Abstract
Host-seeking, ovipositional behavior and mating of insects are controlled mainly by odor perception through sensory organs such as antennae. Antennal chemoreception is extremely important for insect survival. Several antennal chemosensory receptors are involved in mediating the odor detection in insects, especially the odorant receptors (ORs) and ionotropic receptors (IRs), to ensure the specificity of the olfactory sensory neuron responses. In the present study, we identified the chemosensory receptor gene repertoire of the parasitoid wasp Microplitis mediator, a generalist endoparasitoid that infests more than 40 types of Lepidopterous larvae and is widely distributed in the Palaearctic region. By transcriptome sequencing of male and female antennae we identified 60 candidate odorant receptors, six candidate ionotropic receptors and two gustatory receptors in M. mediator. The full-length sequences of these putative chemosensory receptor genes were obtained by using the rapid amplification of cDNA ends PCR (RACE-PCR) method. We also conducted reverse transcription PCR (RT-PCR) combined with real-time quantitative PCR (qPCR) for investigating the expression profiles of these chemosensory receptor genes in olfactory and non-olfactory tissues. The tissue- and sex-biased expression patterns may provide insights into the roles of the chemosensory receptor in M. mediator. Our findings support possible future study of the chemosensory behavior of M. mediator at the molecular level.
Keywords: Microplitis mediator, chemosensory receptor gene, odorant receptor, ionotropic receptor, gustatory receptor, expression profile
Introduction
Parasitic wasps (parasitoids) serve as important natural agents, effectively control major crop pests in the agricultural ecosystem 1. The success of parasitic wasps in suppressing pest populations depends on their ability to locate hosts in the complex chemical environment 1, 2. Like most of the insects, parasitic wasps can also find their hosts in the foraging environments and reproduction occurs through a series of behavioral activities, regulated mainly by semiochemicals 2-5.
It is likely that the semiochemicals are perceived by different types of chemosensory (olfaction and taste) sensilla, which are mostly present on the antennae of parasitic wasps 1, 6, 7. The chemosensory sensilla house olfactory sensory neurons (OSNs) that project their sensory dendrites into sensilla 7-9. On the outer dendrite membrane of OSNs, different chemoreceptors are expressed and form specific receptor binding sites for chemicals 7, 9. Chemoreceptors expressed in insect OSNs are composed to three large and divergent families: the odorant receptors (ORs), gustatory receptors (GRs) and ionotropic receptors (IRs) 10-17.
Insect ORs were the first family of chemosensory receptors discovered in the olfactory organs 12-14. They are seven-transmembrane domain receptors with an inverted membrane topology with an intracellular N-terminus and an extracellular C-terminus compared with vertebrate ORs, which are G protein coupled receptors 18-20. The ORs respond to a variety of volatile chemicals, including pheromones and general odorants 9, 21. The supposed function of insect ORs depends on the presence of a well-conserved odorant receptor coreceptor (Orco), which forms an odorant-gated ion channel with tuning ORs 22-24.
Subsets of OSNs in insect antennae can also express proteins from the GR family 25-27, which share a common ancestor with ORs and encode seven-transmembrane domain receptors 11. However, most GRs are expressed in gustatory organs 15, 25, 28. These GRs mainly sense non-volatile compounds that the insects directly contact with gustatory sensilla. These include sugars, bitter compounds, salts and some gustatory pheromones 8, 29, 30. In addition, one sub-group of the GR family (DmGr21a and DmGr63a) responds to carbon dioxide (CO2) 8, 26.
A large number of OSNs neither express OR genes nor the GR genes 8, 31, but they express IRs that form a third class of chemoreceptors 10, 16, 32. IRs belong to an ancient family of chemosensory receptors that are relatives of ionotropic glutamate receptors (iGluR) and are divided into two subfamilies, the conserved “antennal” IRs and the species-specific “divergent” IRs 10, 17. The antennal IRs, expressed in the antennae of Drosophila melanogaster, are considered to be ligand-gated ion channels that mediate the response of OSNs to a variety of organic acids, amines and aldehydes compounds 17, 33-35. Two IRs, IR8a and IR25a are most closely related to iGluRs, containing a divergent extracellular amino-terminal domain (ATD) and they appear to act as co-receptors. They form complexes with different sets of IRs, which are necessary and sufficient for odor-evoked neuronal responses 10, 17, 33.
The identification of ORs, GRs and IRs in parasitoid wasps is of considerable interest and significance, both to address the mechanisms controlling intraspecific or interspecific chemical communication and for potential applications for the genetic manipulation of parasitoids' behavioral responses to chemical cues used in biological control strategies. The parasitoid wasp Microplitis mediator (Haliday) (Hymenoptera: Braconidae) is a generalist endoparasitoid that is reported to attack approximately 40 different lepidopteran larvae 36 and is widely distributed in the Palaearctic region 37. In China, it has been successfully mass-reared in the laboratory and used as biological control agent 38. Like most parasitoid wasps, M. mediator uses volatile chemicals as olfactory cues for the purpose of foraging for oviposition. It has been shown that the volatile chemicals released by caterpillar-damaged cotton plants can be attractive to M. mediator 39.
In our recent study, 14 OR genes were cloned from M. mediator 40, while additional receptors are remaining to be identified. Following to our previous work on M. mediator chemosensory gene identification, using next-generation sequencing of antennae of both male and female M. mediator, we have identified and annotated large sets of chemoreceptors (ORs, GRs and IRs). By using RACE-PCR, we were able to verify all of the chemoreceptor genes as unique genes. Using reverse transcription PCR (RT-PCR) and real-time quantitative-PCR (qPCR), we analyzed expression profiles of the chemoreceptors. Our results reveal that a number of chemoreceptor genes have specific or enriched expression in the antennae, which may play important functions in the chemoreception system of M. mediator.
Methods
Insect material and RNA extraction
The M. mediator cocoons were obtained from the Institute of Plant Protection, Hebei Academy of Agriculture and Forestry, China. Parasitoid cocoons were reared in a growth chamber maintained at 28 ± 1°C, 60 ± 10 % R.H. and 16L: 8D photoperiod. The emerged adults were fed on 10% sucrose solution. For transcriptome sequencing, 2000 antennal pairs were collected from 1- to 3-d old male and female insects. For RT-PCR, different tissues (male antennae, female antennae, heads without antennae, thoraxes, abdomens, legs and wings) were collected. For qPCR, antennae were dissected at the base of the pedicel and collected in three batches, each consisting of 200 male antennae and 200 female antennae. Fifty body parts (mixture of heads, thoraxes, abdomens, legs, wings) were also collected. All collected tissues were immediately stored in -80 °C for further process. Total RNA was extracted from the antennae or other tissues using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). The quantity of RNA samples was checked by using 1.1% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, DE, USA).
Antennal transcriptome sequencing, assembly and functional annotation
The cDNA library was generated by using a combination of the SMART cDNA Amplification Kit (Clontech, Mountain View, CA, USA) and Ion Xpress™ Plus gDNA Fragment Library Kit (Life Technology) following the manufacturer's protocol. The cDNA with appropriate length (300~400 bp) was purified using the MinElute Gel Recovery Kit (Qiagen, Valencia, CA, USA) and sequenced using the Ion Proton™ System (Life Technology). After removing short or low quality sequences and adaptor sequences using the programs TagDust 41, LUCY 42 and SeqClean 43, male and female reads were assembled separately, and all reads from male and female antennae were also assembled using MIRA3 44 and CAP3 45. The sequence homology search was conducted with BLASTx and BLASTn programs against the Nr (non-redundant protein database) and Nt (non-redundant nucleotide sequence database) in NCBI with an E-value cut-off of 1.0E-5. Gene Ontology terms were extracted from the best hits obtained from the BLASTx against the Nr using the Blast2GO program 46.
