Abstract
Ciliopathies encompass a group of genetic disorders characterized by defects in the formation, maintenance, or function of cilia. Retinitis pigmentosa (RP) is frequently one of the clinical features presented in diverse ciliopathies. RP is a heterogeneous group of inherited retinal disorders, characterized by the death of photoreceptors and affecting more than one million individuals worldwide. The retinitis pigmentosa GTPase regulator (RPGR) gene is mutated in up to 20% of all RP patients. RPGR protein has different interacting partners to function in ciliary protein trafficking. In this review, we specifically focus on RPGR and its two interacting proteins: RPGRIP1 and RPGRIP1L. We summarize the function of the three proteins and highlight recent studies that provide insight into the cellular function of those proteins.
1. Introduction: Cilia and Photoreceptor
1.1. Cilia Architecture
Cilia are tiny, hair-like structures protruding from the cell surface. They are highly conserved organelles and serve a variety of sensory functions in both unicellular and multicellular organisms. Historically, cilia have been classified into two categories: motile and nonmotile cilia (primary cilia), depending on the arrangement of the microtubule triplets, termed the axoneme. The axoneme of motile cilia is built in the classical 9+2 arrangement, where nine outer microtubule doublets surround a central pair of singlet microtubules. The axoneme of primary cilia lacks the central pairs of microtubules and is arrayed in a 9+0 configuration [1]. Primary cilia are immotile and can sense extracellular physical and biochemical signals, acting as a coordination centre of multiple signal transduction pathways [2]. Some epithelial cell surfaces contain a large number of motile cilia, which beat cooperatively to generate fluid movement. For instance, cells lining in the epididymis, the oviducts, the respiratory tract, and ependymal surfaces of the brain have large clusters of motile cilia, which beat in coordinated waves and perform a broad range of functions. The central microtubules within the motile cilia help in the bending motion. Flagella are found on single-celled eukaryotes and sperm cells and are primarily involved in cell locomotion. Prokaryotic flagella are structurally similar to eukaryotic flagella, although there are distinctions made according to the function and the length. Flagella also have a 9+2 axoneme arrangement which is similar to that of motile cilia.
1.2. Protein Transport in Cilia
All protein synthesizing machineries are restricted to the cytoplasm, so continued elongation of the cilium requires the selective import and transport of ciliary proteins by intraflagellar transport (IFT), which mediates bidirectional movement of multiprotein loaded particles along the axoneme. IFT was initially discovered in flagella of Chlamydomonas reinhardtii, in which anterograde and retrograde movements of particles were observed through differential interference contrast microscopy [3]. IFT polypeptides are found in all ciliated organisms and are highly conserved in the evolution, ranging from C. elegans to primates [4, 5]. The transport of IFT particles is powered by motors that include the heterotrimeric kinesin for anterograde movement [4, 6] and the cytoplasmic dynein for retrograde movement [7, 8]. Based on directionality of IFT's movement along the axoneme, IFT particles are organized into two complexes: IFT complex A and IFT complex B. In vertebrate at least six proteins (IFT43, IFTA-1, IFT122, IFT139, IFT140, and IFT144) and ten different proteins (IFT20, 27, 46, 52, 57, 74, 80, 81, 88, and 172) are in complex A and complex B, respectively [9]. IFT complex A is responsible for retrograde transport and complex B organizes anterograde transport towards the microtubule plus-ends of the ciliary tip. The bidirectional movement of IFT particles is continuous without reversing. Although the anterograde movement is slower than the retrograde movement, there is no accumulation of IFT particle in the distal tip of the cilium, suggesting cargo loading and release, IFT particle turn-around, and motor exchange are well regulated at the base and tip of cilia [10]. Most of these cargoes are derived from the Golgi body vesicles, which could explain why cilia usually form apically to the trans-Golgi network. IFT is essential for assembling all eukaryotic cilia and flagella, and defects in IFT can cause a variety of diseases and abnormal developments.
1.3. Photoreceptors
Photoreceptors are the photosensitive cells of the retina responsible for converting incident light into electric signals that are transmitted to the brain via the optic nerves and are interpreted by the nervous system. Rods and cones are the two types of photoreceptors, classified according to their shape and function. The human retina contains over 120 million rods and 6 million cones. Rods and cones are unevenly distributed across the retina. Some organisms, such as mouse, also have a rod dominant retina; others, such as zebrafish and squirrel, have a cone dominant retina. In humans, the central region of the retina is the macula. The central part of the macula is referred to as the “fovea,” which only contains cones and is the region showing highest visual acuity [11]. Rods are responsible for vision at low light levels (scotopic or night vision) and are very sensitive even under dim conditions. Cones are responsible for colour or day vision (photopic vision). They are much less sensitive than rods and generate signals at higher levels of light.
