al. 1987; Boore 2000), whereas the inverted transposition can be described through the combination of these two mechanisms. The global rearrangement pattern can be further analysed through a tandem duplication and random loss (TDRL) mechanism (Bernt et al. 2007; Bernt and Middendorf 2011). According to Bernt and Middendorf (2011), TDRL involves a tandem duplication of a continuous segment of genes such that the original segment and its copy are consecutive followed by the loss of one copy of each of the redundant genes.
Inversion, inversed transposition, and transposition are “reversible” rearrangements because they do not assign the changes a chronological order and, thus, cannot be used alone to define the plesiomorphic/apomorphic status of opposite transformational pathways (i.e., GO1–GO2 vs. GO2–GO1) occurring between two GOs. These rearrangements must be analysed in combination with phylogenetic information using out-group comparison (e.g., Perseke et al. 2008). Conversely, TDRLs generate “generally irreversible” rearrangements and can be used to identify which of the transformational pathways listed above are plesiomorphic or apomorphic (Perseke et al. 2008).
The 37 standard animal mitochondrial genes can be arranged in an astonishing number of GOs (i.e., 37! = 1.367 × 1043 or 38! if the CR is also included) provided that movement of every gene is equally probable (Cameron 2014). The huge potential number of different combinations means that the probability of the same GO occurring in distinct animal lineages is extremely low (Boore 2006). According to this assumption, the chance of two mtDNAs, obtained from distantly related taxa, to share the same derived genome organization is only one in 2,664 (Dowton et al. 2002).
Different GOs have proven to be highly diagnostic in defining animal groups at various taxonomic ranks. This aspect is well documented in the Chordata, a phylum for which a huge amount of data exists on mtDNAs. For example, GOs with diagnostic capability exist for Vertebrata, Marsupialia, and Crocodylidae, to name a few (Pääbo et al. 1991; Kumazawa and Nishida 1995; Boore 1999; supplementary fig. S1, Supplementary Material online). In general, alternative GOs are potentially powerful molecular signatures with high diagnostic and phylogenetic capabilities (Boore and Brown 1998; Boore 2006).
At the onset of mtDNA genomics, the monophyly of Arthropoda received strong corroboration through the study of GOs (Boore et al. 1995). In addition, the sister-taxon relationship between Crustacea and Hexapoda (i.e., the clade Pancrustacea) was strongly supported by their exclusively shared GO (hereafter named PanGO; fig. 1; Boore et al. 1998).
Insects form the biggest group of living beings with a million species described and 1–5 millions still undescribed (Gullan and Cranston 2014). They have a very long history, having emerged 410–420 Ma at the boundaries between Devonian and Silurian (Grimaldi and Engel 2005). Currently they are split into 30 orders (Trautwein et al. 2012), and the relationships among these orders are not fully resolved (see Trautwein et al. [2012] and references therein). A consensus phylogeny for the whole class Insecta including the currently known distribution of GOs in the different orders is provided in figure 1. The PanGO is broadly distributed among insects. Indeed, 15 orders have only PanGO, while in seven orders, PanGO is still well represented or the most observed GO type (e.g., Diptera, Coleoptera; Cameron 2014; and references therein). Hemiptera, or rather the Homoptera suborder, departs from this pattern; other alternative GOs are the dominant type (e.g., Thao et al. 2004; Zhang et al. 2013, 2014). The same is true for Neuroptera in a manner described in more detail below. For Zoraptera, no data are available. Finally, seven orders only present GOs that are different from PanGO. They are listed in red in figure 1. For Dermaptera, Embioptera, Thysanoptera, and Strepsiptera, the available data are restricted to single or few mtDNAs that are complete or nearly complete (e.g., Shao and Barker 2003; Carapelli et al. 2006; Komoto et al. 2012; Wan et al. 2012). Thus, a better taxon sampling could reveal the presence of PanGO in these orders. Conversely, multiple mtDNAs have been sequenced for Psocoptera, Phtiraptera, and Hymenoptera, but PanGO has not yet been found (e.g., Cameron et al. 2011; Wei et al. 2012; Jiang et al. 2013; this article; see also Supplementary Material online for further references dealing with Hymenoptera).
The class Insecta presents a broad array of alternative GOs other than PanGO that have a very uneven distribution (fig. 1; Cameron 2014). Many of these GOs are considered to be molecular signatures for various lineages (Cameron 2014). Several examples of these GOs, with a particular focus on those known for the mega-diverse clade Holometabola that encompasses the vast majority (∼85%) of Insecta (Grimaldi and Engel 2005), are described below (figs. 3 and 4 and supplementary fig. S2, Supplementary Material online).
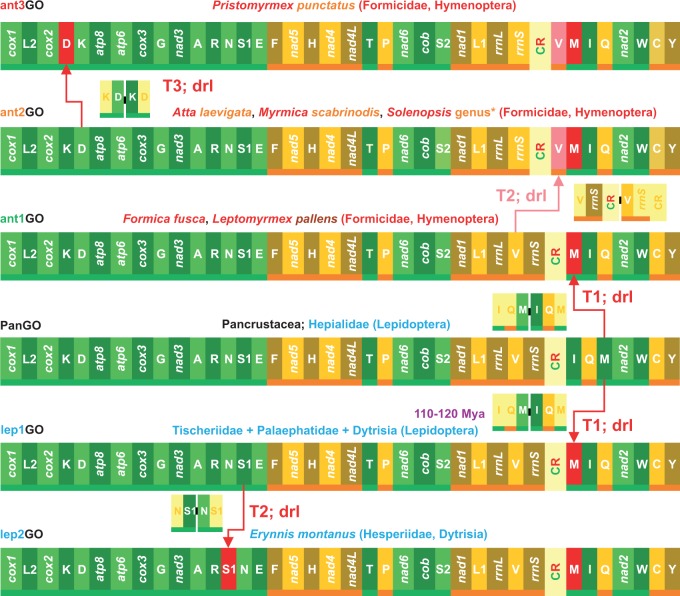
Fig. 3.—
Pairwise comparisons and GO evolution in ants and Lepidoptera mtDNAs. Rearrangements in the GOs of Lepidoptera and ants are investigated and depicted with respect to PanGO. T1–T3, transposition events; dlr, duplication random loss, mechanism producing the observed re-arrangement. *, related to the species of Solenopsis analysed in this article (supplementary table S1, Supplementary Material online). The time dating is expressed in millions of years and was obtained from Grimaldi and Engel (2005). The genomic and genetic nomenclature, as well as the color scheme, are the same as in figure 1. The genes that changed position relative to PanGO are shown with a pink/red background.
The clade Dytrisia accounts for more than 98% of the approximately 158,000 named species of Lepidoptera (i.e., butterflies and moths; van Nieukerken et al. 2011). All sequenced dytrisian mtDNAs (e.g., Salvato et al. 2008; Timmermans et al. 2014) exhibit a transposition of trnM on the α-strand that creates the MIQ versus IQM arrangement (lep1GO in fig. 3). Lep1GO is also present in Tischeriidae and Palaephatidae, two nondytrisian families that are considered to be sister taxa of Dytrisia (Timmermans et al. 2014). Conversely, the moths of the family Hepialidae, a nondytrisian group not closely related to Dytrisia, retain the PanGO (fig. 3; Cao et al. 2012). Thus, taking into account the fossil records for the lepidopteran taxa listed above (Grimaldi and Engel 2005), the lep1GO emerged at least 120–110 Ma in the Cretaceous Period. A transposition of trnC on the β-strand creates a CW versus WC arrangement and is present in a major clade of Neuroptera, while other species retain the PanGO (Negrisolo et al. 2011; Zhao et al. 2013). Transposition of trnD on the α-strand, producing the DK versus KD arrangement, characterizes the suborder Caelifera within the Orthoptera (Flook et al. 1995a). Transposition of trnR on the α-strand, generating the RA versus AR arrangement, differentiates the Culicidae mosquitos from other species of Diptera (Beard et al. 1993; Mitchell et al. 1993; Behura et al. 2011). Finally, the transposition on the β-strand of trnP downstream of nad6 is characteristic of the beetles in the superfamily Dryopoidea (Timmermans and Vogler 2012). All of the GOs listed above are determined by the transposition of a single tRNA. Thus, movements are not equally probable among the mtDNA genes, as tRNAs are the most mobile. This behavior was recognized very early (Moritz et al. 1987) and has been corroborated by successive studies. Different tRNAs move at different paces, and some of them moved along the same path in distinct phyletic lineages generating homoplastic rearrangements. The first reported example involved the Locusta migratoria (Orthoptera Caelifera) and Apis mellifera (Hymenoptera) that exhibit the same DK versus KD arrangement (Flook et al. 1995b). However, when the complete GOs of the grasshopper and bee are compared, they are quite different (supplementary fig. S2, Supplementary Material online). Within Hymenoptera, the DK versus KD gene arrangement has appeared repeatedly in separate subclades (Dowton and Austin 1999) that exhibit otherwise different GOs (see below and fig. 4). In the Braconidae (Hymenoptera), inversion of trnH from the β- to the α-strand occurred multiple times (Dowton 2002). Species of Hymenoptera, one the four major insect orders including more than 150,000 species of ants, wasps, bees, and many others (Aguiar et al. 2013), are involved in all of the homoplastic rearrangements described above. This behavior reflects a high plasticity for rearrangements exhibited by the whole order that is considered to be a hotspot for mtDNA GOs (Dowton and Austin 1999; Dowton et al. 2003; Dowton, Cameron, Dowavic, et al. 2009; this article).
