Abstract
There is an unmet clinical need to identify biomarkers for breast cancer neoadjuvant chemotherapy. Here, using miRNA TaqMan Low-Density Arrays (TLDA), we analyzed the miRNA expression profile in pre-treatment needle aspiration tumor samples from patients who received taxane-anthracycline-based neoadjuvant chemotherapy. Although, in an unsupervised hierarchical cluster analysis, the total miRNA expression profile could not generate a tree with clear distinction between pathologic complete response (pCR) and non-pCR classes, we found that elevated expression of miR-125b and miR-141 was associated with non-pCR. In vitro experiments indicated that inhibition of miR-125b and miR-141 expression reduced cellular survival in response to taxane-anthracycline treatment. Furthermore, co-transfection with miR-125b and miR-141 mimics increased resistance of MCF7 and BT549 cells to taxane-anthracycline induced cytotoxicity. Pathway analyses indicated that many of the target proteins of miR-125b are involved in apoptotic pathways and cell cycle control. Together, we provide evidence that elevated miR-125b and 141 expression predicts a poor clinical responsiveness of taxane-anthracycline-based neoadjuvant chemotherapy.
Keywords: breast cancer, microRNA, neoadjuvant chemotherapy, pathologic complete response, predictive biomarkers.
Introduction
Neoadjuvant chemotherapy for breast cancer is an important therapeutic modality that can reduce the tumor mass, eliminate distant micro-metastases, improve operability, and even allow breast-conserving surgery to take place in some cases 1. The taxane-anthracycline regimen is commonly used in neoadjuvant chemotherapy. Pathologic complete response (pCR), in which no invasion, and no in-site residuals are present in breast and nodes after treatment, is the criteria used to discriminate between patients with favorable and unfavorable outcomes of taxane-anthracycline-based neoadjuvant chemotherapy 2, 3. However, a portion of the patient population does not achieve pCR after this particular combination chemotherapy, subsequently affecting their overall clinical outcome 2, 3. The underlying molecular mechanisms of chemoresistance remain unclear, making it critical to identify molecular biomarkers that can predict the outcome of neoadjuvant chemotherapy.
MicroRNAs (miRNAs) are small noncoding RNAs of ~22 nucleotides that negatively regulate protein expression, primarily by complementary binding to the 3′ untranslated region (UTR) of target messenger RNA (mRNA), resulting in mRNA cleavage or translation repression 4. About 50% of annotated human miRNA genes are located in chromosomal fragile sites that are susceptible to structural genomic alterations during tumor development 5, suggesting that there are tumor specific miRNA profiles. Indeed, aberrant expression of miRNAs has been shown to be associated with many types of cancers 6-11. MiRNAs also regulate the responses of tumor cells to therapeutic agents, as many of them are critical determinants of the DNA damage response (DDR), a pro-survival mechanism in the presence of DNA damage 12 .
In order to define a potential role of specific miRNAs as biomarkers in the chemotherapeutic responses to taxane-anthracycline-based neoadjuvant chemotherapy, we conducted miRNA TaqMan Low-Density Array (TLDA), and determined that the elevated expression of miR-125b and miR-141 correlated with chemoresistance in breast cancer patients receiving neoadjuvant chemotherapy.
Materials and methods
Cell lines
MCF7 and BT549 cells (Obtained from American Type Culture Collection, Manassas, VA, USA ) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS (Invitrogen, Carlsbad, CA) and 50 mg/ml of penicillin/streptomycin (Invitrogen) in humidified 37°C chambers with 5% CO2. Taxane and anthracycline were obtained from Sigma (St. Louis, MO).
Clinical Cases
Written informed consent was obtained from all patients, and the study was approved by the institutional review board. Formalin-fixed paraffin-embedded (FFPE) cancer tissues were collected from breast cancer patients by needle aspiration, and were stored at the Tianjin Medical University Cancer Institute and Hospital (Table 1). Only patients who received at least four cycles of systemic neoadjuvant chemotherapy treatment were included in the study. The histological response to neoadjuvant chemotherapy was characterized as a pathologic complete response (pCR) when there was no evidence of residual invasive tumor in the breast, or axillary lymph nodes. None of the patients included in this study received radiation or chemotherapy prior to needle aspiration.
Table 1.