Chemosensory receptor identification
A FASTA file of the non-redundant contigs was created from a local nucleotide database file using the BioEdit Sequence Alignment Editor 7.1.3.0 47. The local tBLASTn program was performed, using available sequences of OR, GR and IR proteins from Nasonia vitripennis and Apis mellifera as “query” 10, 48, 49 to identify candidate genes encoding putative ORs, GRs and IRs in M. mediator.
RACE-PCR
The unigene from the transcriptome did not always represent full-length transcripts. Some non-full-length transcripts may be parts of independent genes or parts of the same genes. To verify the unigene status and for better resolution of phylogenetic analyses, most of the candidate chemosensory receptor genes were extended using RACE-PCR. The 5' and 3' RACE-Ready cDNAs were synthesized from the adult antennae RNA by using the SMARTer™ RACE cDNA amplification kit (Clontech, Mountain View, CA, USA) following the manufacturer's instructions. Primers were designed manually and further adjusted using the Primer Premier software (PREMIER Biosoft International) (see Supplementary Material: Table S1).
The PCR was done with AdvantageTM 2 polymerase mix (Clontech, Mountain View, CA, USA) under the following conditions: denaturation at 95 °C for 2 min, 40 cycles of denaturation at 94 °C for 30 s, annealing at 66-68 °C, depending on primer pairs, for 30 s and extension at 72 °C for 2 min. The final extension step was at 72 °C for 5 min. The RACE-PCR products were subcloned into the pEasy-T3 vector (TransGen, Beijing, China) and were sequenced.
Sequence and phylogenetic analysis
The OR, IR and GR phylogenetic trees were built based on Hymenoptera data sets. The OR data set contained 54 amino acid sequences from M. mediator, together with a selected subset from N. vitripennis and A. mellifera, representing major subfamilies within the Hymenoptera ORs 48-50. The GR data set contained 2 amino acid sequences from M. mediator, together with sequences from N. vitripennis and A. mellifera 48, 49. The IR data set contained 6 amino acid sequences from M. mediator, together with sequences from N. vitripennis and A. mellifera 10. Because D. melanogaster IRs were well annotated and putative IRs could be distinguished from iGluRs, IR and iGluR sequences from D. melanogaster were also included in the data set 10.
Amino acid sequences were aligned using the program ClustalW 51 and further edited using the BioEdit Sequence Alignment Editor 7.1.3.0 47. The neighbor-joining tree was constructed using the MEGA 5.0 program 52. Bootstrapping was performed by re-sampling amino acid positions of 1000 replicates.
RT-PCR analysis
The expression profiles of chemosensory receptor transcripts in different tissues (male antennae, female antennae, heads without antennae, thoraxes, abdomens, legs and wings) were analyzed by RT-PCR. cDNAs from various tissues were synthesized using the Fast Quant RT kit (TIANGEN, Beijing, China). The gene-specific sense and anti-sense primers in RT-PCR were designed with the Primer 3 program (http://frodo.wi.mit.edu/) and listed in Supplementary Material: Table S2. Each reaction (25 μL volume) contained 200 ng cDNA from different tissues as a template. The cycling conditions were set at 95 °C for 4 min with 38 cycles as following: 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step of 5 min at 72 °C. A primer pair for β-actin (GenBank accession number: KC193266) of M. mediator was employed to assess the cDNA integrity. The PCR products were checked by using 1.2% agarose gels and the target genes were further confirmed by direct DNA sequence analysis. Amplification of each target gene was performed three times with three biological samples.
qPCR measurement
The relative transcript abundance of chemosensory receptor genes in male antennae, female antennae and other body parts (mixture of heads, thoraxes, abdomens, legs and wings) were further explored by qPCR using the ABI Prism 7500 Fast Detection System (Applied Biosystems, Carlsbad, CA, USA). cDNAs from male antennae, female antennae and the body parts were synthesized using the Fast Quant RT kit (TIANGEN, Beijing, China). The reference gene β-actin was used for normalization. The primers of the target and reference genes were designed with the Primer 3 program (http://frodo.wi.mit.edu/) and listed in Supplementary Material: Table S2. The specificity of each primer set was validated by melt-curve analysis and efficiency was calculated by analyzing standard curves with a five-fold cDNA dilution series. The qPCR reaction was conducted in a 20 μL mixture containing 10 μL of 2 × SuperReal PreMix Plus (TianGen, Beijing, China), 0.6 μL of each primer (10 μM), 0.4 μL of 50 × Rox Reference Dye, 1 μL of sample cDNA, 7.4 μL of sterilized H2O. The qPCR cycling parameters consisted of 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s, melt curves stages at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. Negative controls without either template were included in each experiment. Each reaction was performed with three biological replicates and each biological replicate was assessed three times. The comparative 2-ΔΔCT method 53 was used to calculate the relative transcript levels in each tissue. Data analysis was performed using SPSS Statistics 18.0 software (SPSS Inc., Chicago, IL, USA). An ANOVA and Duncan's new multiple range test (P < 0.05) were used to compare the expression of each target gene among various tissues.
Results
An overview of the antennal transcriptome
To identify chemosensory receptor genes from M. mediator, two transcriptomes of the male and female M. mediator antennae were created by high-throughput pyrophosphate sequencing of antennal cDNA libraries. A total of 19388952 (mean length 129 bp) and 11012901 clean reads (mean length 114 bp) were produced from the female and male antennae samples, respectively, and assembled into 48015 (mean length 551 bp) and 44428 (mean length 570 bp) transcripts, respectively. Furthermore, we assembled all clean reads from male and female antennae together and generated 48075 transcripts with 545 bp average length.
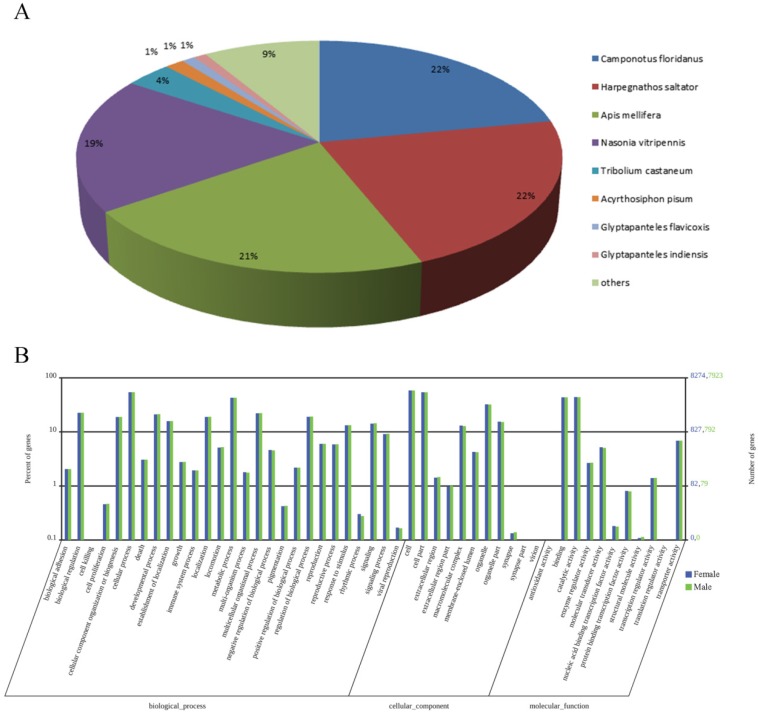
Through annotation by BLASTx and BLASTn program with the E-value cut-off of 1.0E-5, 17440 of the 48075 transcripts (36.3%) had BLASTx hits in the Nr databases and 9235 (19.2%) had BLASTn hits in the Nt databases. Among the annotated unigenes, 15044 of the 17440 Nr-hit unigenes (86.3%) had a best blast match to Hymenopteran sequences, primarily Camponotus floridanus (21.9%), Harpegnathos saltator (21.9%), A. mellifera (21.5%) and N. vitripennis (18.9%) (Figure 1A).
Figure 1.