In this study, the complete mtDNA genomes of two ants, Formica fusca Linnaeus, 1758 (Formicidae Formicinae) and Myrmica scabrinodis Nylander, 1846 (Formicidae Myrmicinae), were sequenced and their GOs were compared with those known for the Hymenoptera as well as for other Insecta. These analyses provided interesting and surprising findings. In particular, the GO of F. fusca was identical to the GO of Dytrisia (the largest clade of Lepidoptera), which represents the first documented case of an identical GO shared by distinct groups of Insecta. Taking into account the time of appearance of this homoplastic rearrangement in ants and Lepidoptera, the event documented here is the oldest known example of convergent evolution in GOs for the whole animal kingdom. An analysis of GOs performed using new bioinformatic tools combined with a phylogenetic approach allowed for exploration into the evolution of the GO of Hymenoptera. Finally, we show that local and complete homoplastic rearrangements are common in the GOs of Insecta and, more generally, in animals. In addition, we provide suggestions for using GOs as molecular signatures. All of these findings are described in detail in the following sections.
Materials and Methods
Samples Collection Details and DNA Extraction
Multiple specimens (20–30) of F. fusca and M. scabrinodis were collected from single nests (NIDO SCU-34, F. fusca; NIDO SCU-30, M. scabrinodis) at: Prà bestemà 850 m.a.s.l., Prada, Monte Baldo, Verona, Italy (45 °38′50.27″N; 10 °45′24.98″E; 10-VIII-2012) by Antonio Scupola who also identified them. The samples were preserved in pure ethanol at 4 °C until DNA extraction.
Total DNA was extracted using the ZR Genomic DNA-Tissue Midiprep (Zymo Research corp.) Kit. DNA quality was assessed through electrophoresis. The DNA concentration was determined using the (high sensitivity) Qubit DNA quantification kit (Invitrogen, USA).
Mitochondrial Genome Sequencing
The total DNAs, at a concentration of at least 100 ng/µl, were sent to the UCDAVIS Genome Center (http://dnatech.genomecenter.ucdavis.edu/, last accessed December 10, 2014; Davis University, California) to be sequenced using next-generation sequencing (NGS) Illumina HiSeq 2500 Rapid Run Mode—PE100 paired technology (see the UCDAVIS Genome Center for further details on the sequencing strategy). After the sequencing process 22,076,120 paired sequences were obtained for F. fusca and 27,105,798 sequences in pairs were obtained for M. scabrinodis.
Genome Assembly and Identification of the Full Length Mitochondrial Genome
Global assembly of the NGS reads obtained for F. fusca and M. scabrinodis was accomplished with the CLC-BIO program: CLC Genomics Workbench v7.0.4 (http://www.clcbio.com, last accessed December 10, 2014). After a BLAST search (Altschul et al. 1990; Tatusova and Madden 1999), the sequences that had a high score match with mitochondrial genes (E 10−20) were fully annotated using the strategy described in the next section. Afterwards a single sequence for both F. fusca and M. scabrinodis covering at least 95% of the final full length mtDNA (see below) was selected as the template for successive assembly performed using the MITObim program (Hahn et al. 2013). This second analysis provided a final assembly encompassing the full length mitochondrial genome (mtDNA) for both F. fusca and M. scabrinodis. Statistics on the final assemblies were calculated with Tablet software (Milne et al. 2013).
The full length sequences of both mtDNAs can be accessed from the EBI/GenBank (F. fusca, LN607805; M. scabrinodis, LN607806).
Mitochondrial Genome Annotation
The annotation strategy applied to the newly sequenced mtDNAs was based on the guidelines provided by Lavrov et al. (2000). The nomenclature of genes and strands follows Negrisolo et al. (2004).
Initially, the mtDNA sequence was translated into putative proteins using the Transeq program available on the EBI website. The identity of these polypeptides was verified using the BLAST program (Altschul et al. 1990; Tatusova and Madden 1999) available at the NCBI website. The boundaries of genes were determined as follows. The 5′-ends of protein-coding genes (PCGs) were defined as the first legitimate in-frame start codon (ATN, GTG, TTG, GTT) in the open reading frame that was not located within an upstream gene encoded on the same strand. The only exception was atp6, which has been previously demonstrated to overlap with its upstream gene atp8 in many mtDNAs (Wolstenholme 1992). The PCG terminus was defined as the first in-frame stop codon that was encountered. When the stop codon was located within the sequence of a downstream gene encoded on the same strand, and a truncated stop codon (T or TA) adjacent to the beginning of the downstream gene was designated as the termination codon. This codon was thought to be completed by polyadenylation, thereby producing a complete TAA stop codon after transcript processing. Finally, pairwise comparisons with orthologous proteins were performed using the ClustalW (Thompson et al. 1994) to better define the limits of the PCGs.
Regardless of the real initiation codon, a formyl-Met was assumed to be the starting amino acid for all proteins as has been previously shown for other mitochondrial genomes (Smith and Marcker 1968; Fearnley and Walker 1987).
Transfer RNA genes were identified using the tRNAscan-SE program (Lowe and Eddy 1997) or recognized manually as sequences having the appropriate anticodon and capable of folding into the typical cloverleaf secondary structure of tRNAs (Wolstenholme 1992). The validity of these predictions was further enhanced by comparison, based on multiple alignment and structural information, to published orthologous counterparts.
The boundaries of the ribosomal rrnL and rrnS genes were determined by comparison to the orthologous counterparts present in the mtDNAs of the Hymenoptera species already sequenced, as well as structural information implied by direct modeling (data not shown).
Data Set Construction
All partial or complete mtDNAs published or publically available, used in this article, were downloaded from GenBank or MetAmiga (http://amiga.cbmeg.unicamp.br/, last accessed December 10, 2014; Feijao et al. 2006) and reannotated following the approach described above to produce very high-quality annotations.
Currently, fewer than 50 partial or complete mtDNAs of species belonging to the major lineages of Hymenoptera are available in GenBank (release August 25, 2014). For 26 mtDNAs (partial or complete) it was possible to unambiguously determine their complete GOs. Of these, 23 were used in all genomic comparisons, while the two Scelionids species, (Idris sp., KF696670 and Ceratobaeus sp. KF696669) (Hymenoptera; Apocrita, Platygastroidea, Scelionidae; Mao and Dowton 2014) and the sawfly Allantus luctifer (Hymenoptera; Tenthredinoidea Tenthredinidae) (KJ713152; Wej, Niu, et al. 2014), became available too late (July 18 and 29, 2014, respectively) to be fully considered in this article. However, their GOs were included in some pairwise comparisons with the GOs observed in ants.
The mtDNAs of a broad spectrum of Hexapoda were also analysed in this study (see supplementary table S1, Supplementary Material online). However, most of these were used exclusively for the identification of the placement of trnN in ants (see following paragraph).
Placement of trnN in the mtDNAs of Ants
Before this study, nearly complete or full length mtDNAs were available for six ant species (Formicidae, Hymenoptera, supplementary table S1, Supplementary Material online; Gotzek et al. 2010; Hasegawa et al. 2011; Berman et al. 2014; Rodovalho et al. 2014). Five of them belong to the subfamily Myrmicinae, that is, Atta laevigata, Pristomyrmex punctatus, Solenopsis geminata, Solenopsis invicta, and Solenopsis richteri. Conversely, Leptomyrmex pallens is a member of the subfamily Dolichoderinae. For At. laevigata and P. punctatus, the original descriptions of the mtDNA placed the trnN in the α-strand between trnR and trnS1, that is, the standard position in the PanGO arrangement (figs. 1 and 2; Hasegawa et al. 2011; Rodovalho et al. 2014). Conversely, in the species of the genus Solenopsis (Gotzek et al. 2010) a new placement for the trnN was identified in the β-strand between rrnS and CR. Analogously, in L. pallens the trnN was identified in the α-strand between rrnS and CR (Berman et al. 2014). The trnN genes of Solenopsis and L. pallens exhibit a legitimate putative anticodon ATT but do not have homologous counterparts (Gotzek et al. 2010).