Patient Clinical Features
| No. | Age(years) | ER (%)† | PgR (%)‡ | HER-2§ | pCR status¶ |
|---|---|---|---|---|---|
| 1 | 43 | 15 | <1 | - | pCR |
| 2 | 58 | 40 | 30 | + | pCR |
| 3 | 48 | <1 | <1 | - | pCR |
| 4 | 46 | 65 | 45 | - | pCR |
| 5 | 53 | <1 | <1 | ++ | pCR |
| 6 | 25 | 70 | 20 | ++ | pCR |
| 7 | 49 | 90 | <1 | - | pCR |
| 8 | 45 | 20 | <1 | + | pCR |
| 9 | 36 | <1 | <1 | - | pCR |
| 10 | 62 | <1 | <1 | ++ | pCR |
| 11 | 51 | 60 | 45 | + | non-pCR |
| 12 | 57 | <1 | 20 | +++ | non-pCR |
| 13 | 65 | 70 | 10 | - | non-pCR |
| 14 | 59 | <1 | 5 | ++ | non-pCR |
| 15 | 37 | 70 | 70 | + | non-pCR |
| 16 | 23 | 95 | 25 | + | non-pCR |
| 17 | 43 | <1 | <1 | ++ | non-pCR |
| 18 | 35 | 70 | <1 | - | non-pCR |
| 19 | 48 | <1 | <1 | +++ | non-pCR |
| 20 | 57 | <1 | <1 | +++ | non-pCR |
| 21 | 43 | <1 | <1 | +++ | non-pCR |
† ER,estrogen receptor
‡ PgR, progesterone receptor
§HER-2, human epidermal growth factor receptor 2
¶ pCR ,pathologic complete response
RNA Extraction
Above 5 sections (10 μm in thickness) of each FFPE tissue were taken. The areas for sampling were circled on the H&E-stained slides by pathologists and then marked on 10 μm-thickness sections to achieve high tumor content (>70%). For FFPE samples, total RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, USA), following the manufacturer's protocol.
MiRNA Profiling Using the TaqMan Low-Density Array
The TaqMan Low-Density Array (TLDA) v3.0 (Applied Biosystems, Foster City, CA) were utilized for miRNA profiling. We analyzed each sample with an A & B card for detection of a total of 754 miRNAs, along with endogenous and negative controls. We then carried out qRT-PCR on an Applied Biosystems 7900HT thermocycler. SDS software v2.4 (threshold, 0.1) was used to analyze real-time PCR data and the GeneSpring software v12.1 (Agilent Technology) was used for relative miRNA levels.
Target Gene Analysis
Using TargetScan, the list of mRNAs predicted to be targeted by the candidate miRNAs were obtained 13. The predicted target mRNAs were analyzed for different signaling pathways by MAS 3.0 (Molecule Annotation System) server with default setting (bioinfo.capitalbio.com/mas3).
Cell Viability Assay
A total of 5×103 cells/well was seeded in 96-well plates. 24 hours later, miR-125b and miR-141 mimics were co-transfected into MCF7 or BT549 cells. 48 hours after transfection, taxane and anthracycline were added into cell culture medium. Cell viability was determined 24 hours after transfection using CCK-8 (Cell Counting Kit-8, Dojindo Laboratories, Japan).
Transfection
Anti-miR inhibitors and miR-mimics were purchased from Shanghai GenePharma, including has-miR-125b, has-miR-141, anti-miR-125b oligonucleotides, anti-miR-141 oligonucleotides ,an anti-miR-inhibitor negative control, and an miR-mimic negative control. Inhibitors and mimics were diluted to a working solution of 20µM in nuclease free water, and transfected into cells either individually at a final concentration of 25nM, or in combination at a final concentration of 12.5nM for each construct for a total of 25nM.
Statistical Analysis
For the qRT-PCR data, the relative expression level of each target miRNA (Log2 relative level) was calculated according to the difference in CT values between the target miRNA and internal control (∆CT). Using unpaired t test, ∆CTs were analyzed by GeneSpring software to find out the differentially expressed miRNAs between the pCR and non-pCR groups. T test was performed to analyze the CCK-8 assay data using SPSS 18.0. A p value <0.05 was considered statistically significant.