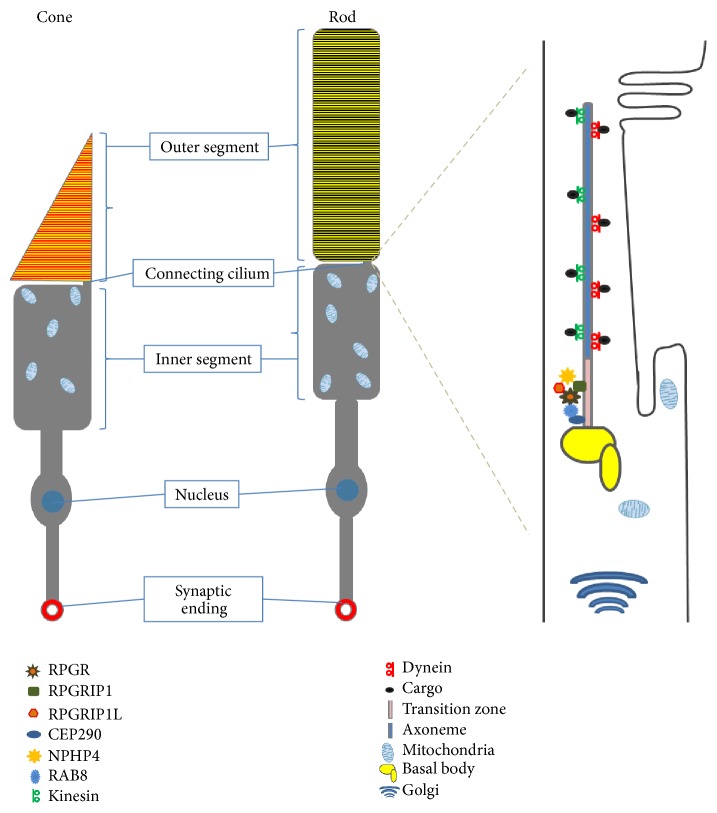
Photoreceptors consist of four distinct components: a synaptic terminal, an outer segment (OS), an inner segment (IS), and a connecting cilium (CC) that connects the IS and OS (Figure 1). The OS is a highly modified primary cilium that contains numerous light sensitive stacks. These stacked lamellae are actual sites of photo transduction. The CC is a structural homologue to the transition zone of cilia [12]. Structurally rods and cones share a similar architecture. However, there is a difference in the way their outer segments are built. In rods, there are large numbers of membrane discs stacked on top of each other unconnected to the ciliary membrane, while the discs in cones are invaginations of the plasma membrane. The inner segment contains cellular organelle such as mitochondria, endoplasmic reticulum, and Golgi body so that they do not interfere with the outer segment biochemical reactions. The visual pigment in both rods and cones consists of opsin proteins. Rods are able to operate in dim light using only one type of opsin, rhodopsin, whereas cones contain several types of opsins thereby detecting different colours. The human eye contains three different types of cones for perception of different colours: blue cones (short wavelength or S cones), green cones (middle wavelength or M cones), and red cones (long wavelength or L cones) which allow us to respond to different wavelengths of light [13]. Many animals including birds, reptiles, and fish have four different types of cones. Cones have a longer lifespan than rods and do not undergo circadian phagocytosis by RPE cells [14]. The molecular processes like IFT or vesicle trafficking observed in primary cilia are also conserved in photoreceptor outer segments (Figure 1).
Figure 1.
The photoreceptor primary cilia. The distinct compartments of both rod and cone cells (left side). Protein trafficking in photoreceptor connecting cilium through intraflagellar transport (IFT) is carried out by motor proteins, kinesin and dynein (right side). CEP290, centrosomal protein 290 kDa; NPHP4, nephronophthisis 4; RPGR, retinitis pigmentosa GTPase regulator; RPGRIP1, RPGR interacting protein 1; RPGRIP1L, RPGR interacting protein 1-like (right side).
2. Ciliopathies
The cilium proteome contains hundreds of different proteins involved in cilia protein trafficking, structure, and signal transductions [15, 16]. Deficiency of one of these proteins may be enough to produce cilia defects, giving rise to a broad spectrum of genetic disorders termed ciliopathies. Wide ranges of extracellular signals are sensed by the cilia and then transduced into decisions required for proliferation, differentiation, polarity, development, and tissue maintenance. A broad range of signals like photosensation, thermosensation, mechanosensation, hormone sensation, and olfactory sensation are received and propagated by specific ciliary receptors [17]. So, ciliary dysfunction can manifest as a variety of clinically overlapping features including primarily retinal degeneration, kidney diseases, and brain anomalies [18].
Phenotypic manifestations of ciliopathies can range from isolated blindness or renal disease to multiorgan system disorders such as retinitis pigmentosa, Meckel-Gruber syndrome (MKS), Joubert syndrome, or Bardet-Biedl syndrome (BBS). Defects of ciliary proteins can give rise to broad range of defects, including retinitis pigmentosa, hepatic, pancreatic, and kidney cyst formation, polydactyly, situs inversus, brain malformations, encephalocele, hydrocephalus, sensory defects, and skeletal abnormalities. However, mutations in the same ciliary gene can give rise to heterogeneous clinical phenotypes with different levels of severity [19]. Conversely, mutations in distinct genes can lead to the same phenotype, for example, mutations in at least eleven genes causing nephronophthisis (NPHP) [20]. In the wide phenotypic spectrum, NPHP is considered to be the mildest, BBS is intermediate in severity, and MKS is lethal. Retinal degeneration is often observed in diverse ciliopathies.