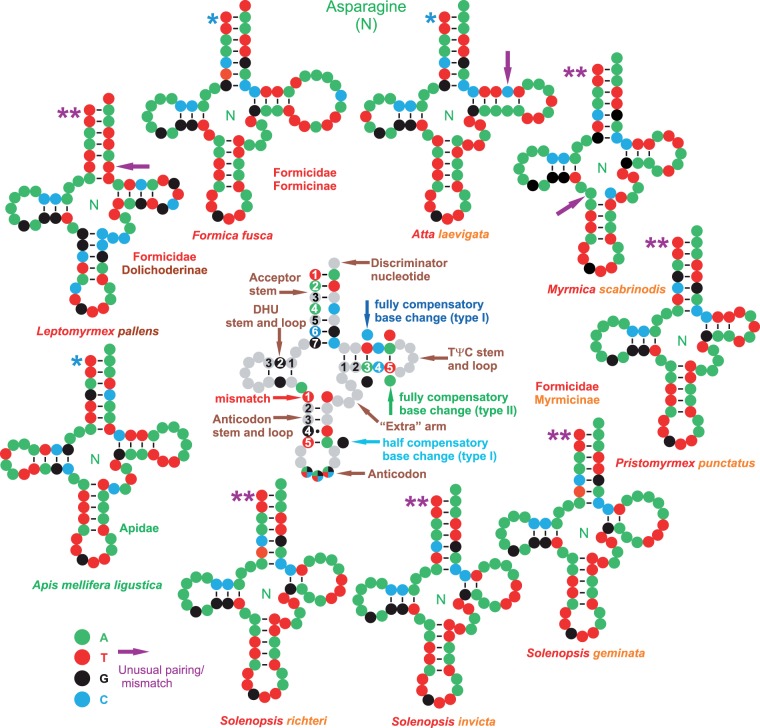
Fig. 2.—
Secondary structures of the ant trnNs. Fully compensatory base changes are substitutions that do not disrupt base pairing in the stem. They are classed here as type 1 (purine—pyrimidine vs. purine—pyrimidine, and vice versa) and type 2 ((purine—pyrimidine vs. pyrimidine—purine, and vice versa). A half compensatory change implies the substitution of a single base without the disruption of the base pairing in the stem (e.g., A-T vs. G-T). A mismatch implies the disruption of the pairing in the stem. See the supplementary multiple alignment, Supplementary material online, for examples. *, trnN structure predicted by tRNAscan-Se program; **, trnN structure produced through homology modeling.
The reannotation of the mtDNAs of the ants allowed the identification of the correct position of trnN through comparisons based on both secondary structure and sequence analysis with a broad array of homologous counterparts (see Results and Discussion).
GO Analysis 1: A Pairwise Approach Using the CREx Program
Pairwise comparisons between different GOs were performed with the CREx program (Bernt et al. 2007). This software analyses genomic rearrangement pathways using common intervals (Bernt et al. 2007, 2008; Bernt and Middendorf 2011). A common interval is a set of genes that appear consecutively in the two GOs being investigated. Two common intervals A and B are said to commute if either A ⊂ B, B ⊃ A, or A ∩ B = Ø. A common interval is defined a strong common interval if it commutes with every common interval. When two strong common intervals are compared, their intersection is either empty or one is totally contained within the other. The CREx program considers only strong common intervals in its analyses. From a mathematical point of view, each GO can be viewed as a signed permutation. Thus, a pair of GOs represents two signed permutations. In other words, genes located on the opposite strands of a linear chromosome, the type of data analysed by CREx, are individually marked with either a + or − sign. The analysis is performed using a unique type of graph called a strong interval tree (SIT). In a GO1 versus GO2 comparison, the SIT is a graph where the root node is an interval containing a whole permutation (GO1 or GO2), and the terminal nodes are the single genes arranged in the other GO. The internal nodes are the strong common intervals shared by the two GOs. Internal nodes (strong common intervals) are connected by branches according to a minimal inclusion relation among the intervals (i.e., there is a branch between node c and c′ if c′ ⊂ c and there is no node c″ with c′ ⊂ c″ ⊂ c). Along the branches of the SIT, the different types of rearrangements can be analysed. Given that CREx performs its analyses on GOs obtained from linear chromosomes, the GOs determined for the various mtDNAs must be first linearized starting from a reference gene (here, usually cox1).
The CREx program models rearrangements involving transpositions, inversions, inverse transpositions as well as TDRLs (Moritz et al. 1987; Boore 2000; Dowton and Campbel 2001). CREx produces transformational pathways in which the common strong intervals, shared by the pairs of GOs, are preserved in all intermediate steps. Once the whole set of strong common intervals has been determined for a pair of GOs (e.g., GO1 and GO2) CREx heuristically identifies the most parsimonious transformational pathways that connect GO1–GO2 and vice versa.
The number of shared common intervals (NSCI) is a genomic distance that can be used to compare the level of similarity or dissimilarity of two GOs. Identical GOs exhibit the highest NSCIs whereas highly divergent GOs have low NSCIs. Pairwise NSCI-based genomic distances were calculated for the Hymenoptera GOs (see Results and Discussion). The placement of the CR was also taken into account in the GOs during the analysis.
GO Analysis 2: A Phylogenetic Approach Using the TreeREx Program
When multiple and highly variable GOs are analysed, it is necessary to apply the phylogenetic approach, implemented in the program TreeREx, for inferring the evolutionary pathways leading to the observed diversity of GOs (Bernt et al. 2008). A fully bifurcating rooted reference tree is necessary. On this tree, the pairwise scenarios computed by CREx are mapped along the branches using TreeREx software that can also infer the putative GOs at the internal nodes. The strategy used by TreeREx is described below. To simplify this description, consider a three-taxon rooted tree ([A, B], C) with A+B branching from the internal node M. To determine the rearrangement scenario M→A occurring along the branch connecting M to A: 1) first, the B→A and C→A transformational scenarios are pairwise computed using CREx; and 2) then, the M→A scenario is constructed considering only the rearrangements common to B→A and C→A. The same approach is applied to the M→B scenario that is derived from the intersection of the A→B and C→B transformations. To verify the correctness of the inferred M→A and M→B scenarios, the rearrangements determined along the M–A branch are applied, starting from M node and those inferred from the X–B branch are applied starting from B. If the two resulting GOs are identical, they represent the ancestral GO computed for node M, and the node is said to be consistent. In the TreeREx output every consistent node is green colored (see Results and Discussion). Now, consider a four-taxon rooted tree ([A,B], [C,D]) with A+B branching from the internal node M and C+D from the internal node N. In this case, the M→A scenario is formed by the intersections of the common rearrangements identified by CREx for the B→A, C→A and D→A scenarios. If one or more scenarios computed by CREx is incorrect, or if the phylogenetic tree is wrong, the intersection may be empty. Thus, an M→A scenario cannot be determined in this way. To circumvent this limit TreeREx attempts to produce nonempty intersections by ignoring one of the pairwise scenarios. Therefore, a first (M→A)a scenario is produced from the intersection of the B→A and C→A scenarios (D→A, excluded) and then a second (M→A)b scenario is made through the intersection of B→A and D→A (C→A excluded). If one of these alternative scenarios is consistent (i.e., matches the definition provided above), then the corresponding GO is assigned as an ancestral GO to the M node. If both (M→A)a and (M→A)b are consistent, the most parsimonious of the two is used to infer the ancestral GO for node M. Every node for which the GO is determined by this procedure is referred to as 1-consistent and has a yellow background in the TreeREx output. If also this strategy fails a third reconstruction approach named fallback mode is applied. Consider again the three-taxon topology; first, all of the intermediate GOs that can be obtained from A to B and vice versa are computed. Then, TreeREx selects an intermediate GO for node M that minimizes the number of rearrangements necessary to pass from the M-GO to the C-GO. In the case of the four-taxa tree, the GOs attributed to M and N nodes represent intermediates GOs with a minimum distance, one for each pair (AB or CD). The nodes determined with the fallback method are colored red in the output. TreeREx works in a bottom-up manner through the iterative analysis of triplets or quadruplets of GOs to determine the whole set of GOs for the entire tree.
In the TreeREx analysis, the consistent nodes are considered to be the most reliable, the 1-consistent nodes exhibit an intermediate level of certainty, and the fallback nodes have the highest level of uncertainty. For further details on how TreeREx works, see Bernt et al. (2008) and Duò et al. (2012).
The TreeREx settings in the various analyses were the default settings suggested at the website: -s (strong consistency method applied) -w (weak consistency method applied) -W (parsimonious weak consistency method applied) -o (get alternative base pair scenario for prime nodes; applied or not in different analyses) -m = 0 (maximum number of reversion + TDRL scenarios considered). (http://pacosy.informatik.uni-leipzig.de/185-0-TreeREx.html, last accessed December 10, 2014; Bernt et al. 2008). To summarize the meaning of these settings as they apply here, we applied a global strategy to search for alternative rearrangements scenarios. In doing so, we used algorithms of varying stringency in one analysis such that every node of the reference phylogenetic tree was defined by a GO, regardless of the certainty level for that node.
GO Analysis 3: A Phylogenetic Tree for Hymenoptera
As mentioned above, the reference tree used by TreeREx is very important for reliable reconstruction of the evolutionary scenario that produced the studied GOs (see also following paragraph). The phylogeny of Hymenoptera, a mega order encompassing more than 150,000 species (Aguiar et al. 2013), is not well established, and alternative hypotheses exist at both high and low taxonomic levels on how the different lineages should be grouped (e.g., Rasnitsyn 1988; Brothers 1999; Ronquist et al. 1999; Vilhelmsen 2001; Sharkey and Roy 2002; Sharkey, 2007; Dowton, Cameron, Austin, et al. 2009; Vilhelmsen et al. 2010; Heraty et al. 2011; Peters et al. 2011; Sharkey et al. 2012; Johnson et al. 2013; Klopfstein et al. 2013; Wei, Li, et al. 2014). One view is presented in figure 4 (see also supplementary fig. S3, Supplementary Material online). In this topology, Aculeata and Icheneumonoidea are sister taxa, as are Ceraphronoidea and Evaniodea. Conversely, Orussidae and Cephidae are taxa branching off early from the base of Hymenoptera. All ant species analysed in this article belong to the Formicoid clade, while data exist neither for the Poneroid clade nor for the subfamily Leptanillinae that is the sister taxon of the Formicoid and Poneroid clades (Moreau et al. 2006). The phylogenetic relationships within the Formicoid clade were derived from Moreau et al. (2006). For the genus Apis, we followed Lo et al. (2010).