Results
miR-125b and miR-141 are upregulated in non-pCR breast cancer patients
In order to identify a potential miRNA signature that could serve as a biomarker in breast cancer neoadjuvant chemotherapy, we tested miRNA profiles of 21 patients with breast cancer who had also received neoadjuvant anthracycline-taxane-based chemotherapy (Table 1). The median age of the patients at time of study entry was 48 years (range, 23 to 65 years). Of the patients participating, 10 patients achieved pCR following standard neoadjuvant chemotherapy. Tumors stained positive for estrogen receptor (ER) in 11 (52.4%), for progesterone receptor (PgR) in 9 patients (42.9%), and for human epidermal growth factor receptor 2 (HER2) in 14 patients (66.7%) (Table 1). Among these 21 patients, there are 2 cases (9.5%) of triple negative breast cancer (TNBC) which do not express the ER,PR and HER2 receptors.
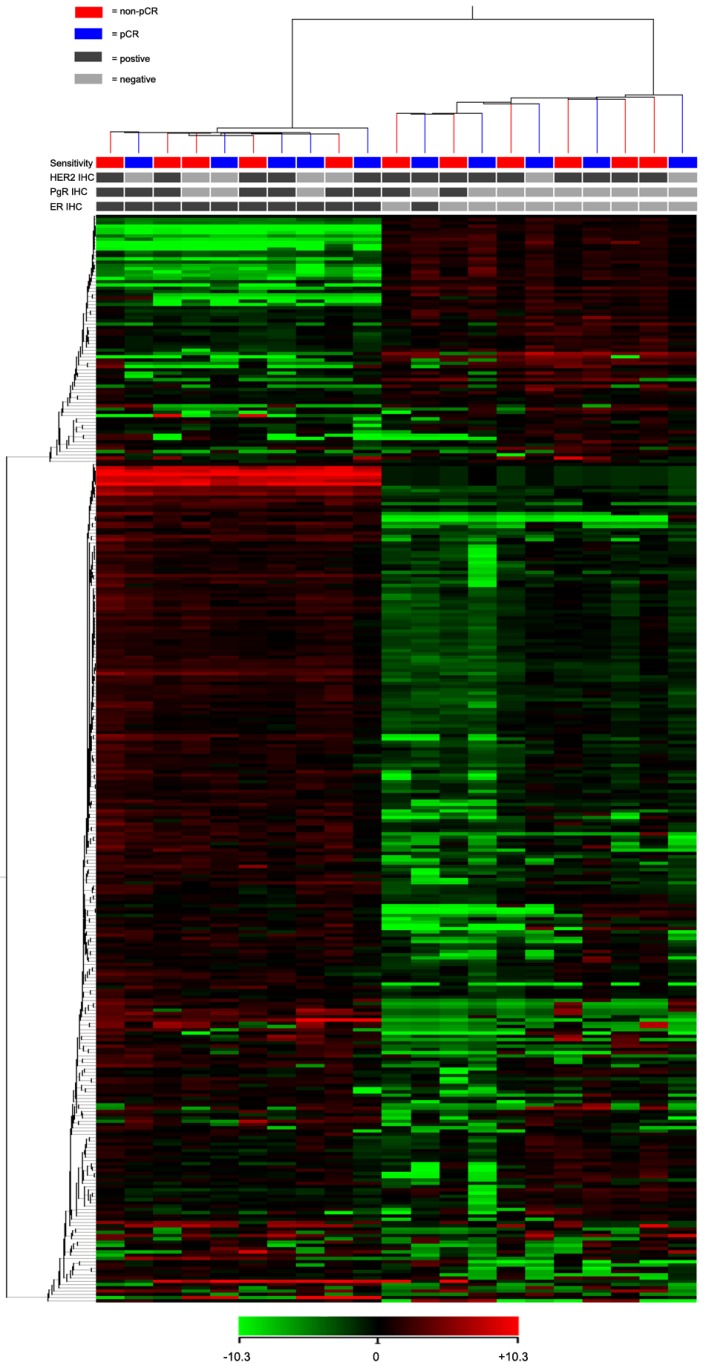
For the TLDA analysis, miRNAs with the threshold cycle (CT) value lower than 35 in at least 10 patients were included into the study. Of the 754 miRNAs incorporated in the array, 333 miRNAs met the inclusion criteria. Using an unsupervised hierarchical cluster analysis of these selected miRNAs, a hierarchical tree was generated with no clear distinction in the miRNA expression levels between pCR and non-pCR classes (Fig. 1). However, this hierarchical tree could clearly separate ER positive and negative patients with only one exception.
Fig 1.