Summary for the annotation of M. mediator antenna transcripts. (A) Species distribution of best BLASTx hits of M. mediator antenna transcripts. (B) Distribution and comparison of the female and male M. mediator antennal transcripts annotated at GO level 2.
With the GO classification, 8274 female antennal unigenes and 7923 male antennal unigenes were classified into 3 functional categories: molecular function, biological process and cellular component (Figure 1B). GO analyses showed that the numbers of each GO category were highly similar between the male and female antennal transcriptomes. In molecular function, the genes expressed in the antennae were mostly linked to binding (3614 female antennal unigenes and 3460 male antennal unigenes) and catalytic activity (3651 female antennal unigenes and 3473 male antennal unigenes). In terms of the biological process, the most represented biological processes were cellular processes (4487 female antennal unigenes and 4299 male antennal unigenes) and metabolic processes (3547 female antennal unigenes and 3387 male antennal unigenes). In the cellular component terms, cell (4843 female antennal unigenes and 4628 male antennal unigenes) and cell part (4476 female antennal unigenes and 4378 male antennal unigenes) constitute the most abundant categories (Figure 1B).
Identification of OR, GR and IR genes
Bioinformatic analyses indicated that a total of 82 cDNA sequences with amino acid homology to known insect ORs were identified in the M. mediator antennal transcriptome. Only one of the transcripts represented the full-length gene, which is an orthologue of olfactory coreceptor Orco. All of the other transcripts were incomplete fragments with a mean length of 374 bp. Among the candidate ORs, we re-identified all of the previously described ORs except MmedOR4 and MmedOR9. The exact number of candidate ORs was difficult to confirm because some of the sequences were too short (<300 bp) and did not show sufficient sequence overlap in a multiple sequence alignment. By RACE-PCR, we extended 40 sequences in both the 3' and 5' directions. After that we entered all candidate ORs as a data set. Sixty candidate genes were verified as unique ORs by using sequence assembly as well as multiple sequence alignment. We named these genes using the conventions followed by Ma et al. (MmedOR14~MmedOR61) 40. MmedOR14~53 encodes complete or nearly complete ORs (260 residues or larger). The completeness of MmedOR14~53 was judged by amino acid alignment with our previously identified OR sequences (Supplementary Material: Figure S1). All of them showed complete sequences except OR24 and OR39. The predicted translations of the complete OR transcripts ranged from 423 (MmedOR46) to 369 amino acids (MmedOR22). All of these proteins exhibited the presence of predicted multiple transmembrane domains with an inside or outside N-terminus, as usually observed for insect ORs (Table 1). MmedOR14~53 were analyzed in a subsequent experiment and submitted to GenBank with the accession numbers listed in Table 1, while other sequences (MmedOR54~61) are provided in Supplementary Material: Table S3.
Table 1.
Odorant receptors in M. mediator antennae.
| Gene name | Acc. number | Length (bp) | ORF (aa) | Completeness | Tm nb | Blast P hit | E-value | % Identify |
|---|---|---|---|---|---|---|---|---|
| MmedOR14 | KM979230 | 1690 | 411 | Complete | 7i | gb|AGS43050.1| odorant receptor Or2a [Cephus cinctus] | 9e-83 | 34% |
| MmedOR15 | KM979231 | 1381 | 385 | Complete | 7i | gb|AGG17939.1| olfactory receptor 6 [Microplitis mediator] | 2e-68 | 34% |
| MmedOR16 | KM979232 | 1293 | 378 | Complete | 8o | ref|NP_001177491.1| odorant receptor 44 [Nasonia vitripennis] | 4e-69 | 36% |
| MmedOR17 | KM979233 | 1720 | 402 | Complete | 8o | gb|AGG17945.1| olfactory receptor 12 [Microplitis mediator] | 3e-84 | 36% |
| MmedOR18 | KM979234 | 1930 | 390 | Complete | 7i | ref|NP_001164394.1| odorant receptor 81 [Nasonia vitripennis] | 2e-76 | 35% |
| MmedOR19 | KM979235 | 1352 | 396 | Complete | 7i | gb|AGG17940.1| olfactory receptor 7 [Microplitis mediator] | 3e-165 | 57% |
| MmedOR20 | KM979236 | 1556 | 392 | Complete | 7i | ref|NP_001177576.1| odorant receptor 204 [Nasonia vitripennis] | 2e-77 | 35% |
| MmedOR21 | KM979237 | 1443 | 372 | Complete | 7i | gb|AGG17945.1| olfactory receptor 12 [Microplitis mediator] | 1e-75 | 38% |
| MmedOR22 | KM979238 | 1281 | 369 | Complete | 5i | ref|XP_003696363.1| putative odorant receptor 13a-like [Apis florea] | 3e-59 | 34% |
| MmedOR23 | KM979239 | 1408 | 373 | Complete | 7i | ref|NP_001177491.1| odorant receptor 44 [Nasonia vitripennis] | 2e-76 | 36% |
| MmedOR24 | KM979240 | 1109 | 260 | Internal exon missing | 4o | gb|EGI68890.1| Odorant receptor 46a, isoform A [Acromyrmex echinatior] | 4e-81 | 51% |
| MmedOR25 | KM979241 | 1416 | 376 | Complete | 8o | ref|NP_001177491.1| odorant receptor 44 [Nasonia vitripennis] | 5e-67 | 33% |
| MmedOR26 | KM979242 | 1349 | 397 | Complete | 7i | gb|AGG17940.1| olfactory receptor 7 [Microplitis mediator] | 7e-165 | 55% |
| MmedOR27 | KM979243 | 1329 | 403 | Complete | 5i | gb|AGG17936.1| olfactory receptor 3 [Microplitis mediator] | 3e-154 | 54% |
| MmedOR28 | KM979244 | 1521 | 402 | Complete | 7i | gb|AGS43067.1| odorant receptor Or3h, partial [Cephus cinctus] | 0 | 61% |
| MmedOR29 | KM979245 | 1551 | 413 | Complete | 6i | gb|AGG17935.1| olfactory receptor 2 [Microplitis mediator] | 1e-168 | 56% |
| MmedOR30 | KM979246 | 1295 | 383 | Complete | 6i | ref|NP_001177491.1| odorant receptor 44 [Nasonia vitripennis] | 3e-64 | 33% |
| MmedOR31 | KM979247 | 1602 | 375 | Complete | 7i | gb|EGI65843.1| Odorant receptor 49b [Acromyrmex echinatior] | 5e-67 | 36% |
| MmedOR32 | KM979248 | 1474 | 420 | Complete | 8o | gb|AEQ62577.1| odorant receptor 1 [Apis cerana] | 2e-159 | 58% |
| MmedOR33 | KM979249 | 1384 | 384 | Complete | 7i | gb|AGG17939.1| olfactory receptor 6 [Microplitis mediator] | 2e-67 | 33% |
| MmedOR34 | KM979250 | 1394 | 395 | Complete | 5i | gb|AGG17946.1| olfactory receptor 13 [Microplitis mediator] | 1e-170 | 60% |