GO Analysis 4: Alternative Topologies Tested
In the TreeREx analyses, 23 species of Hymenoptera were considered (fig. 4 and supplementary fig. S3, Supplementary Material online). Cephus cinctus (Cephidae) and Orussus occidentalis (Orussidae) belong to lineages that branch off near the root of the Hymenoptera tree, and their placement is undisputed in the analysed data set. All other taxa are members of the Apocrita, a very well supported clade containing most of the Hymenoptera (Aguiar et al. 2013). The apocritan species considered here belong to four distinct lineages. Conostigmus sp. (Megaspilidae) is a member of the superfamily Ceraphronoidea. Evania appendigaster (Evaniidae) and Pristaulacus compressus (Aulacidae) belong to the superfamily Evanioidea. Cotesia vestalis and Spathius agrili, both members of the family Braconidae, and Diadegma semiclausum (Ichneumonidae) are included in the superfamily Ichneumonoidea. All remaining species belong to the very well-defined clade Aculeata. Abispa ephippium is included in Vespidae, whereas Wallacidia oculata is a member of Mutillidae. The eight species of ants belong to Formicidae. Philanthus triangulum is contained in Crabronidae, whereas Bombus ignitus, Apis cerana, A. florea, and A. mellifera ligustica are members of the family Apidae. The phylogenetic relationships among the major lineages of Apocrita are not well established, and different views exist (see references listed earlier). This uncertainty also affects the relationships identified among Aculeata, Ceraphronoidea, Evanioidea, and Ichneumonoidea. Within the Aculeata, a critical point is the monophyly of the superfamily Vespoidea (sensu Brothers 1999), represented here by Ab. ephippium, W. oculata, and the ant species. Thus, nine different topologies depicting all of the plausible alternative phylogenetic arrangements (those obtained in previous studies) were created and used in TreeREx analyses to test their effect on the GOs reconstruction (data not shown). The different topologies did not alter the overall results. Minor changes were observed on the GOs reconstructed for a limited number of intermediate nodes. However, they did not involve the pivotal rearrangements described in detail below and may be viewed as the result of the uncertainty that inevitably affects these analyses. Thus, these alternative topologies were not further considered.
GO Analysis 5: Dating the Pivotal Nodes
To estimate the time of appearance of the key rearrangements observed in the GOs analysed in this study, we obtained estimates of geological dating from the reference book of Grimaldi and Engel (2005). Thus, our dating system was based exclusively on estimates derived from fossil records and represents a conservative approach to determine the appearance times of pivotal taxa. In the case of the clade Dytrisia + Tischeriidae + Palaephatidae (Lepidoptera), this dating time was 110–120 Ma. For the evolution of Hymenoptera GOs, the pivotal points in the reference tree were: 1) the origin of Hymenoptera Apocrita (an undisputed clade) 200–205 Ma; 2) the origin of Aculeata (another undisputed clade) 150–160 Ma; and 3) the origin of ants (Formicidae) 110–120 Ma.
We attempted neither to produce a phylogeny of Hymenoptera using the mtDNAs sequences, nor did we try to use a molecular clock due to very limited taxon sampling.
Results and Discussion
Essential Features of the mtDNAs of F. fusca and M. scabrinodis
In this study, the complete mtDNAs of the ants F. fusca (Formicidae Formicinae) and M. scabrinodis (Formicidae Myrmicinae) were sequenced and fully annotated. The Next-Gen final assembly of F. fusca was 16,673 bp long and contained 87,975 reads. The other statistics for this assembly were: base coverage = 100%; mismatch = 0%; average coverage depth = 525,995; maximum coverage depth = 9,842. The Next-Gen final assembly of M. scabrinodis was 15,310 bp long and contained 119,239 reads. The other statistics for this assembly were: base coverage = 100%; mismatch = 0%; average coverage depth = 778,344; and the maximum coverage depth was 1,324. The mtDNAs of F. fusca and M. scabrinodis contain the full set of 37 genes found in insect mtDNAs. A detailed description of these genomes, except for the analysis of GOs below, will not be presented here.
The Placement of trnN in Ant mtDNAs
As noted in the Materials and Methods section, two placements have been postulated for the trnN in the mtDNAs of the ants. The reannotation of published ant mtDNAs confirmed the presence of trnNs in the standard PanGO position in At. laevigata; and P. punctatus. The same situation was true for the newly determined mtDNAs of F. fusca and M. scabrinodis. Conversely, the reannotation of the mtDNAs of the Solenopsis species as well as L. pallens revealed trnNs in the standard PanGO placement in these ants as well. These reannotated trnNs are in the following places in the original genomes: 1) L. pallens, base 4,673–base 4,738; 2) S. geminata, base 4,657–base 4,724; 3) S. invicta, base 4,680–base 4,747; and 4) S. richteri, base 4,682–base 4,748. For all of the aforementioned trnNs, the secondary structure was inferred using the program tRNAscan-SE or through a homology modeling process (fig. 2). Secondary structures were successfully obtained for all of the trnNs. The trnN of L. pallens exhibited a mismatch in the sixth base pair of the acceptor stem, while in all of the species of Solenopsis, the TΨC stem was formed by only 2 bp and had a large TΨC loop. A mismatch was also present in the first pair of the anticodon stem in M. scabrinodis. Finally, At. laevigata exhibited a mismatch in the fourth pair of the TΨC stem. The multiple alignment containing the ant trnNs and their homologous counterparts exhibited a high level of conservation. Furthermore, the reannotated trnNs of Solenopsis and L. pallens share the GTT anticodon with all of their counterparts (see the supplementary multiple alignment of trnNs, Supplementary Material online). Mismatches were observed in several Hymenoptera taxa included in this multiple alignment that affected all of the stems.
The “alternatively placed” trnTs, originally designed for Solenopsis and L. pallens, did not return any putative orthologous counterpart in a BLAST search against Genbank. They do not align with typical insect trnNs. Finally, these supposed trnNs possess a putative ATT anticodon instead of the fully conserved GTT anticodon present in standard hexapoda trnNs. We do not see any reason to maintain these trnNs and consider them to be the result of incorrect annotation.
Globally, the reannotated trnNs of the Solenopsis species and of L. pallens 1) can be folded into the expected cloverleaf secondary structure, 2) can be aligned with the orthologous counterparts of a broad selection of Hexapoda with a high level of conservation, 3) share the GTT anticodon with all of their counterparts, and 4) exhibit the expected PanGO placement.
It is true that some mismatches are present in the stems, but this is a well-known phenomenon for the mtDNAs of insects (e.g., Salvato et al. 2008). These types of mismatch are frequently observed even in well conserved tRNAs (e.g., Negrisolo et al. 2011). The mismatches are apparently corrected through various editing processes or represent non-Watson/Crick base pairings (Negrisolo et al. 2011).
We conclude that there is overwhelming evidence that in the ant mtDNAs sequenced thus far, the true trnN exhibits the standard PanGO placement. This finding affects the GO of Solenopsis species and of L. pallens (see below).
The GOs of Ants and a Comparison to the GOs of Butterflies
The GOs of F. fusca and M. scabrinodis were compared with those known for Hymenoptera as well as for other Insecta. Compared with PanGO, the mtDNA of F. fusca exhibits a transposition of trnM on the α-strand that generated the ant1GO depicted in figure 3. The dolichoderinae ant L. pallens shares the ant1GO with F. fusca (see paragraph above). Conversely, M. scabrinodis exhibits an ant2GO shared with other myrmicinae ants, that is, At. laevigata and species of the genus Solenopsis (fig. 3). Ant2GO differs from ant1GO by a second transposition involving trnV on the β-strand. Finally, the myrmicine P. punctatus exhibits an ant3GO that differs from the ant2GO by the presence of a third transposition involving the trnD on the α-strand. Thus, the most parsimonious and simplest explanation is that the evolutionary pathway that generated the antGOs started from PanGO, and through three successive rounds of duplication random loss events, ended in ant3GO (fig. 3). Within Hymenoptera the ant1GO-ant3GO are currently known only for the ant species listed above. Very surprisingly, the ant1GO was identical to the lep1GO found in most of Lepidoptera, thus demonstrating the occurrence of convergent evolution between these distinct lineages of insects (figs.1 and 3).