Heat map and cluster dendrogram of detected miRNAs. miRNA expression patterns of 21 pre-treatment needle aspiration tumor FFPE samples were examined using TLDA arrays. A total of 333 miRNA were expressed at a significant level (Ct < 35 in at least 10 samples). ER, PgR, HER2 and neoadjuvant response for each patient were presented. Using Pearson centered distance matrix and the centroid-linkage method, delta Ct of miRNAs were subjected to hierarchical clustering analysis.
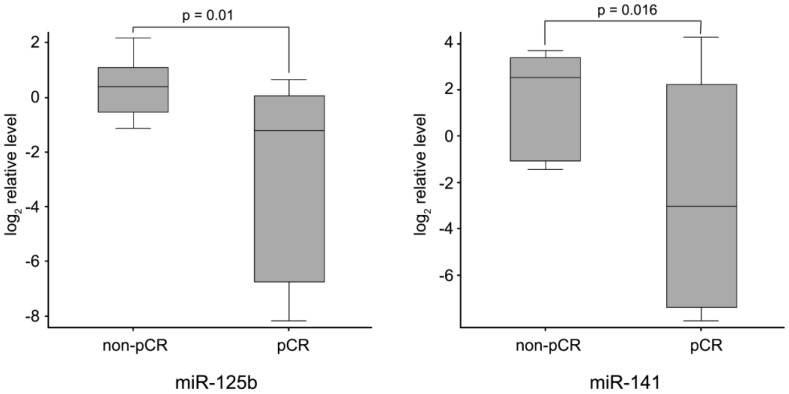
Finally, to identify neoadjuvant chemotherapy sensitivity associated miRNAs, we followed two inclusion criteria to distinguish differential expression of miRNAs between pCR and non-pCR groups: (1) miRNA levels exhibiting > 1.5-fold difference between the two groups; and (2) p value of t-test being < 0.05 as statistically significant. As shown in Fig. 2, miR-125b and miR-141 were found significantly elevated in non-pCR patients compared with pCR patients (miR-125b, p = 0.01, fold change 8.14; miR-141, p = 0.016, fold change 18.96).
Fig 2.
miR-125b and miR-141 levels in non-pCR and pCR pre-treatment needle aspiration FFPE tumor samples. Expression levels of the miRNAs are normalized to U6 snRNA (Log2 relative level).
Overexpression of miR-125b and miR-141 results in chemosensitivity in vitro
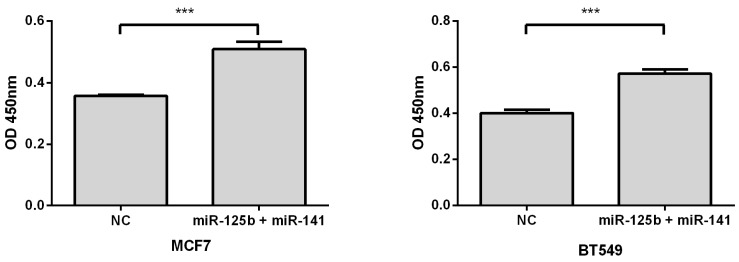
To validate whether elevated expression of miR-125b and miR-141 is associated with chemoresistance, we overexpressed these miRNAs, along with negative controls, in MCF7 or BT549 breast cancer cell lines. Compared with the control, overexpression of miR-125b and miR-141 markedly inhibited taxane-anthracycline induced cell cytotoxicity in MCF7 and BT549 cells, as measured by OD450 (Fig. 3).
Fig 3.