| MmedOR35 | KM979251 | 1295 | 377 | Complete | 7i | gb|AGG17941.1| olfactory receptor 8 [Microplitis mediator] | 3e-75 | 35% |
| MmedOR36 | KM979252 | 1350 | 382 | Complete | 7i | gb|AGG17939.1| olfactory receptor 6 [Microplitis mediator] | 2e-82 | 36% |
| MmedOR37 | KM979253 | 1322 | 384 | Complete | 7i | gb|AGG17939.1| olfactory receptor 6 [Microplitis mediator] | 7e-73 | 34% |
| MmedOR38 | KM979254 | 1299 | 388 | Complete | 7i | ref|NP_001164395.1| odorant receptor 82 [Nasonia vitripennis] | 5e-75 | 35% |
| MmedOR39 | KM979255 | 1725 | 349 | N-terminus missing | 5o | ref|NP_001177576.1| odorant receptor 204 [Nasonia vitripennis] | 3e-58 | 34% |
| MmedOR40 | KM979256 | 1321 | 406 | Complete | 8i | ref|NP_001229918.1| odorant receptor 115 [Apis mellifera] | 1e-97 | 39% |
| MmedOR41 | KM979257 | 1611 | 384 | Complete | 9o | ref|NP_001177491.1| odorant receptor 44 [Nasonia vitripennis] | 2e-71 | 34% |
| MmedOR42 | KM979258 | 1287 | 381 | Complete | 7i | gb|AGG17939.1| olfactory receptor 6 [Microplitis mediator] | 2e-73 | 34% |
| MmedOR43 | KM979259 | 1634 | 377 | Complete | 8o | ref|NP_001177603.1| odorant receptor 260 [Nasonia vitripennis] | 2e-60 | 35% |
| MmedOR44 | KM979260 | 1426 | 395 | Complete | 5i | gb|AGG17946.1| olfactory receptor 13 [Microplitis mediator] | 4e-147 | 51% |
| MmedOR45 | KM979261 | 1381 | 393 | Complete | 7i | ref|NP_001177567.1| odorant receptor 191 [Nasonia vitripennis] | 7e-56 | 31% |
| MmedOR46 | KM979262 | 2138 | 423 | Complete | 6o | ref|NP_001177488.1| odorant receptor 37 [Nasonia vitripennis] | 3e-46 | 29% |
| MmedOR47 | KM979263 | 1394 | 404 | Complete | 8i | ref|NP_001177467.1| odorant receptor 10 [Nasonia vitripennis] | 4e-92 | 38% |
| MmedOR48 | KM979264 | 1399 | 401 | Complete | 5i | gb|AGG17946.1| olfactory receptor 13 [Microplitis mediator] | 8e-160 | 56% |
| MmedOR49 | KM979265 | 1353 | 393 | Complete | 6o | gb|AGG17946.1| olfactory receptor 13 [Microplitis mediator] | 2e-166 | 58% |
| MmedOR50 | KM979266 | 1320 | 395 | Complete | 6o | gb|AGG17938.1| olfactory receptor 5 [Microplitis mediator] | 8e-87 | 37% |
| MmedOR51 | KM979267 | 1295 | 410 | Complete | 7i | gb|AGG17942.1| olfactory receptor 10 [Microplitis mediator] | 2e-65 | 34% |
| MmedOR52 | KM979268 | 1314 | 406 | Complete | 7o | ref|NP_001229918.1| odorant receptor 115 [Apis mellifera] | 1e-89 | 37% |
| MmedOR53 | KM979269 | 1539 | 387 | Complete | 7i | gb|AGG17941.1| olfactory receptor 8 [Microplitis mediator] | 1e-106 | 44% |
Note: Transmembrane domains(Tm) were predicted using HMMTop (http://www.enzim.hu/hmmtop/), iN-terminus inside and oN-terminus outside.
The partial sequences of two candidate GRs were also identified in M. mediator antennae transcriptomes. Both candidates were cloned as full-length coding sequences by using RACE-PCR and deposited in the GenBank database with accession numbers KM979270 and KM979271. We compared the candidate full-length GRs (MmedGR6 and MmedGR64f) against the Nr database at NCBI using BLASTp and revealed a 41 % and 60 % identity with GR6 of N. vitripennis and GR64f of H. saltator, respectively. Then we confirmed that all candidate GRs belonged GR family (Table 2).
Table 2.
Gustatory receptors and ionotropic receptors in M. mediator antennae.
| Gene name | Acc. number | Length (bp) | ORF (aa) | Tm nb |
Blast P hit | E-value | % Identify |
|---|---|---|---|---|---|---|---|
| MmedGR6 | KM979270 | 1598 | 410 | 7i | ref|NP_001177425.1| gustatory receptor 6 [Nasonia vitripennis] | 1e-82 | 41% |
| MmedGR64f | KM979271 | 1517 | 452 | 8o | gb|EFN77950.1| Putative gustatory receptor 64f [Harpegnathos saltator] | 1e-180 | 60% |
| MmedIR8a | KM979274 | 3116 | 898 | 4i | ref|XP_003697873.1| glutamate receptor, ionotropic kainate 2-like [Apis florea] | 0 | 59% |
| MmedIR25a.1 | KM979275 | 3072 | 938 | 3o | ref|XP_008206981.1| glutamate receptor 2-like [Nasonia vitripennis] | 0 | 59% |
| MmedIR25a.2 | KM979277 | 3219 | 899 | 5o | ref|XP_003703813.1| glutamate receptor, ionotropic kainate 1-like [Megachile rotundata] | 0 | 45% |
| MmedIR64a | KM979276 | 2133 | 650 | 4i | ref|XP_008208508.1| glutamate receptor ionotropic, kainate 2-like [Nasonia vitripennis] | 0 | 49% |
| MmedIR75u | KM979272 | 2039 | 586 | 3o | gb|EGI70316.1| Glutamate receptor delta-2 subunit [Acromyrmex echinatior] | 0 | 52% |
| MmedIR76b | KM979273 | 2414 | 577 | 2o | ref|XP_003400706.1| glutamate receptor, ionotropic kainate 2-like isoform 2 [Bombus terrestris] | 4e-131 | 40% |
Note: Transmembrane domains(Tm) were predicted using HMMTop (http://www.enzim.hu/hmmtop/), iN-terminus inside and oN-terminus outside.
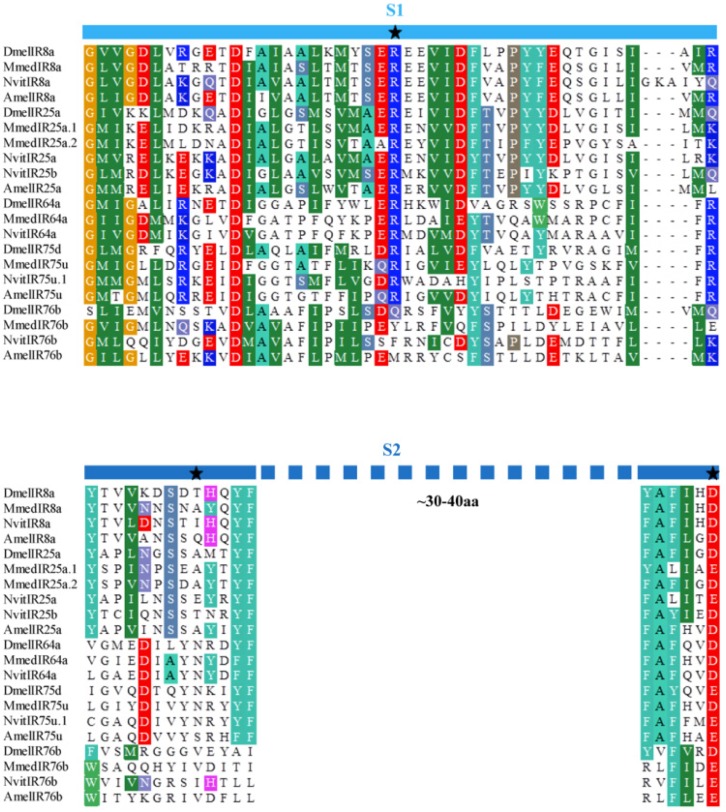
Six putative IR genes in the M. mediator antennal transcriptome were identified according to their similarity to IR sequences of other insects. By using RACE-PCR, full length sequences of these MmedIRs were obtained. The MmedIRs were named MmedIR8a, MmedIR25a.1, MmedIR25a.2, MmedIR64a, MmedIR75u and MmedIR76b, respectively, according to their similarities with IR genes in N. vitripennis and A. mellifera 10. The predicted translations of the complete MmedIR transcripts ranged from 577 (MmedIR76b) to 938 amino acids (MmedIR25a.1). All the MmedIR genes with GenBank accession numbers were listed in Table 2. By using HMMTop prediction program we found that there were two or more transmembrane domains presented in all of these genes (Table 2). The protein secondary structure analysis showed that all the putative MmedIRs had similar structures as insect IRs, comprising an extracellular N terminus, one ion channel pore (P), a bipartite ligand-binding domain with two lobes (S1 and S2) and a short cytoplasmic C terminus (data not shown). However, there was a lack of one or several key amino acids in the predicted glutamate binding domains (Figure 2). In the S1 lobe, the arginine (R) residue was conserved in all putative MmedIRs except MmedIR76b. In the first half of the S2 domain, the threonine (T) residue was not conserved in any putative MmedIRs. In the second half of the S2 domain, an aspartate (D) or glutamate (E) was only conserved in the coreceptor IR25a /8a orthologues.