Tracing of the Evolutionary Scenarios Leading to the Hymenoptera and Lepidoptera GOs
The transposition of trnM is the only rearrangement known for most of Dytrisia. A major exception is represented by Erynnis montanus (Wang et al. 2014), which exhibits a second transposition of trnS1 on the α-strand (fig. 3). Contiguous copies of single tRNAs are also known (e.g., trnS1a–trnS1b in the butterfly Coreana raphaelis; Kim et al. 2006), but they do not have a deep impact on the study of the GO evolution in Lepidoptera and are not further considered here. Pairwise comparisons with PanGO were sufficient to establish a plausible order of appearance of the rearrangements that characterized the evolution of the Lepidoptera GOs (fig. 3). Conversely, numerous and diverse GOs are present in Hymenoptera. In this latter case, a phylogenetic approach was necessary to identify a reasonable order of appearance of the rearrangements that occurred during the evolution of the hymenopteran GOs. The available hymenopteran mtDNAs represent a very sparse taxonomic sampling. Furthermore, uncertainties exist with respect to the phylogenetic relationships among various lineages. Despite these limits, a consensus phylogeny of Hymenoptera was produced, and the evolution of the different GOs was tracked along this tree using the TreeREx program that was developed for this specialized task (see Materials and Methods). Results of this analysis are presented in figure 4 (see also supplementary fig. S3, Supplementary Material online) and show that a GO identical to the ant1GO emerged very early in the evolution of Hymenoptera at the onset of the lineage Apocrita (node 4), that is, the clade encompassing the vast majority of Hymenoptera. Taking information from the fossil record into account, the appearance of the ant1GO can be placed in the early Jurassic period (200–205 Ma; fig. 4). Using a more stringent approach in terms of consistent GO reconstruction and phylogenetic certainty (node 6, Aculeata an undisputed clade), the ant1GO occurred at least 160–150 Ma in the late Jurassic period. Finally, if a very stringent approach is used (node 8, the ant clade) the ant1GO appeared at least 110–120 Ma in the Cretaceous period. In the analysed taxa, the MIQ arrangement is still present in D. semiclausum (a parasitic wasp of the superfamily Ichneumonoidea), in Pristaulacus compressus (a parasitic wasp of the superfamily Evanioidea), and in all ants sequenced to date. It is also conserved in the partial mtDNA of Enicospilus sp., again an ichneumonid wasp (Dowton Cameron, Dowavic, et al. 2009; see supplementary fig. S6, Supplementary Material online). MIQ was certainly present at the base of the Aculeata clade (node 6; wasp, ants, bees, and many more), but was successively disrupted in the highly rearranged mtDNAs of bees (Apis genus and B. ignitus) wasps Ab. ephippium (a eumenid wasp), and W. oculata (a mutillid wasp).
The levels of structural divergence among the known hymenopteran GOs and PanGO were expressed in terms of the NSCI, a measure of genomic similarity calculated by the CREx program (table 1). Ant1GO proved to be the most similar to PanGO (NSCI: 1,258 over 1,400), whereas ant2GO was the next most similar (NSCI: 1,124 over 1,440).
Table 1.
Shared Common Intervals Calculated in Pairwise Comparisons Performed with the CREx Program
| N | Taxon | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pancrustacea GO | 1,400 | |||||||||||||||||||||
| 2 | Cephus cinctus GO | 1,190 | 1,400 | ||||||||||||||||||||
| 3 | Orussus occidentalis GO | 880 | 958 | 1,400 | |||||||||||||||||||
| 4 | Diadegma semiclausum GO | 838 | 780 | 536 | 1,400 | ||||||||||||||||||
| 5 | Cotesia vestalis GO | 232 | 238 | 142 | 120 | 1,400 | |||||||||||||||||
| 6 | Spathius agrili GO | 512 | 518 | 372 | 334 | 438 | 1,400 | ||||||||||||||||
| 7 | Conostigmus sp. GO | 234 | 232 | 154 | 212 | 42 | 120 | 1,400 | |||||||||||||||
| 8 | Pristaulacus compressus GO | 892 | 882 | 952 | 574 | 128 | 358 | 148 | 1,400 | ||||||||||||||
| 9 | Evania appendigaster GO | 666 | 666 | 668 | 486 | 74 | 286 | 148 | 662 | 1,400 | |||||||||||||
| 10 | Abispa ephippium GO | 1,006 | 1,058 | 888 | 648 | 138 | 378 | 184 | 946 | 652 | 1,400 | ||||||||||||
| 11 | Wallacidia oculata GO | 1,060 | 1,000 | 790 | 726 | 264 | 518 | 236 | 766 | 614 | 818 | 1,400 | |||||||||||
| 12 | Leptomyrmex pallens ant1GO | 1,258 | 1,258 | 968 | 856 | 266 | 556 | 236 | 1,010 | 670 | 1,066 | 1,062 | 1,400 | ||||||||||
| 13 | Formica fusca ant1GO | 1,258 | 1,258 | 968 | 856 | 266 | 556 | 236 | 1,010 | 670 | 1,066 | 1,062 | 1,400 | 1,400 | |||||||||
| 14 | Atta laevigata ant2GO | 1,124 | 1,120 | 950 | 750 | 206 | 476 | 236 | 1,000 | 656 | 1,060 | 994 | 1,258 | 1,258 | 1,400 | ||||||||
| 15 | Myrmica scabrinodis ant2GO | 1,124 | 1,120 | 950 | 750 | 206 | 476 | 236 | 1,000 | 656 | 1,060 | 994 | 1,258 | 1,258 | 1,400 | 1,400 | |||||||
| 16 | Pristomyrmex punctatus ant3GO | 1,060 | 1,056 | 892 | 700 | 210 | 538 | 226 | 940 | 608 | 998 | 934 | 1,190 | 1,190 | 1,328 | 1,328 | 1,400 | ||||||
| 17 | Solenopsis genus ant2GO | 1,124 | 1,120 | 950 | 750 | 206 | 476 | 236 | 1,000 | 656 | 1,060 | 994 | 1,258 | 1,258 | 1,400 | 1,400 | 1,328 | 1,400 | |||||
| 18 | Philanthus triangulum GO | 998 | 938 | 698 | 774 | 232 | 534 | 192 | 668 | 528 | 714 | 1,060 | 998 | 998 | 874 | 874 | 818 | 874 | 1,400 | ||||
| 19 | Bombus ignitus GO | 380 | 442 | 260 | 344 | 100 | 218 | 156 | 244 | 204 | 328 | 356 | 382 | 382 | 322 | 322 | 344 | 322 | 492 | 1,400 | |||
| 20 | Apis florea GO | 360 | 358 | 244 | 320 | 88 | 202 | 178 | 250 | 170 | 310 | 300 | 364 | 364 | 314 | 314 | 336 | 314 | 396 | 520 | 1,400 | ||
| 21 | Apis cerana GO | 358 | 358 | 244 | 318 | 88 | 202 | 178 | 248 | 170 | 308 | 300 | 362 | 362 | 312 | 312 | 334 | 312 | 396 | 520 | 1,328 | 1,400 | |
| 22 | Apis mellifera ligustica GO | 360 | 358 | 244 | 320 | 88 | 202 | 178 | 250 | 170 | 310 | 300 | 364 | 364 | 314 | 314 | 336 | 314 | 396 | 520 | 1,400 | 1,328 | 1,400 |
Note.—N, number identifying the taxon in the pairwise comparisons. A graphical representation of the different GOs is provided in figure 4.
The evolutionary pathway postulated by TreeREx for the most basal nodes of the Hymenoptera tree (fig. 4) implied a greater number of rearrangements than the most parsimonious scenario to produce the ant1GO arrangement presented in figure 2. In the TreeREx analysis, trnM was initially transposed upstream to the CR (node 2), thus producing a GO identical to that observed in C. cinctus. The successive step generated an arrangement identical to that observed in O. occidentalis (node 3), and only later did the ant1GO emerge (node 4). However, this part of the TreeREx reconstruction exhibited a high level of uncertainty, such that the reconstructions generated by the software are no better than the alternative scenarios (see Materials and Methods and supplementary fig. S3, Supplementary Material online). Therefore, we can postulate an alternative scenario where the ant1GO appeared at node 2 and remained unchanged in node 3. Both C. cinctus and O. occidentalis GOs can be transformed in ant1GO by a single event of transposition of trnM. Thus, the TreeREx Scenario and our alternative scenario are equally parsimonious in term of rearrangements. Indeed, both require three events of transposition to move from PanGO (node 1) to the ant1GO (node 4), and both fully describe the observed GOs.
The TreeREx scenario is not supported by the NSCI values that show that both C. cinctus (NSCI: 1,190 over 1,440) and O. occidentalis (NSCI: 880 over1,400) GOs are structurally different from PanGO more than ant1GO and even ant2GO. Thus, the TreeREx scenario favored an implausible pathway, implying an early appearance of more apomorphic GOs and a successive reversion to the most plesiomorphic state (see also below). Conversely, our alternative scenario was supported by the NSCI scores. Thus, the available evidence suggests the possibility that the ant1GO emerged near to or even at the root of the Hymenoptera tree. This hypothesis needs to tested through a dense sampling of mtDNAs obtained from lineages branching off near to or from the base of the Hymenoptera root.
The mtDNAs of the Apocrita Scelionidae Idris sp. and Ceratobaeus sp. and that of the sawfly Al. luctifer became available too late to be fully analysed in this study (see Materials and Methods). However, we were able to compare their GOs with our results.
