Abstract
The gypsy moth, Lymantria dispar, is an important economic pest that causes large-scale damage to forests worldwide. Because of its important role in initiating and controlling insect behavior, olfaction—and olfaction-based pest management—has drawn increasing attention from entomologists. In this study, we identified the gene that encodes the olfactory receptor co-receptor (OrCo). Through amino acid sequence alignment, we found that LdisOrCo shares high identity with other OrCo proteins from different insect orders. Next, we performed RNA-interference (RNAi) to assess the role of OrCo in olfaction. Electroantennographic assays showed that after RNAi, the average value of males' response to sex pheromones was 0.636 mV, significantly lower than that of the positive control (average = 1.472 mV). Females showed no response to sex pheromones before or after RNAi. Finally, quantitative PCR showed a strong decrease in the expression of OrCo after RNAi, by ~74% in males and by 23% in females relative to the positive controls. These results indicate that OrCo is not only critical to odor recognition, but it may also represent a new target for development of semiochemicals that can influence insect behavior.
Keywords: Lymantria dispar, RNA interference, OrCo, EAG, qPCR
Introduction
Insects are among the most abundant and diverse multicellular animals, with approximately one million species identified to date, and they play an important role in the biosphere. Numerous insect pests can cause severe damage to agricultural crops, forests, and humans. This paper focuses on the gypsy moth, Lymantria dispar, an economically important insect that can severely disrupt forested ecosystems worldwide. The gypsy moth population cycle rises and falls, alternating between low numbers and large-scale outbreaks; during outbreaks, the moths cause extensive damage to plant foliage that can lead to massive tree death 1. Most insect behaviors, such as mating, feeding, and nesting, are closely related to olfaction. Odorant-binding proteins and olfactory receptors (Ors) are primarily involved in semiochemical transduction 2-7. Olfactory receptor complexes of unknown stoichiometry consist of a conserved ion channel named olfactory receptor co-receptor (Orco) 8-11 and a diverse ligand-binding olfactory receptor molecule (Ors) 2, 8, 12. OrCo proteins have been found in Lepidoptera, Coleoptera, Hymenoptera, and Diptera 13, 14. OrCo amino acid sequences are highly conserved among insects, in contrast to traditional olfactory receptors, suggesting that these proteins have specific, key functions in different insect species. Knockout of Ors has revealed that OrCo proteins are required for insect olfaction 15-17.
While there is agreement that Orco locates and maintains Ors on the dendritic membranes of olfactory receptor neurons, there is no consensus on the physiological function of Orco in the olfactory signal transduction cascade. There are three theories about Orco function. The first is that Ors and Orco form a ligand-gated olfactory receptor ion channel complex that underlies ionotropic odor transduction only in insects 18. The second theory suggests a mixed ionotropic and metabotropic odor transduction cascade with Ors coupling to G-proteins 19. The final theory suggests that Orco is not involved in ionotropic odor transduction but rather forms an ion channel that controls spontaneous activity, sensitivity, and the time course of odor transduction 17.
Here, we first identified the OrCo gene in the gypsy moth, and we analyzed evolutionary relationships between OrCo genes in different species. Next, we knocked down OrCo genes with RNAi. The response of mutant moths to sex pheromones, measured with the EAG method, was compared to that of wild-type moths. OrCo expression was then verified with qPCR, and its role in the olfactory mechanism in gypsy moths was further clarified.
Materials and methods
Insect samples
Male and female gypsy moths were obtained from the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry, Beijing, China. Antennae were stored in RNAlater® RNA Stabilization Solution (Invitrogen) at -80 °C.
RNA extraction and cDNA synthesis
Frozen antennae from males and females were ground under liquid nitrogen. Then, following the protocols for an RNA simple Total RNA Kit (Invitrogen, Germany), total RNA was eluted in 40 µL RNase-free water and stored at -80 °C.
The first strand of complementary DNA (cDNA) was synthesized using a Quantscript RT Kit. Total RNA solution, the template, was transcribed into cDNA in a 20-µL reaction volume containing 2 µL (10 μM) oligo-dT15 primer, 2 µL 10× RT Mix, 2 µL (2.5 mM each) Super Pure dNTP, 1 µL Quant reverse transcriptase, and 3 µL RNase-free ddH2O. This synthesis was performed for 60 min at 37 °C followed by storage at -20 °C.