Restoring the expression of miR-125b and miR-141 inhibited taxane-anthracycline induced cell cytotoxicity in MCF7 and BT549 cells. MCF7 or BT549 cells cotransfected with miR-125b and miR-141 were seeded into 96-well plate at the density of 5×103 cells/well and then treated with taxane-anthracycline for 24 h (48 h after transfection). Results of the means of ten assays with standard deviation of the mean are presented ***, p < 0.001
Inhibition of miR-125b and miR-141 reduces chemoresistance in vitro
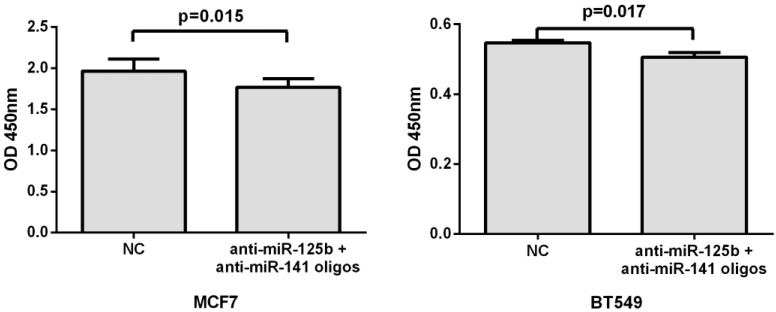
To further validate the correlation of miR-125b and miR-141 expression with chemosensitivity, we also transfected anti-miR-125b and anti-miR-141 oligonucleotides into the MCF7 or BT549 cells and analyzed the change in chemosensitivity to taxane and anthracycline treatment. As shown in Fig. 4, the miR-125b and miR-141 inhibitors significantly increased the sensitivity of MCF7 and BT549 cells to taxane and anthracycline when compared to control cells.
Fig 4.
Inhibition of miR-125b and miR-141 reduces chemoresistance in vitro. MCF7 or BT549 cells cotransfected with anti-miR-125b and anti-miR 141 oligonucleotides were seeded into 96-well plate at the density of 5×103 cells/well and then treated with taxane-anthracycline for 24 h (48 h after transfection). Results of the means of ten assays with standard deviation of the mean are presented.
miR-125b mediated apoptotic and DNA damage response pathways likely contribute to chemoresistance
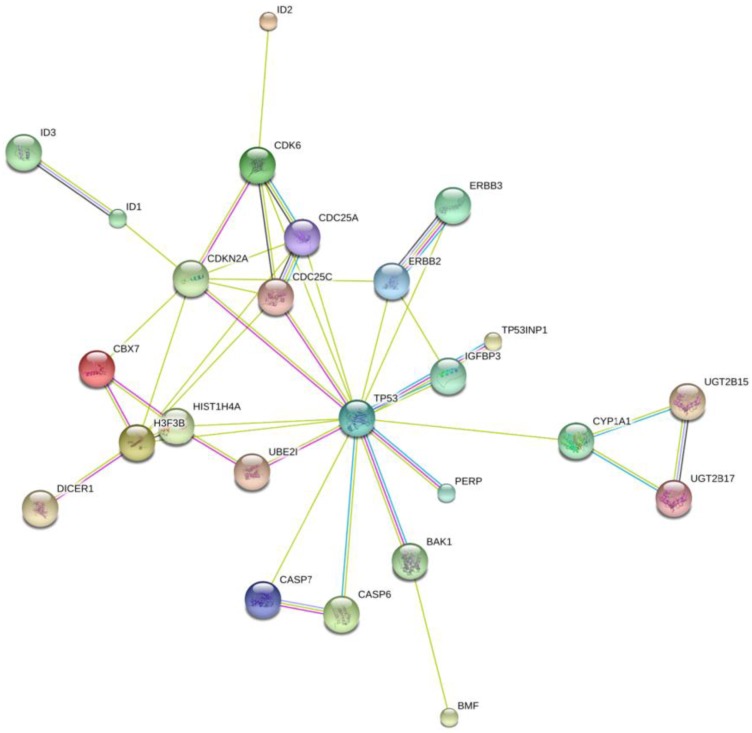
To further investigate the possible mechanism of miR-125b and miR-141 in chemosensitivity, we conducted KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis to identify potential upstream and downstream targets that might be associated with chemoresistance. We found that one potential upstream regulator of miRNA -125b biogenesis is the DNA damage response protein kinase ataxia-telangiectasia mutated (ATM). The induction of miRNA biogenesis upon DNA damage is dependent on ATM kinase activity. This regulation can be linked to the direct binding and phosphorylation of KSRP (KH-type splicing regulatory protein) by ATM. KSRP is a component of the Drosha and Dicer complexes, and also regulates the maturation of a handful of miRNAs involved in cell proliferation and apoptotic pathways 14. One of the miRNAs regulated by KSRP is miR-125b. It is highly likely that the elevated levels of miR-125b correlate with hyper-activated ATM in the tissues, and the subsequent phosphorylated KSRP in the presence of DNA damage. Most of the downstream targets of miR-125b, shown in Fig. 5, are validated targets which are involved in apoptotic pathways, as well as in and cell cycle regulation.
Fig 5.