Figure 2.
Amino acid alignment of part of the S1 and S2 ligand binding domains of MmedIRs with IR sequences from other insects. The three typical iGluR glutamate-interacting residue positions are marked with asterisks at the top.
Sequence similarity and phylogenetic analysis
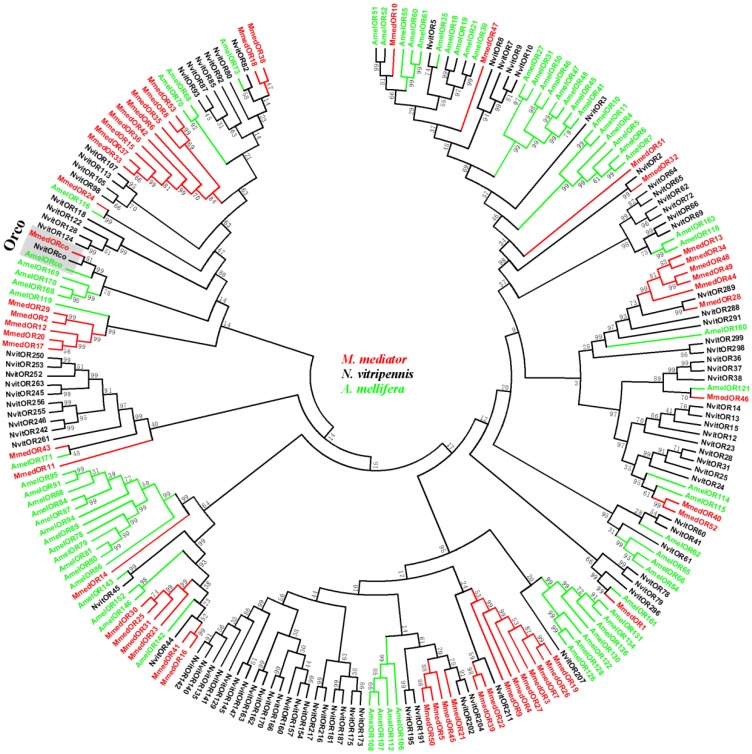
As illustrated in a phylogenetic tree constructed with sequences from three Hymenoptera insects, ORs are extremely divergent between species and most ORs from the same species formed monophyletic groups (Figure 3). In the phylogenetic tree, seven MmedORs (OR3, OR4, OR7, OR9, OR19, OR26 and OR27) and nine other MmedORs (OR6, OR8, OR15, OR33, OR35, OR36, OR37, OR42 and OR53) clustered in a species-specific subgroup, respectively. In addition, several other MmedORs did not cluster in species-specific clades. They grouped with the N. vitripennis or A. mellifera ORs. MmedOR24 clusters with AmelOR116 and MmedOR32 is most closely related to NvitOR2. Interestingly, MmedOR1 clusters with AmelOR161 and NvitOR296, which have been classified as I subfamily in the Hymenoptera OR family 50. This I subfamily showed a limited number of genes, two in N. vitripennis and one in A. mellifera 50. There is no suspense that the olfactory Orco subfamily is highly conserved and clustered in one branch with clear orthologous relationships in three species.
Figure 3.
Phylogenetic relationship of candidate odorant receptor (OR) genes from M. mediator and other Hymenoptera insects. A neighbour-joining tree was constructed using MEGA5 which was made based on a sequence alignment using ClustalW, the same as below. Amel, A. melifera; Mmed, M. mediator; Nvit, N. vitripennis.
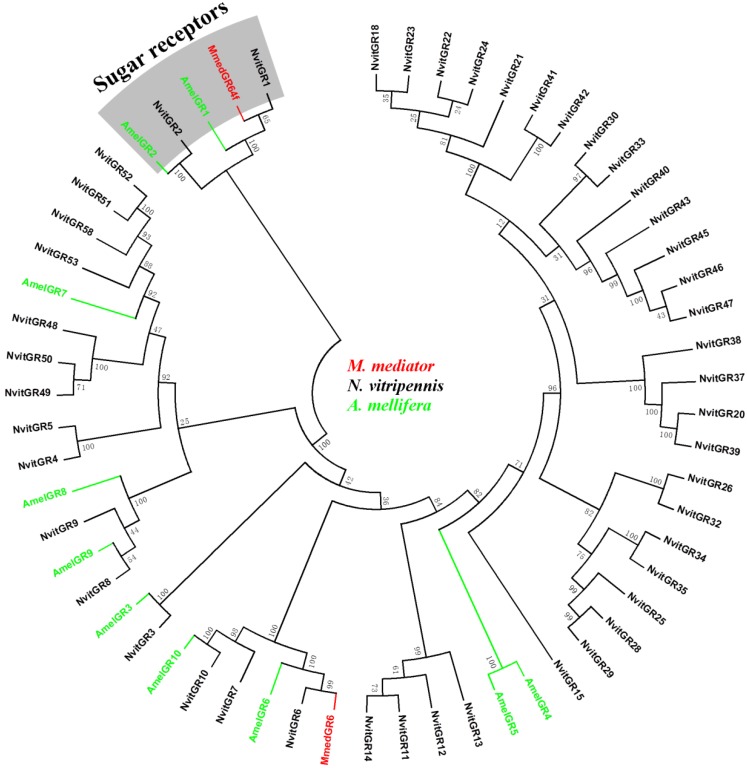
In the GR phylogeny (Figure 4), MmedGR64f was classified into the N. vitripennis GR1 orthologue subgroup, which has orthologue of the known sugar receptors (GR1 and GR2) 49, 54, 55. Another is similar to a member of GR6 family (MmedGR6).
Figure 4.
Phylogenetic relationship of candidate gustatory receptor (GR) genes from M. mediator and other Hymenoptera insects. Amel, A. melifera; Mmed, M. mediator; Nvit, N. vitripennis.
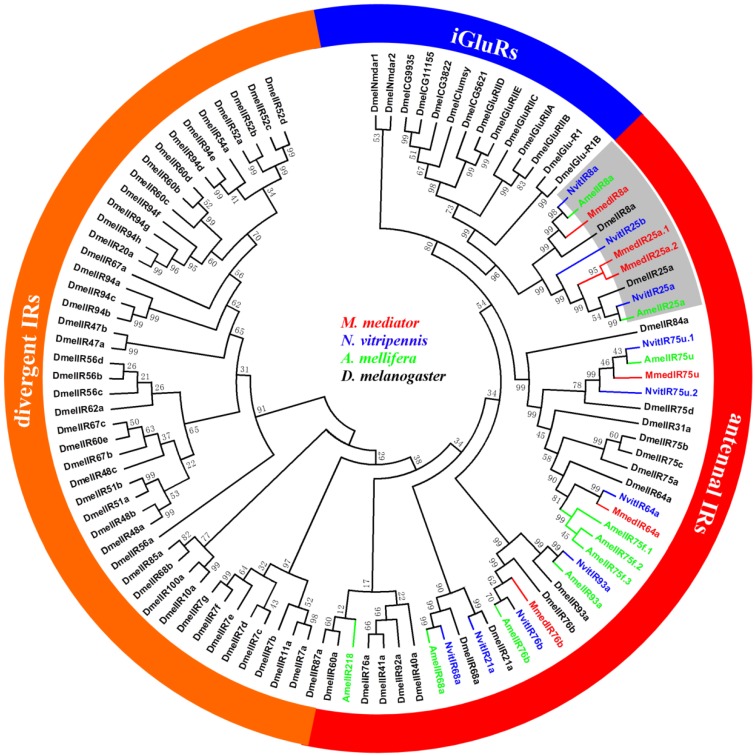
The neighbor-joining tree of IRs suggests that the putative MmedIR sequences did not cluster with iGluRs, and they generally tend to group with other IRs (Figure 5). All of the MmedIRs show clear homology to antennal IRs in N. vitripennis. Among the six antennal IRs, three putative MmedIRs (MmedIR8a, MmedIR25a.1 and MmedIR25a.2) clustered in the highly conserved IR8a and IR25a subfamilies. Comparative analysis revealed that the amino acid identity of the IR8a is between 38.0 % and 56.6 %. Likewise, the pair-wise sequence identity of the IR25 ranged from 35.3 % to 54.6 % among the species (NvitIR25b, which has a short N-terminus, was not included). Additionally, MmedIR25a.1 and MmedIR25a.2 share 54.6 % of their amino acids. Alignment of MmedIR8a and MmedIR25a amino acid sequences with related proteins in the other species are shown in Figure 6 and Figure 7.