3. Retinitis Pigmentosa
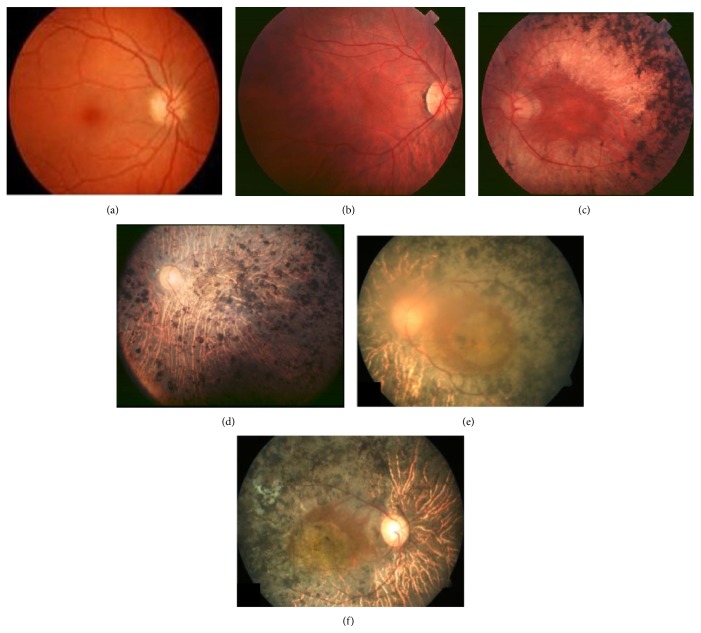
Retinitis pigmentosa (RP) encompasses a group of inherited retinal degenerations that show progressive loss of photoreceptors with highly variable clinical and genetic heterogeneity. It is one of the most heterogeneous genetic disorders known in man [21], resulting from a mutation in one or more genes. It affects 1 in 4000 individuals worldwide. RP is caused by mutations in more than 50 genes, including 23 genes for autosomal dominant RP, 36 for autosomal recessive RP, and 3 for X-linked RP (XLRP) (http://www.sph.uth.tmc.edu/Retnet/). Symptoms of RP include accumulation of intraretinal pigment-like deposits, retinal vessel attenuation, and characteristic changes in electroretinogram (ERG) patterns [22]. The typical signs of early disease are night blindness and tunnel vision with progressive constriction of the visual field. The progression of typical RP can be divided into 3 stages. The early stage of the disease is within the first decade or second decade of life. The typical signs of early disease are night blindness, although at this stage visual acuity and fundus examinations appear normal; at the midstage, rod photoreceptors degeneration starts at the periphery of the retina and then progresses towards the central region during the third or fourth decade, eventually causing tunnel vision with progressive constriction of the visual field; at this stage the presence of bone spicule-shaped pigment deposits is observed in the midperiphery of the retina. At the end-stage, complete loss of peripheral vision (classical tunnel vision) is sufficiently disabling to make independent movement difficult. There is a clear deposition of pigment at different retinal regions and a clear reduction of retinal vessel thickness. At this stage cone degeneration also takes place, leading to central vision impairment (Figure 2) [23].
Figure 2.
Fundus of an RP patient at different stages. (a) Image of a normal healthy eye. (b) Fundus of an RP patient at early stage. (c) Midstage of the disease showing midperipheral pigment deposits. (d) End-stage fundus showing pigment deposit present all over retina with thin retinal vessels and pale optic disc, modified from Hamel [23]. (e-f) End-stage fundus of an RP patient with an RPGR mutation c.852c>G (p.S284X), modified from Yang et al., 2014 [73].
4. The Retinitis Pigmentosa GTPase Regulator (RPGR)
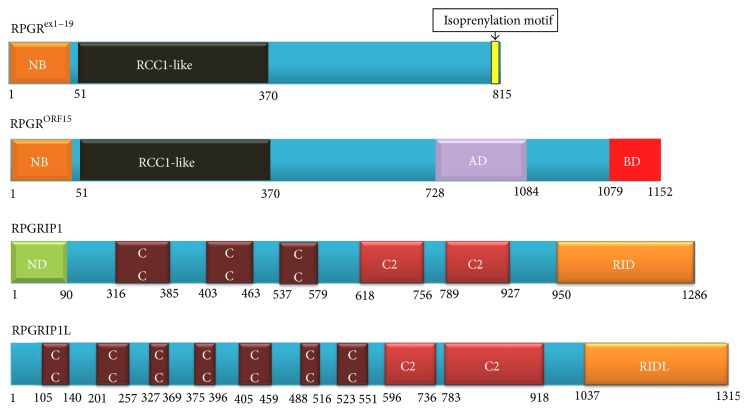
Mutations in the RPGR gene are the major cause of RP, accounting for more than 70% of XLRP and over 20% of nonsyndromic RP in North American families [24, 25]. RPGR mutations are responsible for 8.8% of Japanese RP patients [26] and 4.26% of Chinese patients with cone-rod dystrophy [27]. The RPGR gene is located in chromosomal region Xp21.1, spanning 172 kilobases. It was first cloned in 1996 and was initially reported to consist of 19 exons [28, 29]. Later, Vervoort and colleagues identified an additional alternatively spliced C-terminal exon, called open reading frame (ORF) 15, which is highly expressed in photoreceptors [30]. The RPGR ex1–19 transcript contains 19 exons, coding a protein with 815 amino acids with an isoprenylation motif at the C-terminus; the RPGR ORF15 has 15 exons, encoding a protein with 1152 amino acids. It shares exons 1–14 with RPGR ex1–19 plus the exon ORF15, encoding 567 amino acids with a repetitive glycine and glutamic acid-rich domain and a conserved basic C-terminal domain (Figure 3). In addition to these two major transcripts of the gene, RPGR encodes complex alternative spliced transcripts and many novel tissue-specific exons have been reported [31, 32]. All of the transcripts encode an amino (N)-terminal RCC1-like domain that is structurally similar to the RCC1 protein, a guanine nucleotide exchange factor for the small GTP-binding protein, Ran. Mutations have been reported in exon 2 to exon ORF15, where most mutations are taking place in the latter, which is regarded as a mutation hot spot [30, 33]. More than 300 mutations have been identified to cause X-linked forms of RP (Figures 2(e) and 2(f)), cone-rod, cone, and macular dystrophies, or syndromal forms of XLRP with hearing loss and primary ciliary dyskinesia [25].