Network Map of proteins that are validated targets of miR-125b. All of these proteins are involved in apoptotic pathways and cell cycle control, with the exception of the ID proteins. Those are involved in DNA binding. The UGT proteins are involved in the clearance of toxic substances.
Unlike miR-125b, there is no direct link between the activation of ATM and the up-regulation of miR-141. The up-regulation of miR-141, a member of the miR-200a family, is most likely due to stress induced senescence, which would result from the activation of the DNA damage repair pathway 15. A String pathway analysis of the list of known targets of this particular miRNA indicates that there is a direct link to cell proliferation and apoptosis pathways, but not directly linked to DNA damage response pathways.
Discussion
Breast cancer neoadjuvant treatment serves to reduce the tumor size so that a breast-conserving surgery can be performed. Biomarkers that have predictive values for tumor responses would provide critical information to identify a subset of patients that could benefit from the neoadjuvant therapy. In this report, we identified that the expression levels of two miRNAs (miRNA-125b and miRNA-141) can predict the clinical response to taxane-anthracycline-based neoadjuvant chemotherapy.
In the clinical setting, pCR has been used to predict long-term outcome of patients who received neoadjuvant chemotherapy and is therefore a potential surrogate marker for survival. However, for a long time there has been little agreement regarding the precise definition of what constitutes a pCR. Von Minckwitz et al attempted to standardize the method of reporting a true pCR by examining more than 6,000 patients treated in a series clinical trials of neoadjuvant anthracycline and taxane based chemotherapy 2, 3. They compared multiple existing definitions of pCR to determine how robust each definition serves as a surrogate for patient survival outcomes and found that the least residual cancer after treatment in the breast and nodes was highly correlated with better survival 2, 3. This standard of pCR was used in the present study that the biopsy before neoadjuvant chemotherapy is suitable for identifying treatment-sensitivity associated miRNAs. Our findings may help select patients that will eventually benefit from taxane-anthracycline -based neoadjuvant chemotherapy.
Alteration of miRNA frequently has been observed in breast cancer. Dysregulated miRNAs affect not only carcinogenesis progress, but also can have a direct impact on sensitivity to chemotherapy and radiotherapy 16. The two miRNAs identified in this study are no exception. miR-125b has been reported to be overexpressed in breast cancer. Zhou et al found that miR-125b was up-regulated in Taxol-resistant breast cancer cells and could confer Taxol resistance through the suppression of BAK1 expression 17. Another report from the same group showed that overexpression of Snail in breast cancer cells dramatically increases the expression of miR-125b through the Snail-activated Wnt/β-catenin/TCF4 axis 18. Moreover, miR-125b was also shown to be involved in chemotherapy resistance in other caners 19, 20. For example, miR-125b promoted leukemia cell resistance to daunorubicin (an anthracycline family member) by inhibiting apoptosis 20. On the other hand, miR-141 is overexpressed in cisplatin resistant ovarian, gastric and esophageal squamous cancer cells by targeting KEAP1 and YAP1 21-23.
Pathway analysis conducted has indicated potential mechanisms of miRNA-125b in chemoresistance. For example, miR-125b is linked to KSRP mediated biogenesis, which is also dependent on the ATM kinase. Elevated levels of miR-125b might correlate with ATM activity. This is consistent with our recent findings regarding ERα regulation of ATM expression in breast cancer tissues 24. Furthermore, activation of ATM-Snail pathway in the DNA damage response also echoes the link of miR-125b in the chemoresistant pathway 25, 26.
In conclusion, using Taqman Human miRNA Low-Density Array, global miRNA profiling of pCR and non-pCR tissues has identified differentially expressed miRNAs. Among them, miR-125b and miR-141 were found to be up-regulated in pre-treatment needle aspiration tumor samples in non-pCR patients. Our study revealed the global patterns of miRNA in pCR and non-pCR tumor tissues, and has led to the identification of miR-125b and miR-141 as potential molecular biomarkers that can predict the clinical outcome in breast cancer patients who will receive taxane-anthracycline -based neoadjuvant chemotherapy.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (30930038, 31400673, 81202101 and 81302292); Specialized Research Fund for the Doctoral Program of Higher Education [20131202120002]; Tianjin Research Program of Application Foundation and Advanced Technology [14JCQNJC09800]; China Postdoctoral Science Foundation [20110490788]; and the Alabama Innovation Fund to Southern Research Institute.