Figure 5.
Phylogenetic relationship of candidate ionotropic receptor (IR) genes from M. mediator and other insects. Amel, A. melifera; Dmel, D. melanogaster; Mmed, M. mediator; Nvit, N. vitripennis.
Figure 6.
Amino acid Alignment of the MmedIR8a with IR8a sequences in other insects. Black shadings indicate amino acids which show at least 75% identity between sequences. Positions of the amino-terminal domain (ATD), the ligand-binding domain lobes (S1 and S2), the pore loop (P) and the transmembrane segments (M1, M2, M3) are marked with different colors bars according to their position in DmelIR8a. Amel, A. melifera; Dmel, D. melanogaster; Mmed, M. mediator; Nvit, N. vitripennis.
Figure 7.
Amino acid Alignment of the MmedIR25a.1/25a.2 with IR25a sequences in other insects. Black shadings indicate amino acids which show at least 75% identity between sequences. Positions of the amino-terminal domain (ATD), the ligand-binding domain lobes (S1 and S2), the pore loop (P) and the transmembrane segments (M1, M2, M3) are marked with different colors bars according to their position in DmelIR25a. Amel, A. melifera; Dmel, D. melanogaster; Mmed, M. mediator; Nvit, N. vitripennis.
Tissue- and sex-specificity of the chemosensory receptor transcripts
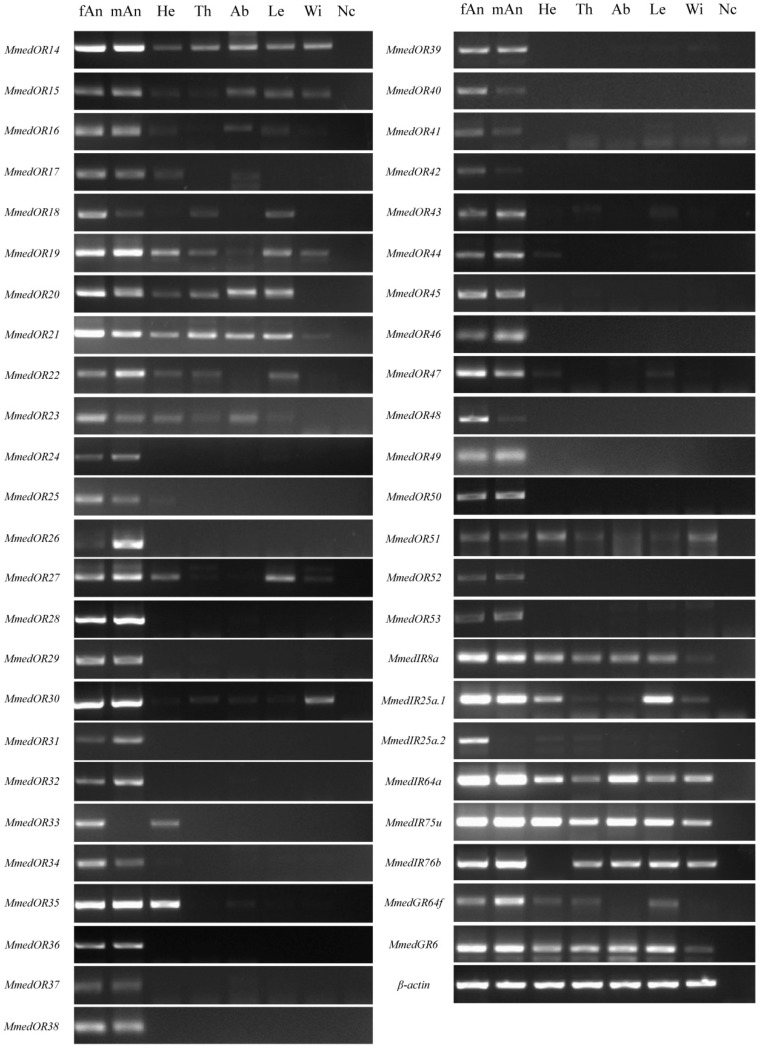
We explored the expression profiles of chemosensory receptors (ORs, GRs and IRs) in different tissues of M. mediator by using RT-PCR. The OR transcripts display different patterns in various tissues (Figure 8). The results showed that the transcripts of MmedOR24, MmedOR26, MmedOR28, MmedOR29, MmedOR31, MmedOR32, MmedOR34, MmedOR36-OR43, MmedOR45, MmedOR46, MmedOR48, MmedOR49, MmedOR50, MmedOR52 and MmedOR53 were only expressed in male and female antennae. MmedOR33 was only presented in the female antennae and head. However, the remaining ORs showed a wide range of expression patterns. Among them, the MmedOR25, MmedOR35, MmedOR44 and MmedOR47 were predominantly expressed in antennae. Compared with ORs, IRs and GRs showed a ubiquitous expression pattern except the MmedIR25a.2 which was present predominantly in the female antennae. Others appeared to be expressed in all tested tissues.
Figure 8.
Tissue- and sex- specific expression profiles of M. mediator OR, IR and GR genes. fAn: female antenna, mAn: male antenna, He: heads, Th: thoraxes, Ab: abdomens, Le: legs, Wi: wings, Nc: no template control. The β-actin was used as control for the integrity of each cDNA template.