Cloning of OrCo sequences from Lymantria dispar
cDNA were prepared as described above. Because of the conservation of OrCo in other Lepidopterans, degenerate primer pairs were designed according to the conserved sequence as follows:
OR1F, 5'-YTHATYGARGAGAGYTCATC-3',
OR1R, 5'-GYTGBAYCAAYACCATGAAG-3',
OR2F, 5'-GVWCHGCBATMAARTAYTGGG-3',
OR2R, 5'-TCCATHACBGATGARCTCTC-3',
OR3F, 5'-GAVGTCAAYGADCTVACVGC-3',
OR3R, 5'-ACGACATGCTTATGCCTCTC-3',
PCR amplification was performed with the following conditions: a denaturing step at 95 °C for 5 min, followed by 35 cycles (95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s), and a final step of 7 min at 72 °C. The 200-bp PCR product was purified and ligated into a pUC 19-T vector (Takara, China). After transformation, the competent cells were plated on LB/agar containing ampicillin, isopropyl thio-β-D-galactoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-Gal). White colonies were then grown in liquid LB/ampicillin and analyzed for the presence of the positive clone. More than 10 independent clones were sequenced.
Based on the sequence of the fragment obtained by reverse transcription-polymerase chain reaction (RT-PCR), gene-specific primers were synthesized and used for rapid amplification of cDNA ends (RACE). RACE was performed using the RLM-RACE kit (Ambion) according to the manufacturer's protocol. A first 35-cycle nested PCR was performed using the 5' RACE outer primer 5'-CATGGCTACATGCTGACAGCCTA-3' in combination with the specific primer 5'1, CCCAGTGTATAACCGAGATAGCCG. The second 35-cycle nested PCR was performed using the sequence 5'-CGCGGATCCACAGCCTACTGATGATCAGTCGATG-3' as the 5' RACE inner primer in combination with sequence 5'2, CAGAGTAGTACCGTATGTGTCTCC. For 3' RACE, a first 35-cycle nested PCR was performed using the 3' RACE outer primer 5'-TACCGTCGTTCCACTAGTGATTT-3' in combination with the specific primer 3'1, GGAGACACATACGGTACTACTCTG. The second 35-cycle nested PCR was performed using the sequence 5'-CGCGGATCCTCCACTAGTGATTTCACTATAGG-3' as the 3' RACE inner primer in combination with sequence 3'2, CGGCTATCTCGGTTATACACTGGG. Each of the 35 cycles consisted of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C. The amplified product was purified, cloned, and sequenced on both strands.
Sequence analysis
We conducted homology analysis on the NCBI website, sequence analysis on DNAMAN and multiple aligned sequences homology analysis on CLC Sequence Viewer 6. The TMHMM program (http://www.cbs.dtu.dk/services/TMHMM) was used to analyze transmembrane domains of OrCo. The phylogenetic tree of insect odorant receptors was built using Mega5.1 using the neighbor-joining method with Bootstrap 20.
RNAi of OrCo genes
The cDNA products were prepared, the double-stranded RNA (dsRNA) of OrCo genes was generated using the cDNA template and MEGAscript RNAi kit, and the new primers were designed using Primer Premier 5.0 (Table 1). The concentration of dsRNA (three measurements) was 615 ng/µL for females and 650 ng/µL for males. Approximately 0.5 µL (400 ng) of dsRNA was injected through the thorax membranes into the pupae with a microinjector.
Table 1.
Primers for generating dsRNA of OrCo genes.
| Primer | Sequence (5'-3') |
|---|---|
| OrCo-Forward | TAATACGACTCACTATAGGGAGAATAACCCTAATGGACTTTCA |
| OrCo-Reverse | TAATACGACTCACTATAGGGAGAAAATAAGTTACCACAGCACC |
Four treatments were tested using the injection volumes shown in Table 2 and an injection depth of 2-3 mm. Each kind of dsRNA was injected into 15 moths. All pupae were injected once every 2 days until emergence. The antennae of 2- or 3-day-old adults were removed; one antenna was used to analyze the response to the pheromone using EAG, and the other was collected for relative qRT-PCR.