References
- 1.Chia S, Swain SM, Byrd DR, Mankoff DA. Locally advanced and inflammatory breast cancer. J Clin Oncol. 2008;26:786–790. doi: 10.1200/JCO.2008.15.0243. doi:10.1200/JCO.2008.15.0243. [DOI] [PubMed] [Google Scholar]
- 2.Schott AF, Hayes DF. Defining the benefits of neoadjuvant chemotherapy for breast cancer. J Clin Oncol. 2012;30:1747–1749. doi: 10.1200/JCO.2011.41.3161. doi:10.1200/JCO.2011.41.3161. [DOI] [PubMed] [Google Scholar]
- 3.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA. et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. doi:10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. doi:10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings Of the National Academy Of Sciences Of the United States Of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. doi:DOI 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC. et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. doi:10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J. et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. doi:10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. doi:10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S. et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. doi:10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Boohaker RJ, Xu B. The versatile functions of ATM kinase. Biomed J. 2014;37:3–9. doi: 10.4103/2319-4170.125655. doi:10.4103/2319-4170.125655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. doi:DOI 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A. et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. doi:10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Quirantes R, Segura-Carretero A, Micol V. et al. Metformin lowers the threshold for stress-induced senescence: a role for the microRNA-200 family and miR-205. Cell Cycle. 2012;11:1235–1246. doi: 10.4161/cc.11.6.19665. doi:10.4161/cc.11.6.19665. [DOI] [PubMed] [Google Scholar]
- 16.Mulrane L, McGee SF, Gallagher WM, O'Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. doi:10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y. et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–21507. doi: 10.1074/jbc.M109.083337. doi:10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Liu H, Desai S, Schmitt DC, Zhou M, Khong HT. et al. miR-125b functions as a key mediator for snail-induced stem cell propagation and chemoresistance. J Biol Chem. 2013;288:4334–4345. doi: 10.1074/jbc.M112.419168. doi:10.1074/jbc.M112.419168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haemmig S, Baumgartner U, Gluck A, Zbinden S, Tschan MP, Kappeler A. et al. miR-125b controls apoptosis and temozolomide resistance by targeting TNFAIP3 and NKIRAS2 in glioblastomas. Cell Death Dis. 2014;5:e1279.. doi: 10.1038/cddis.2014.245. doi:10.1038/cddis.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Bai H, Wang C, Wei D, Qin Y, Xu X. microRNA125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis. Mol Med Rep. 2014;9:1909–1916. doi: 10.3892/mmr.2014.2011. doi:10.3892/mmr.2014.2011. [DOI] [PubMed] [Google Scholar]
- 21.van Jaarsveld MT, Helleman J, Boersma AW, van Kuijk PF, van Ijcken WF, Despierre E. et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32:4284–4293. doi: 10.1038/onc.2012.433. doi:10.1038/onc.2012.433. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Su J, Zhu L, Zhang G. Helicobacter pylori modulates cisplatin sensitivity in gastric cancer by down-regulating miR-141 expression. Helicobacter. 2014;19:174–181. doi: 10.1111/hel.12120. doi:10.1111/hel.12120. [DOI] [PubMed] [Google Scholar]
- 23.Imanaka Y, Tsuchiya S, Sato F, Shimada Y, Shimizu K, Tsujimoto G. MicroRNA-141 confers resistance to cisplatin-induced apoptosis by targeting YAP1 in human esophageal squamous cell carcinoma. J Hum Genet. 2011;56:270–276. doi: 10.1038/jhg.2011.1. doi:10.1038/jhg.2011.1. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Yang C, Qian X, Lei T, Li Y, Shen H. et al. Estrogen receptor alpha regulates ATM Expression through miRNAs in breast cancer. Clin Cancer Res. 2013;19:4994–5002. doi: 10.1158/1078-0432.CCR-12-3700. doi:10.1158/1078-0432.CCR-12-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL. et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol. 2012;4:304–315. doi: 10.1093/jmcb/mjs048. doi:10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boohaker RJ, Cui X, Stackhouse M, Xu B. ATM-mediated Snail Serine 100 phosphorylation regulates cellular radiosensitivity. Radiother Oncol. 2013;108:403–408. doi: 10.1016/j.radonc.2013.06.017. doi:10.1016/j.radonc.2013.06.017. [DOI] [PubMed] [Google Scholar]