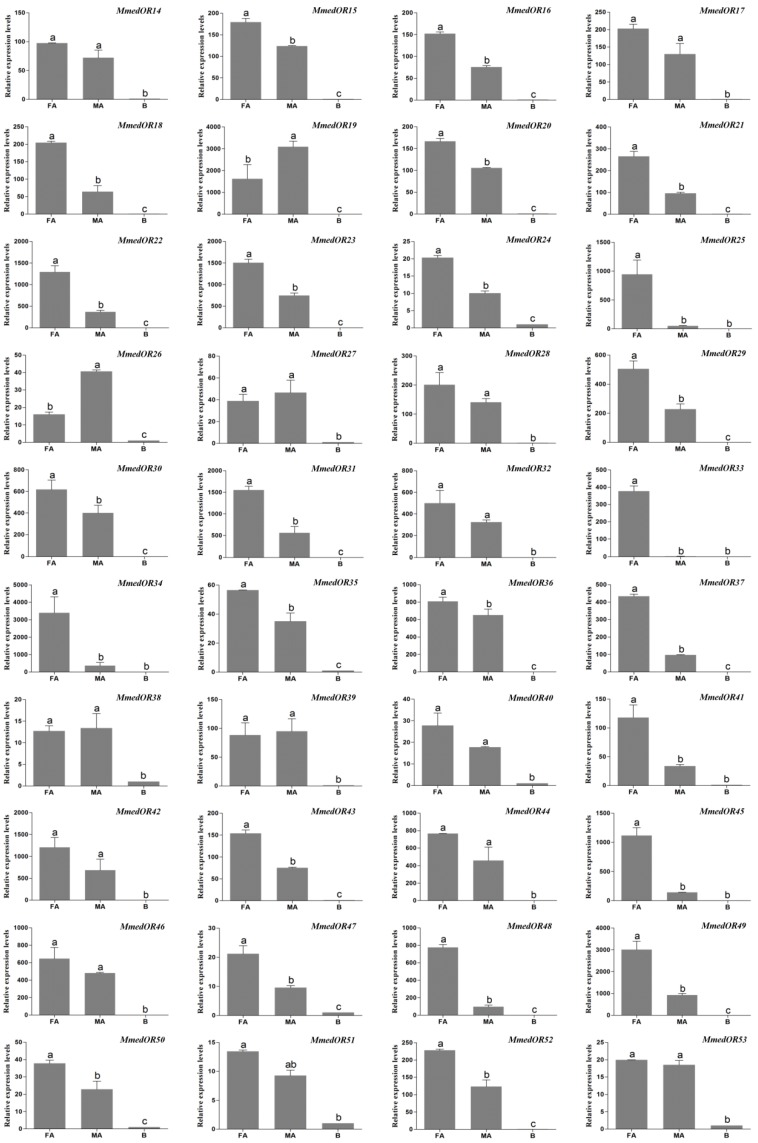
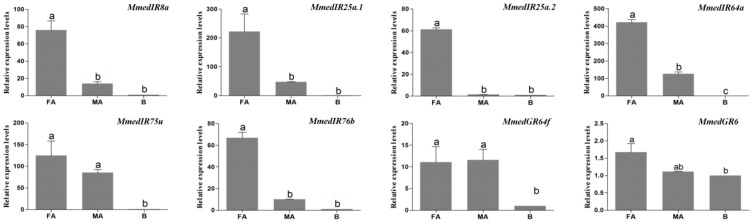
The qPCR measurement was performed to investigate the chemosensory receptor transcript abundances in the female antennae, male antennae and body tissues. By comparing expression levels in female antennae, male antennae and body tissues, we found all of the 40 putative OR genes were mainly expressed in the antennae. In addition, 25 OR genes (OR15, OR16, OR18, OR20, OR21, OR22, OR23, OR24, OR25, OR29, OR30, OR31, OR33, OR34, OR35, OR36, OR37, OR41, OR43, OR45, OR47, OR48, OR49, OR50, OR52) were more highly expressed in female antennae, whereas two ORs, OR19 and OR26, were more highly expressed in male antennae. The remaining 13 OR genes (OR14, OR17, OR27, OR28, OR32, OR38, OR39, OR40, OR42, OR44, OR46, OR51, OR53) had similar expression levels between the antennae of two sexes (Figure 9). MmedGR64f was highly expressed in the female antennae, whereas MmedGR6 was expressed at a similar level in all three tested tissues. Six IR genes had higher expression levels in antennae than that in body parts. The MmedIR8a, MmedIR25a.1, MmedIR25a.2, MmedIR64a and MmedIR76b showed higher expression levels in female antennae than those in male antennae. In addition, MmedIR75u had similar expression levels between the antennae of both sexes (Figure 10).
Figure 9.
Transcript abundances of M. mediator OR genes. FA: female antennae, MA: male antennae, B: body. Transcription levels of OR genes were normalized by β-actin, and normalized transcript levels to that of wasp bodies. The error bar represents standard error and the different small letters above each bar indicate significant differences in transcript abundances (p < 0.05). The same as below.
Figure 10.
Transcript abundances of M. mediator IR and GR genes. FA: female antennae, MA: male antennae, B: body.
Discussion
In the present study, we focused on three crucial families of chemosensory receptor (ORs, GRs and IRs) due to their significance as potential target genes for manipulating parasitoid wasps in order to improve biological pest control. From our transcriptome results, we identified 68 chemosensory receptor genes, including 60 ORs, two GRs and six IRs. Now, a total of 62 ORs (one co-receptor, Orco, and 61 odor-specific ORs) were identified in the antennae of M. mediator combined with our previous work 40. The amount of identified ORs in M. mediator is less than in A. mellifera and N. vitripennis, which have a total of 170 and 301 OR genes, respectively 48, 49. In this work, OR genes were identified only from the antennae transcriptome. The ORs expressed in other tissues might be difficult to identify from the antennae. Moreover, the lab-reared wasps have no opportunity to exposure to the odors from host insects or related plants in surroundings. Hence, we speculated that there was difficult to identify the additional ORs with low expression level or without exposing to the odors from host insects or related plants. In our previous study, we found that some OR genes of M. mediator were up-regulated after contacted with host odors (unpublished data). A recent study demonstrated that the olfactory receptor gene expression in A. mellifera was modulated by scent conditioning 56. Moreover, the factors of physiology-dependent expression may also affect the amount of identified ORs in the antennae of M. mediator. One of the putative Anopheles gambiae odorant receptors (AgOr1) was down-regulated after blood feeding and the transcript of AgOr1 was not detectable in antennae after blood feeding 57. Overall, all of the factors mentioned above may influence the identification of the repertoires of ORs in M. mediator by antennae transcriptome sequencing.
Most ORs in insects are extensively expressed in the antennae 14. In our study, some ORs showed obviously antennal-specific expression profile, others demonstrated ubiquitous expression characteristic but were predominantly expressed in antennae. The ORs with antennal-specific or dominant expression profiles may play crucial roles in olfactory chemoreception of wasps. Similar to the present study, it was reported that some ORs could be expressed in a variety of tissues apart the olfactory organs 58-60. Our previous work also suggests a divergent expression profile of ORs in M. mediator 40. The expression of ORs in non-olfactory tissues suggests that they may have physiological functions in other organs. In A. gambiae, the Orco expressed in the testes is involved in mediating activation of spermatozoa 59. In M. mediator, there are differences in OR genes expression between the antennae of males and females. Twenty five of 40 OR genes are much highly expressed in female antennae, which may be involved in the perception of volatile semiochemicals during host location process.
Because GR expression levels are very low and mainly expressed in gustatory organs 25, 28, only a small number of putative GR-encoding transcripts were identified from the antennae of M. mediator. To our knowledge, this is the first report on the GR genes in wasp antennae, although some studies described the distribution of some gustatory sensilla in wasp antennae 1, 2, 6. The identified GRs included a GR64f orthologue, which clusters with the sugar receptor GR1 subfamily in Hymenoptera, suggesting that the antennae of M. mediator participate in sugar detection. In addition, the high expression level of MmedGR64f in the antennae also indicated a gustatory function of the antennae.
In D. melanogaster, 15 of the 66 IR genes were classified as antennal IRs 10. In our study, all the identified MmedIRs were belonging to the antennal IR subfamily. There was no MmedIRs grouped with the divergent IRs. In the genome of N. vitripennis, there was also no orthologue-divergent IRs 10, 50. In insects, IR8a and IR25a are two most conserved IRs 10. In the N. vitripennis genome, there were two putative IR25a orthologues 10. Interestingly, we also identified two putative IR25a orthologues in M. mediator, which share 54.6% amino acid sequence identity. However, there is no any suggestion about divergent function of IR25a orthologues to date. Compared with Diptera insects, there were very few IR genes identified in Hymenoptera insects. There were only ten IR genes identified in A. mellifera and N. vitripennis, however, there were 66 IRs in D. melanogaster 10. In our work, six IR genes were identified in M. mediator, a number that is close to the amount of AmelIRs or NvitIRs. Most of the IR genes were expressed higher in the female antennae than in male antennae indicating that IR-mediated signaling may play more important roles in host recognition behavior. However, the ubiquitous expression feature of MmedIRs revealed that these genes may be involved in other physiological functions in non-olfactory organs.