Figure 3.
Structures of RPGR and its interacting proteins: RPGRIP1 and RPGRIP1L. The protein sequences of RPGRex1–19 (NP_000319.1), RPGRORF15 (NP_001030025.1), RPGRIP1 (NP_065099.3), and RPGRIP1L (NP_056087.2) were obtained from Ensembl database (http://www.ensembl.org) and domains were identified using InterPro, protein sequence analysis, classification tool (http://www.ebi.ac.uk/interpro/search/sequence-search), and details are from the literature. The isoprenylation motif (CAAX), four peptides at the N-terminal of the RPGRex1–19, is marked in yellow vertical rectangle. AD, acidic domain; BD, basic domain; C2, protein kinase C conserved domain 2; CC, coiled-coil domain; NB, GTP-binding motif; ND, nuclear domain; RID, RPGR interacting domain; RIDL, RPGR interacting domain-like.
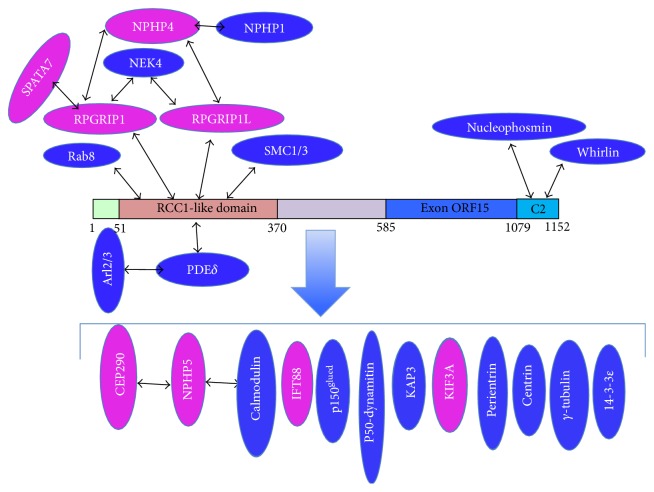
Localization of RPGR in the retina is species dependent. RPGR is localized predominantly in the connecting cilia of mouse photoreceptors, but, in human and bovine photoreceptors, localization has been reported in the outer segments of both rod and cone cells [34, 35]. Outside of the retina, RPGR is also detected in motile cilia of airway epithelia and the centrosomes/basal bodies of cultured cells [34, 36, 37]. RPGR has been reported to interact with different proteins. The first RPGR interacting protein, the delta subunit of the rod cyclic phosphodiesterase (PDEδ), was identified by yeast two-hybrid screening mouse embryonic cDNA library using the RCC1-like domain [38]. Using a similar strategy, three groups independently found the RPGR interacting partner: RPGR interacting protein 1 (RPGRIP1) [39–41]. Coimmunoprecipitation and mass spectrometry analysis showed that SMC1 and SMC3 proteins interact with the RCC1-like domain [42]. The C-terminal of RPGRORF15 interacts with whirlin, whose mutations cause Usher syndrome, and with nucleophosmin, which functions in cell division [36, 43]. Coimmunoprecipitation also found RPGR formed protein complexes with other ciliary proteins: IFT88, 14-3-3ε, γ tubulin, kinesin II subunit KIF3A and KAP3, dynein subunit intermediate chain, dynactin subunits: P150Glued and P50-dynamitin, IQCB1 (NPHP5), and CEP290 (NPHP6) (Figure 4) [42, 44, 45]. Mutations in the IQCB1 gene cause Senior-Loken syndrome (SLS), an inherited disorder characterized by RP and renal diseases [44]. Genetic deficiency in CEP290 is responsible for 15% of Leber congenital amaurosis (LCA) cases and has been implicated in other ciliopathies including SLS, nephronophthisis, Joubert syndrome, Bardet-Biedl syndrome (BBS), and Meckel syndrome (MKSS) [46–48]. IQCB1 physically interacts with CEP290 and both proteins involved in ciliogenesis [49].
Figure 4.
RPGR interacting protein network. Black arrows show direct interaction; the big blue arrow shows proteins in RPGR complex detected by coimmunoprecipitation. Proteins labelled in pink are implicated in retinal degeneration. The RCC1-like domain directly interacts with RPGRIP1, RPGRIP1L, SMC1/3, PDEδ, and Rab8. The end section of the C-terminal directly interacts with nucleophosmin and whirlin. Mutations in RPGRIP1, RPGRIP1L, CEP290, NPHP4, NPHP5, IFT88, KIF3A, and SPATA7 also cause ciliopathies.