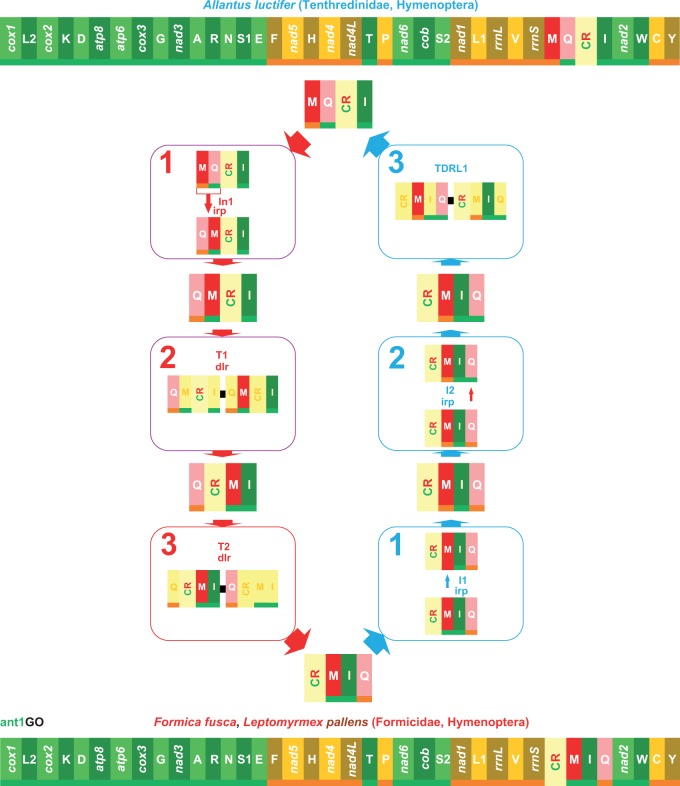
Many tRNAs (10–11) of both scelionid mtDNAs are involved in different types of rearrangements, that is, inversions, transpositions, and inverted transpositions, with respect to PanGO (supplementary fig. S4, Supplementary Material online). The trnM is located on the β-strand upstream to the CR. The Scelionids mtDNAs have highly rearranged GOs, as demonstrated by their NSCI values (Idris sp., NSCI: 562 over 1,400; Ceratobaeus sp., NSCI: 518 over 1,400). Thus, they were not pivotal to understanding the first steps of GO evolution in Hymenoptera, and they were considered no further. More interesting was the GO observed in the mtDNA of Al. luctifer. This species belongs to Tenthredinidae, a lineage branching-off close to the base of Hymenoptera tree. Al. luctifer is a sister group of the Hymenoptera pictured in figure 4 (see Phylogenetic references in the Materials and Methods section). The Al. luctifer GO was similar to PanGO (see supplementary fig. S4, Supplementary Material online) and exhibited an inverted transposition of trnM+trnQ. The high similarity was corroborated by the NSCI score (1,192 over 1,440), which was slightly lower than the 1,258 value calculated for ant1GO (table 1). The Al. luctifer GO was compared with ant1GO using CREx (fig. 5). The transformational pathway going from ant1GO to the Al. luctifer GO required two sequential inversion events involving trnM (I1) and trnQ (I2) and a TDRL move (blue pathway in fig. 5). The alternative pathway (the red pathway in fig. 5) required the simultaneous inversion of trnM and trnQ (I1) followed by two sequential transpositions involving first trnM (T1) and then trnQ (T2). As noted in the introduction, an inversion, a transposition or an inverted transposition observed in a pairwise comparison between two GOs, are reversible rearrangements that do not establish the order of the changes (Perseke et al. 2008). Conversely, a TDRL move is an “irreversible” change that allows the order of the rearrangements to be determined. In other words, the ant1GO is plesiomorphic with respect to Al. luctifer GO, which is an important finding that corroborates our results on the antiquity of ant1GO. If the Al. luctifer GO is the standard GO for Tenthredinidae, this finding will further push back the appearance of the ant1GO very near to or even at the base of the Hymenoptera clade. This supposition requires further corroboration. Regardless of the uncertainties of the early steps of GO evolution, what is evident from the data presented above is that the ant1GO appeared very early in the cladogenetic process leading to the diversification Hymenoptera.
Fig. 5.—
Transformational pathways between ant1GO and Al. luctifer GO. Numbered in blue are the steps of the transformation pathway leading from ant1GO to Al. luctifer GO, whereas numbered in red are the steps of the opposite pathway. T1–T3, transposition events; dlr, duplication random loss, mechanism producing the observed rearrangement; I1-I2, inversion events; irp, intramitochondrial recombination process generating the observed re-arrangement. The genomic and genetic nomenclature, as well as the color scheme, are the same as in figure 1. The genes that changed position relative to PanGO are shown with a pink/red background.
Identification of New Homoplastic Rearrangements among Insect GOs
Partial or complete mtDNAs sequenced for other insects exhibiting GOs different from PanGO were analysed to check for the presence of undetected homoplasies. Four further homoplastic rearrangements were identified (fig. 4 and supplementary figs. S6 and S7, Supplementary Material online).
Transposition of trnP generating the PT versus TP rearrangement occurred independently three times in the mtDNAs of the hymenopteran D. semiclausum, Ph. triangulum, and B. ignitus (fig. 4; Cha et al. 2007; Wei et al. 2009; Kaltenpoth et al. 2012). Transposition of trnC generating the CW versus WC rearrangement found in many Neuroptera (see above) occurred independently in the braconid wasp S. agrii (fig. 4) and in the Hemiptera Delphacidae (Zhang et al. 2013, 2014; supplementary fig. S6, Supplementary Material online). However, Neuroptera, S. agrii, and Delphacidae all have distinct different GOs. Analogously, the transposition of trnR producing the RA versus AR rearrangement that is characteristic of the mosquitos of the Culicidae family arose independently twice in the beetles Peploptera acromialis (Chrysomelidae) and Naupactus xanthographus (Curculionidae; Song et al. 2010; Timmermans et al. 2010). In this latter case, only partial mtDNAs exist for both species; thus, it was not possible to fully compare their GOs to that of mosquitos (supplementary fig. S6, Supplementary Material online). Finally, the transposition of Y on the β-strand created the YC versus CY rearrangement in Ichneumonid D. semiclausum and Enicospilus sp. (Hymenoptera) (Dowton, Cameron, Dowavic, et al. 2009; Wei et al. 2009) and in whiteflies of the Aleyrodidae family (Hemiptera; Thao et al. 2004). Even in this case, comparison of the complete mtDNAs reveals very different GOs for Enicospilus sp., D. semiclausum, and whiteflies (supplementary fig. S7, Supplementary Material online).
Implications for the Study of mtDNA GO Evolution
The analyses presented here shed new light on the evolution of mtDNA GOs in the Hymenoptera. Our study shows that the ant mtDNAs exhibit less divergent rearrangements with respect to PanGO and that the ant1GO appeared at the onset (maybe before) of the Apocrita, that is, the largest phyletic lineage of Hymenoptera, and represents the plesiomorphic condition for this clade.
Until recently, the analysis of GOs was performed through pairwise comparisons due to the absence of suitable bioinformatic tools allowing for more sophisticated investigations. This approach led to an overestimation of the number of rearrangements detected. This point is exemplified by the behavior of trnM, which seemed to have independently generated the MIQ arrangement three times (Dowton, Cameron, Dowavic, et al. 2009), while the reconstruction provided here suggests that this event occurred only once. Our study underscores the necessity of a phylogenetic approach coupled with new sophisticated algorithms, like those implemented in TreeREx, to properly reconstruct the evolution of GOs in mtDNAs that are created by complex patterns of rearrangements.
Fully homoplastic identical GOs had previously been identified only twice in vertebrate taxa (supplementary fig. S5, Supplementary Material online). In birds, two GOs (named here bird1GO, bird2GO) exist, and the bird2GO has appeared repeatedly in separate lineages (Mindell et al. 1998). This interpretation has been questioned (Boore and Brown 1998; Boore 2006) because the arrangements of 37 genes are identical when the CR is not considered. However, the CR encodes the origin of replication for the mtDNA, thus in our view, its placement cannot be ignored. The amphisbaenian reptile Rhineura floridana has a GO identical to the bird1GO, the most widespread GO in birds (Macey et al. 2004). Vertebrata exhibit a limited number of GOs (Boore 1999); thus, convergence could be the result of constraints of limited gene mobility in this peculiar clade.
This study demonstrates that identical GOs can occur even in a clade such as the Insecta that is GO-rich and dynamic in term of rearrangements (Cameron 2014). Our results favor the view that convergent evolution in GOs is a general phenomenon of the animal mtDNA and is not restricted to a particular lineage. The ant1GO/lep1GO represents the oldest documented event of this type of convergence for animal taxa. Both the ant1GO and the lep1GO appeared very early in the cladogenesis (Grimaldi and Engel 2005) of Hymenoptera and Lepidoptera and emerged, respectively, in the Jurassic and Cretaceous periods.