Female M. mediator uses different types of host-related volatile cues for foraging and host location. Host-plant volatiles (green leaf volatiles and herbivore-induced plant volatiles) play an important role in the initial searching behavior of female wasps 2. The high expression of ORs in female antennae may be important in recognition of host volatile chemicals, which provide a starting point for the potential use of ORs as targets to manipulate wasp host location behavior and develop novel crop protection strategies. In further study, we will investigate the binding function of candidate ORs with known host-plant volatiles, such as 3, 7-dimethyl-1, 3, 6-octatriene, (Z)-3-hexenyl acetate and nonanal, which have significant electrophysiological and taxis behavioral effects on M. mediator 39. The findings in this work may help to shed light on the chemoreception mechanisms of M. mediator.
Supplementary Material
Figures S1, Tables S1-S3.
Acknowledgments
This work was supported by the China National Basic Research Program (2012CB114104) and the National Natural Science Foundation of China (31171858, 31272048 and 31321004). YJ Zhang and JJ Zhou acknowledge the financial support from the international joint project between China and the UK (31111130203; JP100849). This manuscript has been edited by the native English-speaking experts of Elsevier Language Editing Services.
References
- 1.Quicke DL. Parasitic wasps. Chapman & Hall Ltd; 1997. [Google Scholar]
- 2.Godfray HCJ. Parasitoids: Behavioral and Evolutionary Ecology. Princeton University Press; 1994. [Google Scholar]
- 3.Vet LEM, Dicke M. Ecology of Infochemical Use by Natural Enemies in a Tritrophic Context. Annual review of entomology. 1992;37:141–72. [Google Scholar]
- 4.Turlings TC, Tumlinson JH, Lewis WJ. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science. 1990;250:1251–3. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- 5.De Moraes CM, Lewis WJ, Pare PW, Alborn HT, Tumlinson JH. Herbivore-infested plants selectively attract parasitoids. Nature. 1998;393:570–3. [Google Scholar]
- 6.Barbarossa IT, Muroni P, Dardani M, Casula P, Angioy AM. New insight into the antennal chemosensory function of Opius concolor (Hymenoptera, Braconidae) Ital J Zool. 1998;65:367–70. [Google Scholar]
- 7.Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139:45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–33. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 9.Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annual review of physiology. 2009;71:307–32. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- 10.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. Plos Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 2):14537–42. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–9. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- 13.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–38. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 14.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–36. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 15.Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Current biology: CB. 2001;11:822–35. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 16.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–62. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. Plos Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundin C, Kall L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR. et al. Membrane topology of the Drosophila OR83b odorant receptor. Febs Lett. 2007;581:5601–4. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV. et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect biochemistry and molecular biology. 2008;38:770–80. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Dahanukar A, Hallem EA, Carlson JR. Insect chemoreception. Current opinion in neurobiology. 2005;15:423–30. doi: 10.1016/j.conb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–6. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 23.Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–11. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 24.Stengl M, Funk NW. The role of the coreceptor Orco in insect olfactory transduction. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2013;199:897–909. doi: 10.1007/s00359-013-0837-3. [DOI] [PubMed] [Google Scholar]
- 25.Scott K, Brady R Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C. et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–73. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 26.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 27.Popescu A, Couton L, Almaas TJ, Rospars JP, Wright GA, Marion-Poll F. et al. Function and central projections of gustatory receptor neurons on the antenna of the noctuid moth Spodoptera littoralis. J Comp Physiol A. 2013;199:403–16. doi: 10.1007/s00359-013-0803-0. [DOI] [PubMed] [Google Scholar]
- 28.Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830–4. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- 29.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Current biology: CB. 2004;14:1065–79. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Freeman EG, Wisotsky Z, Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1598–603. doi: 10.1073/pnas.1311724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montell C. A taste of the Drosophila gustatory receptors. Current opinion in neurobiology. 2009;19:345–53. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rytz R, Croset V, Benton R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect biochemistry and molecular biology. 2013;43:888–97. doi: 10.1016/j.ibmb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Ai M, Blais S, Park JY, Min S, Neubert TA, Suh GS. Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J Neurosci. 2013;33:10741–9. doi: 10.1523/JNEUROSCI.5419-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–67. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS. et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357–75. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mir Khan S. Effectiveness of Microplitis mediator (HYM.: Braconidae) against its hosts Agrotis segetum and A. ipsilon (Lepidoptera: Noctuidae) Pakistan Journal of Biological Sciences. 1999;2(2):344–346. [Google Scholar]
- 37.Mason PGE, M A Youngs, B J. Effects of Parasitism by Banchus flavescens (Hymenoptera: Ichneumonidae) and Microplitis mediator (Hymenoptera: Braconidae) on the Bertha Armyworm, Mamestra configurata (Lepidoptera: Noctuidae)1. Journal of Hymenoptera Research; 2001. p. 10. [Google Scholar]
- 38.Li JC, Yan FM, Coudron TA, Pan WL, Zhang XF, Liu XX. et al. Field release of the parasitoid Microplitis mediator (Hymenoptera: Braconidae) for control of Helicoverpa armigera (Lepidoptera: Noctuidae) in cotton fields in Northwestern China's Xinjiang Province. Environmental Entomology. 2006;35:694–9. [Google Scholar]
- 39.Yu H, Zhang Y, Wyckhuys KA, Wu K, Gao X, Guo Y. Electrophysiological and behavioral responses of Microplitis mediator (Hymenoptera: Braconidae) to caterpillar-induced volatiles from cotton. Environ Entomol. 2010;39:600–9. doi: 10.1603/EN09162. [DOI] [PubMed] [Google Scholar]
- 40.Ma L, Gu SH, Liu ZW, Wang SN, Guo YY, Zhou JJ. et al. Molecular characterization and expression profiles of olfactory receptor genes in the parasitic wasp, Microplitis mediator (Hymenoptera: Braconidae) Journal of insect physiology. 2014;60:118–26. doi: 10.1016/j.jinsphys.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Lassmann T, Hayashizaki Y, Daub CO. TagDust--a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–40. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- 43.Chen YA, Lin CC, Wang CD, Wu HB, Hwang PI. An optimized procedure greatly improves EST vector contamination removal. Bmc Genomics. 2007;8:416. doi: 10.1186/1471-2164-8-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T. et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–59. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 47.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–8. [Google Scholar]
- 48.Robertson HM, Gadau J, Wanner KW. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 2010;19(Suppl 1):121–36. doi: 10.1111/j.1365-2583.2009.00979.x. [DOI] [PubMed] [Google Scholar]
- 49.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Slone JD, Rokas A, Berger SL, Liebig J, Ray A. et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. Plos Genet. 2012;8:e1002930. doi: 10.1371/journal.pgen.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Kent LB, Robertson HM. Evolution of the sugar receptors in insects. Bmc Evol Biol. 2009;9:41. doi: 10.1186/1471-2148-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–16. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claudianos C, Lim J, Young M, Yan S, Cristino AS, Newcomb RD. et al. Odor memories regulate olfactory receptor expression in the sensory periphery. Eur J Neurosci. 2014;39:1642–54. doi: 10.1111/ejn.12539. [DOI] [PubMed] [Google Scholar]
- 57.Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14693–7. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu SH, Sun L, Yang RN, Wu KM, Guo YY, Li XC. et al. Molecular Characterization and Differential Expression of Olfactory Genes in the Antennae of the Black Cutworm Moth Agrotis ipsilon. PloS one. 2014;9:e103420. doi: 10.1371/journal.pone.0103420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pitts RJ, Liu C, Zhou X, Malpartida JC, Zwiebel LJ. Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2566–71. doi: 10.1073/pnas.1322923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson MN, Videvall E, Walden KK, Harris MO, Robertson HM, Lofstedt C. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae) Bmc Genomics. 2014;15:501. doi: 10.1186/1471-2164-15-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1, Tables S1-S3.