5. RPGRIP1 and RPGRIP1L
Mutations in RPGRIP1 lead to LCA, a severe retinal dystrophy causing blindness or severe visual defects at birth or in early childhood [50]. Defects in RPGRIP1L cause either the lethal Meckel syndrome (MKSS) or Joubert syndrome type B (JBTS), which show much broader cilia defects [51, 52]. Human RPGRIP1 gene is located on chromosome 14q11 and encodes a protein with 1286 residues. RPGRIP1L gene is located on chromosome 16q12.2 and encodes a protein of 1315 residues [51]. RPGRIP1 protein has 29% amino acid identity to RPGRIP1L, which possesses similar functional domains. The N-terminals of both proteins contain coiled-coil domain with two leucine zipper motifs; the central regions contain two protein kinase C conserved region 2 (C2) domains; the C-terminal regions have an RPGR-interacting domain (Figure 3). Though the RPGR-interacting domain of RPGRIP1L is less homologous to that of RPGRIP1, the interaction with RPGR was confirmed by yeast two-hybrid and pull-down assays [53, 54]. The expression of RPGRIP1 is restricted to the retina, but RPGRIP1L is detected in retina and other tissues [35, 41, 51, 52]. Both RPGRIP1 and RPGRIP1L have similar localization to that of RPGR in retina and cultured cells [36, 51, 52]. Previous reports predicted there is a putative Ca2+-binding site in RPGRIP1 C-terminal C2 (C2-C) domain, but the recent crystal structure of RPGR-interacting domain of RPGRIP1 did not support the prediction [54, 55]. Both RPGRIP1 and RPGRIP1L interact with NPHP4, whose mutations result in nephronophthisis [51, 55]. Mutations in RPGRIP1, RPGRIP1L, or NPHP4 disrupted the interaction. Coene et al. used tandem affinity purification and mass spectrometry to identify that nek4 serine/threonine kinase also interacts with both RPGRIP1 and RPGRIP1L proteins [56]. More recently, SPATA7, which is responsible for LCA3 and juvenile RP, was identified as an RPGRIP1 interacting partner (Figure 4) [57]. Mislocalization of RPGRIP1 was detected in SPATA7 knockout mouse photoreceptor connecting cilium, suggesting SPATA7 is required for RPGRIP1 targeting to cilia.
6. The Role of RPGR and Its Interacting Proteins in Cilia Defects
Large amounts of data from studies in cell lines and animal models suggest RPGR and interacting proteins play a critical role in cilia genesis, maintenance, and function. Knockdown of RPGR in hTERT-RPE1 cells resulted in defects in ciliogenesis with a reduced number of cilia or shortened cilia [58, 59]. Morpholino knockdown of RPGR in zebrafish embryos led to defects in retinal development, such as defective lamination of retinal cell layers and abnormal photoreceptors. Other ciliary defects in RPGR morphants include shorter Kupffer's vesicle cilia, a shortened body axis, kinked tail, and hydrocephaly [60, 61]. RPGR knockout (KO) mice showed a slow retinal degeneration with photoreceptor death noted by 6 months of age [62]. Although mislocalization of cone opsins was detected as early as postnatal day 20, degenerative changes were found at age of two months. The connecting cilia of KO mice initially appeared normal and the discs of outer segments were well packed, suggesting RPGR is not necessary for the development of photoreceptors.
Knockout of RPGRIP1 in mouse (referred to as RPGRIP1 tm1Tili) resulted in early retinal degeneration with most of photoreceptors degenerating by 3 months of age, resembling the phenotypes in LCA patients. The abnormal photoreceptors were apparent at the age of postnatal day 15 (P15), showing disorganized outer segments and pyknotic nuclei, a sign of ongoing cell death [63]. RPGRIP1 KO mice presented a normal structure of connecting cilia, suggesting RPGRIP1 is not needed for the connecting cilia development and maintenance. RPGR, however, lost localization to connecting cilia in RPGRIP1 KO mice, suggesting RPGR correct localization is dependent on RPGRIP1. A recent N-ethyl-N-nitrosourea-induced RPGRIP1 null mouse model (referred to as RPGRIP1 nmf247) showed a more severe retinal degeneration when compared to that of RPGRIP1 tm1Tili mice. RPGRIP1 tm1Tili mice showed a rapid and progressive photoreceptor cell death. Outer segments were not seen in RPGRIP1 nmf247 mice at P7 and P14. Only 3-4 photoreceptor nuclei layers remained by P21. Rhodopsin mislocalization was detected at P12, the earliest time point examined. Other outer segment proteins, ROM1, transducin, and arrestin, were also mislocalized, but neither transducin nor arrestin mislocalized in RPGRIP1 tm1Tili mice [63, 64]. The phenotype of RPGRIP1 tm1Tili mice suggested that RPGRIP1 is essential for rod outer segment morphogenesis, whereas phenotypes in the RPGRIP1 nmf247 mice suggested RPGRIP1 is essential not only for rod outer segment disc morphogenesis but also for outer segment formation. The phenotypic difference between the two mouse mutant strains was caused by a short alternative splice variant in RPGRIP1 tm1Tili mice, which may have a role in outer segment formation.