The analyses presented here strongly corroborate and extend earlier findings that gene movements in mtDNAs occur preferentially along specific pathways. This characteristic reduces the number of possible arrangements that are likely to be observed and drastically increases the probability of convergent evolution in GOs. Convergence can be limited to the sharing of local homoplastic rearrangements or involve the full rearrangement of a GO. Based on earlier work, tRNAs are likely the most mobile genes (Moritz et al. 1987; Cameron 2014). Finally, the genes most prone to homoplastic rearrangements are contiguous in the genome or located around the origin of replication of the mtDNA (Boore and Brown 1998; Boore 2006; this article).
Our results reveal that local homoplastic rearrangements, and even completely homoplastic GOs, are more common than previously thought. Sequencing of additional mtDNAs, especially through new genomic technologies, is progressing at a very fast pace. It is plausible that the number of observed GOs will increase rapidly, as will the identification of local or global homoplastic rearrangements. However, most of the partly or fully homoplastic GOs will remain as important molecular signatures, provided that they are considered in a phylogenetic and systematic context and that comparisons are made on complete GOs rather than focusing on local arrangements. Even if the Dytrisia + Tischeriidae + Palaephatidae group shares an identical GO with some ants, having the lep1GO is a synapomorphic molecular signature that characterizes this clade and distinguishes its members from the other Lepidoptera taxa. Analogously, the GO found in Culicidae is an apomorphy within Diptera (Beard et al. 1993; Mitchell et al. 1993; Behura et al. 2011) even if these mosquitos share the RA versus AR rearrangement with some beetles (Timmermans et al. 2010). The same reasoning can be applied to the other local homoplastic rearrangements described above.
Therefore, regardless of their level of homoplasy, GOs have a lot to offer. However, to paraphrase the suggestion provided by Boore and Brown (1998), GOs must be wisely evaluated.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank two anonymous reviewers and the Associate Editor Prof. Dr John Archibald for constructive criticism on an earlier version of this work that were very useful in preparing the final manuscript. This research was supported by a grant (PRIN 2009: 2009NWXMXX_004) awarded to Enrico Negrisolo by the Italian Ministry of University and Scientific Research (MIUR) and by a grant (ex60% 2012) assigned to Enrico Negrisolo by the University of Padua.
Literature Cited
- Aguiar AP, et al. Order Hymenoptera. Zootaxa. 2013;3703:051–062. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2:103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Behura SK, et al. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem Mol Biol. 2011;41:770–777. doi: 10.1016/j.ibmb.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman M, Austin CM, Miller AD. Characterisation of the complete mitochondrial genome and 13 microsatellite loci through next-generation sequencing for the New Caledonian spider-ant Leptomyrmex pallens. Mol Biol Rep. 2014;41:1179–1187. doi: 10.1007/s11033-013-2657-5. [DOI] [PubMed] [Google Scholar]
- Bernt M, et al. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
- Bernt M, Merkle D, Middendorf M. An algorithm for inferring mitogenome rearrangements in a phylogenetic tree. In: Nelson CE, Vialette S, editors. Comparative genomics, RECOMB-CG 2008, LNB 5267. Berlin: Springer; 2008. pp. 143–157. [Google Scholar]
- Bernt M, Middendorf M. A method for computing an inventory of metazoan mitochondrial gene order rearrangements. BMC Bioinformatics. 2011;12(Suppl 9):S6. doi: 10.1186/1471-2105-12-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 2000. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In: Sankoff D, Nadeau JH, editors. Comparative genomics. Dordrecht: Kluwer Academic Publishers. p. 133147.
- Boore JL. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol. 2006;21:439–446. doi: 10.1016/j.tree.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM. Deducing arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- Boore JL, Lavrov DV, Brown WM. Gene translocation links insects and crustaceans. Nature. 1998;392:667–668. doi: 10.1038/33577. [DOI] [PubMed] [Google Scholar]
- Brothers DJ. Phylogeny and evolution of wasps, ants and bees (Hymenoptera, Chrysidoidea, Vespoidea and Apoidea) Zool Scr. 1999;28:233–250. [Google Scholar]
- Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera) BMC Genomics. 2011;12:394. doi: 10.1186/1471-2164-12-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YQ, Ma C, Chen JY, Yang DR. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 2012;13:276. doi: 10.1186/1471-2164-13-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapelli A, et al. The mitochondrial genome of the entomophagous endoparasite Xenos vesparum (Insecta: Strepsiptera) Gene. 2006;376:248–259. doi: 10.1016/j.gene.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Cha SY, et al. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae) Gene. 2007;392:206–220. doi: 10.1016/j.gene.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Dowton M. Simultaneous molecular and morphological analysis of braconid relationships (Insecta: Hymenoptera: Braconidae) indicates independent mt-tRNA gene inversions within a single wasp family. J Mol Evol. 2002;54:210–226. doi: 10.1007/s00239-001-0003-3. [DOI] [PubMed] [Google Scholar]
- Dowton M, Austin AD. Evolutionary dynamics of a mitochondrial rearrangement ‘‘hot spot’’ in the Hymenoptera. Mol Biol Evol. 1999;16:98–309. doi: 10.1093/oxfordjournals.molbev.a026111. [DOI] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Austin AD, Whiting MF. Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera—a lineage with both rapidly and slowly evolving mitochondrial genomes. Mol Phylogenet Evol. 2009;52:512–519. doi: 10.1016/j.ympev.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting M. Characterization of 67 mitochondrial tRNA gene rearrangements in the hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol Biol Evol. 2009;26:1607–1617. doi: 10.1093/molbev/msp072. [DOI] [PubMed] [Google Scholar]
- Dowton M, Campbell NJH. Intramitochondrial recombination—is it why some mitochondrial genes sleep around? Trends Ecol Evol. 2001;16:269–271. doi: 10.1016/s0169-5347(01)02182-6. [DOI] [PubMed] [Google Scholar]
- Dowton M, Castro LR, Austin AD. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome “morphology”. Invertebr Syst. 2002;16:345–356. [Google Scholar]
- Dowton M, Castro LR, Campbell SL, Bargon SD, Austin AD. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J Mol Evol. 2003;56:517–526. doi: 10.1007/s00239-002-2420-3. [DOI] [PubMed] [Google Scholar]
- Duò A, Bruggmann R, Zoller S, Bernt M, Grünig CR. Mitochondrial genome evolution in species belonging to the Phialocephala fortinii s.l.—Acephala applanata species complex. BMC Genomics. 2012;13:166. doi: 10.1186/1471-2164-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley IM, Walker JE. Initiation codons in mammalian mitochondria: differences in genetic code in the organelle. Biochemistry. 1987;26:8247–8251. doi: 10.1021/bi00399a034. [DOI] [PubMed] [Google Scholar]
- Feijao PC, Neiva LS, Azeredo-Espin AML, Lessinger AC. AMiGA: the Arthropodan mitochondrial genomes accessible database. Bioinformatics. 2006;22:902–903. doi: 10.1093/bioinformatics/btl021. [DOI] [PubMed] [Google Scholar]
- Flook PK, Rowell CH, Gellissen G. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J Mol Evol. 1995a;41:928–941. doi: 10.1007/BF00173173. [DOI] [PubMed] [Google Scholar]
- Flook PK, Rowell H, Gellissen G. Homoplastic rearrangements of insect mitochondrial tRNA genes. Naturwissenschaften. 1995b;82:336–337. [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. New York: Cambridge University Press; 2005. [Google Scholar]
- Gotzek D, Clarke J, Shoemaker D. Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae) BMC Evol Biol. 2010;10:300. doi: 10.1186/1471-2148-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullan PS, Cranston PJ. 2014 The insects. An outline of entomology. 5th ed. Chichester: Wiley & Sons. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa E, Kobayashi K, Yagi N, Tsuji K. Complete mitochondrial genomes of normal and cheater morphs in the parthenogenetic ant Pristomyrmex punctatus (Hymenoptera: Formicidae) Myrmecol News. 2011;15:85–90. [Google Scholar]
- Heraty J, et al. Evolution of the hymenopteran megaradiation. Mol Phylogenet Evol. 2011;60:73–88. doi: 10.1016/j.ympev.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Jiang H, Barker SC, Shao R. Substantial variation in the extent of mitochondrial genome fragmentation among blood-sucking Lice of Mammals. Genome Biol Evol. 2013;5:1298–1308. doi: 10.1093/gbe/evt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, et al. Phylogenomics resolves evolutionary relationships among ants, bees, and wasps. Curr Biol. 2013;23:2058–62. doi: 10.1016/j.cub.2013.08.050. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, et al. Accelerated evolution of mitochondrial but not nuclear genomes of hymenoptera: new evidence from crabronid wasps. PLoS One. 2012;7:e32826. doi: 10.1371/journal.pone.0032826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, et al. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae) Insect Mol Biol. 2006;15:217–225. doi: 10.1111/j.1365-2583.2006.00630.x. [DOI] [PubMed] [Google Scholar]