RPGRIP1L is implicated in different types of ciliopathies. Inactivation of the RPGRIP1L (Ftm) gene results in the death of mouse at midgestation and recapitulating similar malformation observed in human MKS fetues, including brain, liver, kidney, limb, and eye developmental defects [52, 65]. Morpholino knockdown of RPGRIP1L in zebrafish showed a broad range of ciliary defects, including shortened body axis, curved body, malformed somites, kinked notochord, abnormal tail extension, and hydrocephaly [53, 66]. In mouse cochlear hair cells, RPGRIP1L is required for planar cell polarity. In zebrafish, it is required for convergent extension and polarized positioning of motile cilia in floor plate and is also required for the stability of dishevelled protein (Dvl) at the cilium base [66].
Mislocalization of outer segment proteins has been observed in RPGR or RPGRIP1 KO mice and in a patient carrying an RPGR mutation, suggesting RPGR and its interacting proteins are involved in protein trafficking in cilia [62–64, 67]. RPGR can regulate the ciliary trafficking of prenylated membrane-associated proteins by interacting with PDEδ, which is loaded with farnesylated cargo [68]. Since RPGR targeting to connecting cilia is dependent on RPGRIP1, RPGRIP1 may be involved in this process. RPGRIP1 is also essential for other ciliary protein trafficking. Mislocalization of NPHP4 and SDCCAG8, another ciliopathic protein [69], was observed in the photoreceptors of RPGRIP1 KO mice (RPGRIP1 nmf247) [70]. RPGRIP1 and RPGRIP1L may function as scaffolds to recruit NEK4 to cilia, helping to maintain ciliary stability [56].
The primary cilium is a coordinating hub for different signaling pathways, such as Wnt (canonical and noncanonical), sonic hedgehog (SHH), fibroblast growth factor (FGF), Notch, platelet derived growth factor receptor α (PDGFR α), mTOR, and Hippo signaling pathways [71]. RPGR and its interacting protein may be directly or indirectly involved in these signaling pathways. Silencing of RPGR in hTERT-RPE1 caused defective cilia formation and abnormal remodelling of actin cytoskeleton [59]. The noncanonical Wnt pathway, also referred to as planar cell polarity (PCP) pathway, regulates actin cytoskeleton rearrangement by activating the small GTPase, RhoA, which plays a critical role in actin cytoskeleton dynamics. Our unpublished data suggest RPGR and its interacting protein regulate the stability of the key components of the PCP pathway. RPGRIP1L (Ftm) was shown to be involved in SHH pathway by controlling the production of Gli3 protein, which regulates SHH signaling [65]. RPGRIP1L is also involved in the PCP pathway through targeting Dvl to the cilium base and stabilizing the proteins there [66]. NPHP4, a direct interacting partner of both RPGRIP1 and RPGRIP1L, has been shown to stabilize Dvl [72]. In fact RPGRIP1L and NPHP4 coordinate to modulate dishevelled stability and control the Wnt pathway [66].
7. Conclusion
Mutations in the RPGR gene are the most common cause of RP, frequently associated with ciliopathies. RPGR directly or indirectly interacts with ciliary proteins to form one or more protein complexes. RPGR and its interacting proteins may regulate cilia genesis, maintenance, and function mainly through recruiting ciliary protein to cilia. RPGR protein complex is involved in cilia regulatory signaling pathways, but understanding the complicated RPGR-involved molecular mechanisms remains a challenge.
Acknowledgments
The authors would like to thank the National Eye Research Centre, Rosetrees Trust, Fight for Sight, Tenovus Scotland, the Visual Research Trust, the W. H. Ross Foundation, Yorkhill Children's Charity, and the Carnegie Trust for the Universities of Scotland for supporting this work.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. The primary cilium as a complex signaling center. Current Biology. 2009;19(13):R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singla V., Reiter J. F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313(5787):629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 3.Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(12):5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. The Journal of Cell Biology. 1998;141(4):993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazour G. J., Dickert B. L., Vucica Y., et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. The Journal of Cell Biology. 2000;151(3):709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozminski K. G., Beech P. L., Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. The Journal of Cell Biology. 1995;131(6 I):1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pazour G. J., Dickert B. L., Witman G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. The Journal of Cell Biology. 1999;144(3):473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas . Molecular Biology of the Cell. 1999;10(3):693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholey J. M. Intraflagellar transport motors in cilia: moving along the cell's antenna. Journal of Cell Biology. 2008;180(1):23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nature Reviews Molecular Cell Biology. 2002;3(11):813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson A. Organization of the adult primate fovea. In: Penfold P. L., Provis J. M., editors. Macular Degeneration. Berlin, Germany: Springer; 2005. pp. 1–20. [DOI] [Google Scholar]
- 12.Horst C. J., Johnson L. V., Besharse J. C. Transmembrane assemblage of the photoreceptor connecting cilium and motile cilium transition zone contain a common immunologic epitope. Cell Motility and the Cytoskeleton. 1990;17(4):329–344. doi: 10.1002/cm.970170408. [DOI] [PubMed] [Google Scholar]
- 13.Solomon S. G., Lennie P. The machinery of colour vision. Nature Reviews Neuroscience. 2007;8(4):276–286. doi: 10.1038/nrn2094. [DOI] [PubMed] [Google Scholar]
- 14.Kevany B. M., Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25(1):8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q., Tan G., Levenkova N., et al. The proteome of the mouse photoreceptor sensory cilium complex. Molecular and Cellular Proteomics. 2007;6(8):1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrowski L. E., Blackburn K., Radde K. M., et al. A proteomic analysis of human cilia: identification of novel components. Molecular and Cellular Proteomics. 2002;1(6):451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Mahjoub M. R. The importance of a single primary cilium. Organogenesis. 2013;9(2):61–69. doi: 10.4161/org.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetz S. C., Anderson K. V. The primary cilium: a signalling centre during vertebrate development. Nature Reviews Genetics. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoefele J., Wolf M. T. F., O'Toole J. F., et al. Evidence of oligogenic inheritance in nephronophthisis. The Journal of the American Society of Nephrology. 2007;18(10):2789–2795. doi: 10.1681/asn.2007020243. [DOI] [PubMed] [Google Scholar]
- 20.Wolf M. T. F., Hildebrandt F. Nephronophthisis. Pediatric Nephrology. 2011;26(2):181–194. doi: 10.1007/s00467-010-1585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rattner A., Sun H., Nathans J. Molecular genetics of human retinal disease. Annual Review of Genetics. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- 22.Hartong D. T., Berson E. L., Dryja T. P. Retinitis pigmentosa. The Lancet. 2006;368(9549):1795–1809. doi: 10.1016/s0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 23.Hamel C. Retinitis pigmentosa. Orphanet Journal of Rare Diseases. 2006;1(1, article 40) doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breuer D. K., Yashar B. M., Filippova E., et al. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. The American Journal of Human Genetics. 2002;70(6):1545–1554. doi: 10.1086/340848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu X., Black G. C., Rice J. M., et al. RPGR mutation analysis and disease: an update. Human Mutation. 2007;28(4):322–328. doi: 10.1002/humu.20461. [DOI] [PubMed] [Google Scholar]
- 26.Jin Z. B., Liu X. Q., Hayakawa M., Murakami A., Nao-i N. Mutational analysis of RPGR and RP2 genes in Japanese patients with retinitis pigmentosa: identification of four mutations. Molecular Vision. 2006;12:1167–1174. [PubMed] [Google Scholar]
- 27.Huang L., Zhang Q., Li S., et al. Exome sequencing of 47 chinese families with cone-rod dystrophy: mutations in 25 known causative genes. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0065546.e65546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meindl A., Dry K., Herrmann K., et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nature Genetics. 1996;13(1):35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 29.Roepman R., van Duijnhoven G., Rosenberg T., et al. Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Human Molecular Genetics. 1996;5(7):1035–1041. doi: 10.1093/hmg/5.7.1035. [DOI] [PubMed] [Google Scholar]
- 30.Vervoort R., Lennon A., Bird A. C., et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nature Genetics. 2000;25(4):462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner R., Rosenberg T., Schultz-Heienbrok R., et al. RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Human Molecular Genetics. 1999;8(8):1571–1578. doi: 10.1093/hmg/8.8.1571. [DOI] [PubMed] [Google Scholar]
- 32.Hong D.-H., Li T. Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Investigative Ophthalmology and Visual Science. 2002;43(11):3373–3382. [PubMed] [Google Scholar]
- 33.Shu X., McDowall E., Brown A. F., Wright A. F. The human retinitis pigmentosa GTPase regulator gene variant database. Human Mutation. 2008;29(5):605–608. doi: 10.1002/humu.20733. [DOI] [PubMed] [Google Scholar]
- 34.Hong D. H., Pawlyk B., Sokolov M., et al. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Investigative Ophthalmology and Visual Science. 2003;44(6):2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 35.Mavlyutov T. A., Zhao H., Ferreira P. A. Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species. Human Molecular Genetics. 2002;11(16):1899–1907. doi: 10.1093/hmg/11.16.1899. [DOI] [PubMed] [Google Scholar]
- 36.Shu X., Fry A. M., Tulloch B., et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Human Molecular Genetics. 2005;14(9):1183–1197. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- 37.Shu X., Zeng Z., Eckmiller M. S., et al. Developmental and tissue expression of Xenopus laevis RPGR. Investigative Ophthalmology and Visual Science. 2006;47(1):348–356. doi: 10.1167/iovs.05-0858. [DOI] [PubMed] [Google Scholar]
- 38.Linari M., Ueffing M., Manson F., Wright A., Meitinger T., Becker J. The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boylan J. P., Wright A. F. Identification of a novel protein interacting with RPGR. Human Molecular Genetics. 2000;9(14):2085–2093. doi: 10.1093/hmg/9.14.2085. [DOI] [PubMed] [Google Scholar]
- 40.Roepman R., Bernoud-Hubac N., Schick D. E., et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Human Molecular Genetics. 2000;9(14):2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 41.Hong D.-H., Yue G., Adamian M., Li T. Retinitis pigmentosaGTPase regulator (RPGR)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. The Journal of Biological Chemistry. 2001;276(15):12091–12099. doi: 10.1074/jbc.m009351200. [DOI] [PubMed] [Google Scholar]
- 42.Khanna H., Hurd T. W., Lillo C., et al. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. The Journal of Biological Chemistry. 2005;280(39):33580–33587. doi: 10.1074/jbc.m505827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright R. N., Hong D.-H., Perkins B. RpgrORF15 connects to the usher protein network through direct interactions with multiple whirlin isoforms. Investigative Ophthalmology and Visual Science. 2012;53(3):1519–1529. doi: 10.1167/iovs.11-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto E. A., Loeys B., Khanna H., et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nature Genetics. 2005;37(3):282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 45.Chang B., Khanna H., Hawes N., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Human Molecular Genetics. 2006;15(11):1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayer J. A., Otto E. A., O'Toole J. F., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nature Genetics. 