- Klopfstein S, Vilhelmsen L, Heraty JM, Sharkey M, Ronquist F. The hymenopteran tree of life: evidence from protein-coding genes and objectively aligned ribosomal data. PLoS One. 2013;8:e69344. doi: 10.1371/journal.pone.0069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoto N, Yukuhiro K, Tomita S. Novel gene rearrangements in the mitochondrial genome of a webspinner, Aposthonia japonica (Insecta: Embioptera) Genome. 2012;55:222–233. doi: 10.1139/g2012-007. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Nishida M. Variations in mitochondrial tRNA gene organization of reptiles as phylogenetic markers. Mol Biol Evol. 1995;12:759–772. doi: 10.1093/oxfordjournals.molbev.a040254. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 2000;17:813–824. doi: 10.1093/oxfordjournals.molbev.a026360. [DOI] [PubMed] [Google Scholar]
- Lo N, Gloag RS, Anderson DL, Oldroyd BP. A molecular phylogeny of the genus Apis suggests that the Giant Honey Bee of the Philippines, A. breviligula Maa, and the Plains Honey Bee of southern India, A. indica Fabricius, are valid species. Syst Entomol. 2010;35:226–233. [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genome sequences. Mol Phylogenet Evol. 2004;33:22–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Mao M, Dowton M. Complete mitochondrial genomes of Ceratobaeus sp. and Idris sp. (Hymenoptera: Scelionidae): shared gene rearrangements as potential phylogenetic markers at the tribal level. Mol Bio Rep. 2014;41:6419–6427. doi: 10.1007/s11033-014-3522-x. [DOI] [PubMed] [Google Scholar]
- Milne I, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- Mindell D, Sorenson MD, Dimcheff DE. Multiple independent origins of mitochondrial gene order in birds. Proc Natl Acad Sci U S A. 1998;95:10693–10697. doi: 10.1073/pnas.95.18.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SE, Cockburn AF, Seawright JA. The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome. 1993;36:1058–1073. doi: 10.1139/g93-141. [DOI] [PubMed] [Google Scholar]
- Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the Ants: diversification in the age of Angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Syst. 1987;18:269–292. [Google Scholar]
- Negrisolo E, Babbucci M, Patarnello T. The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC Genomics. 2011;12:221. doi: 10.1186/1471-2164-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrisolo E, Minelli A, Valle G. Extensive gene order rearrangement in the mitochondrial genome of the centipede Scutigera coleoptrata. J Mol Evol. 2004;58:413–423. doi: 10.1007/s00239-003-2563-x. [DOI] [PubMed] [Google Scholar]
- Pääbo S, Thomas WK, Whitfield KM, Kumazawa Y, Wilson AC. Rearrangements of mitochondrial transfer RNA genes in marsupials. J Mol Evol. 1991;33:426–430. doi: 10.1007/BF02103134. [DOI] [PubMed] [Google Scholar]
- Perseke M, et al. Evolution of mitochondrial gene orders in echinoderms. Mol Phylogenet Evol. 2008;47:855–864. doi: 10.1016/j.ympev.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Peters RS, et al. The taming of an impossible child: a standardized all-in approach to the phylogeny of Hymenoptera using public database sequences. BMC Biol. 2011;9:55. doi: 10.1186/1741-7007-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RS, et al. The evolutionary history of holometabolous insects inferred from transcriptome-based phylogeny and comprehensive morphological data. BMC Evol Biol. 2014;14:52. doi: 10.1186/1471-2148-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnitsyn AP. An outline of evolution of the hymenopterous insects. Orient Insects. 1988;22:115–145. [Google Scholar]
- Rodovalho C M, Lyra ML, Ferro M, Bacci M., Jr The mitochondrial genome of the leaf-cutter ant Atta laevigata: a mitogenome with a large number of intergenic spacers. PLoS One. 2014;9:e97117. doi: 10.1371/journal.pone.0097117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Rasnitsyn AP, Roy A, Eriksson K, Lindgren M. Phylogeny of the Hymenoptera: a cladistic reanalysis of Rasnitsyn’s (1988) data. Zool Scr. 1999;28:13–50. [Google Scholar]
- Saito S, Tamura K, Aotsuka T. Replication origin of mitochondrial DNA in Insects. Genetics. 2005;171:1695–1705. doi: 10.1534/genetics.105.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato P, Simonato M, Battisti A, Negrisolo E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae) BMC Genomics. 2008;9:331. doi: 10.1186/1471-2164-9-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two Novel gene boundaries and an extraordinary arrangement of rRNA Genes. Mol Biol Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- Shao R, Kirkness EF, Barker SC. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009;19:904–912. doi: 10.1101/gr.083188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey MJ. Phylogeny and classification of Hymenoptera. Zootaxa. 2007;1668:521–548. [Google Scholar]
- Sharkey MJ, et al. Phylogenetic relationships among superfamilies of Hymenoptera. Cladistics. 2012;28:80–112. doi: 10.1111/j.1096-0031.2011.00366.x. [DOI] [PubMed] [Google Scholar]
- Sharkey MJ, Roy A. Phylogeny of the Hymenoptera: a reanalysis of the Ronquist et al. (1999) reanalysis, emphasizing wing venation and apocritan relationships. Zool Scr. 2002;31:57–66. [Google Scholar]
- Smith AE, Marcker KA. N-formylmethionyl transfer RNA in mitochondria from yeast and rat liver. J Mol Biol. 1968;38:241–243. doi: 10.1016/0022-2836(68)90409-9. [DOI] [PubMed] [Google Scholar]
- Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol. 2010;35:429–448. [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thao ML, Baumann L, Baumann P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha) BMC Evol Biol. 2004;4:25. doi: 10.1186/1471-2148-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJ, et al. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic Acids Res. 2010;38:E197. doi: 10.1093/nar/gkq807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJTN, Lees DC, Simonsen TJ. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 2014;79:169–178. doi: 10.1016/j.ympev.2014.05.031. [DOI] [PubMed] [Google Scholar]
- Timmermans MJTN, Vogler AP. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea) Mol Phylogenet Evol. 2012;63:299–304. doi: 10.1016/j.ympev.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Trautwein MD, Wiegmann BM, Beutel R, Kjer KM, Yeates DK. Advances in Insect phylogeny at the dawn of the postgenomic era. Annu Rev Entomol. 2012;57:449–468. doi: 10.1146/annurev-ento-120710-100538. [DOI] [PubMed] [Google Scholar]
- van Nieukerken EJ, et al. Order Lepidoptera Linnaeus, 1758. In: Zhang ZQ, editor. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Magnolia Press: Zootaxa; 2011. 3148:212–221. [DOI] [PubMed] [Google Scholar]
- Vilhelmsen L. Phylogeny and classification of the extant basal lineages of the Hymenoptera (Insecta) Zool J Linn Soc. 2001;131:393–442. [Google Scholar]
- Vilhelmsen L, Miko I, Krogmann L. Beyond the wasp-waist: structural diversity and phylogenetic significance of the mesosoma in apocritan wasps (Insecta: Hymenoptera) Zool J Linn Soc. 2010;159:22–194. [Google Scholar]
- Wan X, Kim MI, Kim MJ, Kim I. Complete mitochondrial genome of the free-living earwig, Challia fletcheri (Dermaptera: Pygidicranidae) and phylogeny of Polyneoptera. PLoS One. 2012;7:e42056. doi: 10.1371/journal.pone.0042056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AR, et al. Complete mitochondrial genome of the dwarf honeybee, Apis florea (Hymenoptera: Apidae) Mitochondrial DNA. 2013;24: 208–210. doi: 10.3109/19401736.2012.744986. [DOI] [PubMed] [Google Scholar]
- Wang AR, Jeong HC, Han YS, Kim I. The complete mitochondrial genome of the mountainous duskywing, Erynnis montanus (Lepidoptera: Hesperiidae): a new gene arrangement in Lepidoptera. Mitochondrial DNA. 2014;25:93–94. doi: 10.3109/19401736.2013.784752. [DOI] [PubMed] [Google Scholar]
- Wei DD, et al. The Multipartite Mitochondrial Genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS One. 2012;7:e33973. doi: 10.1371/journal.pone.0033973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Niu FF, Du BZ. Rearrangement of trnQ-trnM in the mitochondrial genome of Allantus luctifer (Smith) (Hymenoptera: Tenthredinidae) Mitochondrial DNA. 2014 doi: 10.3109/19401736.2014.919475. Advance Access published May 27, 2014, doi:10.3109/19401736.2014.919475. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Li Q, van Achterberg K, Chen XX. Two mitochondrial genomes from the families Bethylidae and Mutillidae: independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol Phylogenet Evol. 2014;77:1–10. doi: 10.1016/j.ympev.2014.03.023. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Shi M, He JH, Sharkey MJ, Chen XX. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome. 2009;52:308–319. doi: 10.1139/g09-008. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- Zhang KJ, et al. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 2013;14:417. doi: 10.1186/1471-2164-14-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KJ, et al. The complete mitochondrial genome sequence of Sogatella furcifera (Horváth) and a comparative mitogenomic analysis of three predominant rice planthoppers. Gene. 2014;533:100–109. doi: 10.1016/j.gene.2013.09.117. [DOI] [PubMed] [Google Scholar]
- Zhao J, Li H, Winterton SL, Liu Z. Ancestral gene organization in the mitochondrial genome of Thyridosmylus langii (McLachlan, 1870) (Neuroptera: Osmylidae) and Implications for Lacewing Evolution. PLoS One. 2013;8:e62943. doi: 10.1371/journal.pone.0062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.