2006;38(6):674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 47.Valente E. M., Silhavy J. L., Brancati F., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nature Genetics. 2006;38(6):623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 48.Coppieters F., Lefever S., Leroy B. P., de Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Human Mutation. 2010;31(10):1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 49.Barbelanne M., Song J., Ahmadzai M., Tsang W. Y. Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Human Molecular Genetics. 2013;22(12):2482–2494. doi: 10.1093/hmg/ddt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dryja T. P., Adams S. M., Grimsby J. L., et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. The American Journal of Human Genetics. 2001;68(5):1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arts H. H., Doherty D., van Beersum S. E. C., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nature Genetics. 2007;39(7):882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 52.Delous M., Baala L., Salomon R., et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nature Genetics. 2007;39(7):875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 53.Khanna H., Davis E. E., Murga-Zamalloa C. A., et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nature Genetics. 2009;41(6):739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remans K., Bürger M., Vetter I., Wittinghofer A. C2 domains as protein-protein interaction modules in the ciliary transition zone. Cell Reports. 2014;8(1):1–9. doi: 10.1016/j.celrep.2014.05.049. [DOI] [PubMed] [Google Scholar]
- 55.Roepman R., Letteboer S. J. F., Arts H. H., et al. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18520–18525. doi: 10.1073/pnas.0505774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coene K. L. M., Mans D. A., Boldt K., et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Human Molecular Genetics. 2011;20(18):3592–3605. doi: 10.1093/hmg/ddr280. [DOI] [PubMed] [Google Scholar]
- 57.Eblimit A., Nguyen T. T., Chen Y., et al. Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina. Human Molecular Genetics. 2015;24(6):1584–1601. doi: 10.1093/hmg/ddu573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murga-Zamalloa C. A., Atkins S. J., Peranen J., Swaroop A., Khanna H. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Human Molecular Genetics. 2010;19(18):3591–3598. doi: 10.1093/hmg/ddq275.ddq275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gakovic M., Shu X., Kasioulis I., Carpanini S., Moraga I., Wright A. F. The role of RPGR in cilia formation and actin stability. Human Molecular Genetics. 2011;20(24):4840–4850. doi: 10.1093/hmg/ddr423.ddr423 [DOI] [PubMed] [Google Scholar]
- 60.Ghosh A. K., Murga-Zamalloa C. A., Chan L., Hitchcock P. F., Swaroop A., Khanna H. Human retinopathy-associated ciliary protein retinitis pigmentosa GTPase regulator mediates cilia-dependent vertebrate development. Human Molecular Genetics. 2010;19(1):90–98. doi: 10.1093/hmg/ddp469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shu X., Zeng Z., Gautier P., et al. Zebrafish Rpgr is required for normal retinal development and plays a role in dynein-based retrograde transport processes. Human Molecular Genetics. 2009;19(4):657–670. doi: 10.1093/hmg/ddp533. [DOI] [PubMed] [Google Scholar]
- 62.Hong D.-H., Pawlyk B. S., Shang J., Sandberg M. A., Berson E. L., Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3) Proceedings of the National Academy of Sciences of the United States of America. 2000;97(7):3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y., Hong D.-H., Pawlyk B., et al. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: Subserving RPGR function and participating in disk morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3965–3970. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Won J., Gifford E., Smith R. S., et al. RPGRIP1 is essential for normal rod photoreceptor outer segment elaboration and morphogenesis. Human Molecular Genetics. 2009;18(22):4329–4339. doi: 10.1093/hmg/ddp385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vierkotten J., Dildrop R., Peters T., Wang B., Rüther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134(14):2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- 66.Mahuzier A., Gaudé H.-M., Grampa V., et al. Dishevelled stabilization by the ciliopathy protein rpgrip1l is essential for planar cell polarity. Journal of Cell Biology. 2012;198(5):927–240. doi: 10.1083/jcb.201111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adamian M., Pawlyk B. S., Hong D.-H., Berson E. L. Rod and cone opsin mislocalization in an autopsy eye from a carrier of X-linked retinitis pigmentosa with a Gly436Asp mutation in the RPGR gene. American Journal of Ophthalmology. 2006;142(3):515–518. doi: 10.1016/j.ajo.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 68.Wätzlich D., Vetter I., Gotthardt K., et al. The interplay between RPGR, PDEδ and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Reports. 2013;14(5):465–472. doi: 10.1038/embor.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otto E. A., Hurd T. W., Airik R., et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nature Genetics. 2010;42(10):840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patil H., Tserentsoodol N., Saha A., Hao Y., Webb M., Ferreira P. A. Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell Death and Disease. 2012;3(7, article e355) doi: 10.1038/cddis.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oh E. C., Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139(3):443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burcklé C., Gaudé H.-M., Vesque C., et al. Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Human Molecular Genetics. 2011;20(13):2611–2627. doi: 10.1093/hmg/ddr164. [DOI] [PubMed] [Google Scholar]
- 73.Yang L., Yin X., Feng L., et al. Novel mutations of RPGR in Chinese retinitis pigmentosa patients and the genotype-phenotype correlation. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0085752.e85752 [DOI] [PMC free article] [PubMed] [Google Scholar]