Table 2.
Injection volume of dsRNA in each treatment.
| Treatment | Sex | Number of pupae | OrCo (µL) |
ddH2O (µL) |
Total volume (µL) |
|---|---|---|---|---|---|
| ddH2O (Control) | Female | 15 | 0 | 1.75 | 1.75 |
| Male | 15 | 0 | 1.70 | 1.70 | |
| OrCo | Female | 15 | 0.65 | 1.10 | 1.75 |
| Male | 15 | 0.62 | 1.08 | 1.70 |
EAG Assay
The high-efficiency sex pheromone used in the EAG assay was produced by Trece Inc. (Adair, OK). We then used n-hexane to dissolve and dilute the compound in a concentration gradient of 0.00005, 0.0005, 0.005, 0.05, and 0.5 mg. Considering the differential sensitivity of antennae, the response value to 1 mg of sex pheromone was regarded as a reference control (i.e., 100%); all other values were obtained relative to the reference.
Both ends of adult antennae were cut and blocked with a drop of conductive adhesive, and the basal part was linked to a reference electrode while the distal end was connected to the recording electrode. Both electrodes were connected to an amplifier (Syntech, Hilversum, the Netherlands). Charcoal-filtered, humidified air was delivered at a constant flow rate of 150 mL/min; the stimulus flow was 2.5 mL/s, and the time of stimuli was 0.5 s with a 40-s interval. Chemical solutions (10 μL) were added to 1 cm × 2 cm filter paper; the folded papers were put into glass tubes as a stimuli source, and the same volume of n-hexane was added as a solvent control. To verify the more sensitive concentrations, the wild moths were measured with different concentrations of pheromone. The treated antennae were then analyzed with the more sensitive concentration of pheromone, and the antennae were stimulated alternately, three times, with pheromone and solvent. A minimum of three male or female antennae were measured for each treatment, and the reaction values were recorded. The antennae were stimulated three times with 0.5 mg sex pheromone dissolved in n-hexane (ck) before and after pheromone stimulation.
Real-time relative quantitative PCR
We collected another set of gypsy moth antenna that had been treated with dsRNA. RNA extraction and synthesis of first-strand cDNA were performed according to methods described above. The primers used for the OrCo gene and the reference actin gene are shown in Table 3. A Roche LightCycler 480II was used for qPCR.
Table 3.
Primers for qPCR of OrCo and actin genes.
| Primer | Sequence (5'-3') |
|---|---|
| Actin-Forward | GGGAAATCGTGCGTGAC |
| Actin-Reverse | GAAGGAAGGCTGGAAGAG |
| OrCo-Forward | GCAGAGTTATTCCGAGCAT |
| OrCo-Reverse | GTCCATTAGGGTTATTTCCA |
The optimized qPCR conditions (annealing temperature, primer density, and reaction time) consisted of a denaturing step at 94 °C for 5 min; followed by 40 cycles of 94 °C for 40 s, 55 °C for 30 s, 72 °C for 30 s; a melting curve program including 95 °C for 1 min, 65 °C for 1 min, 95 °C for continuous; and a final step of 10 s at 40 °C. Fluorescence signals were measured during the annealing steps of each cycle.
The reaction system was prepared in a 96-well plate; the components are shown in Table 4. Each treatment included three replicates on one 96-well plate, and each treatment was repeated three times.
Table 4.
Components of qPCR reaction system.
| Reagent | Quantity | Final concentration |
|---|---|---|
| SYBR® Premix Ex TaqTM (2×) | 12.5 μL | 1× |
| PCR Forward Primer (10 μM) | 1 μL | 0.4 μM |
| PCR Reverse Primer (10 μM) | 1 μL | 0.4 μM |
| Template (cDNA) | 1 μL | |
| dH2O | 9.5 μL | |
| Total | 25 μL | |
The relative qPCR were analyzed using the Advanced Relative Quantification method. The cycle threshold (CT) was calculated, and the 2-ΔΔCT method was used to show the differential expression.
Results
Identification of olfactory receptors (Ors)
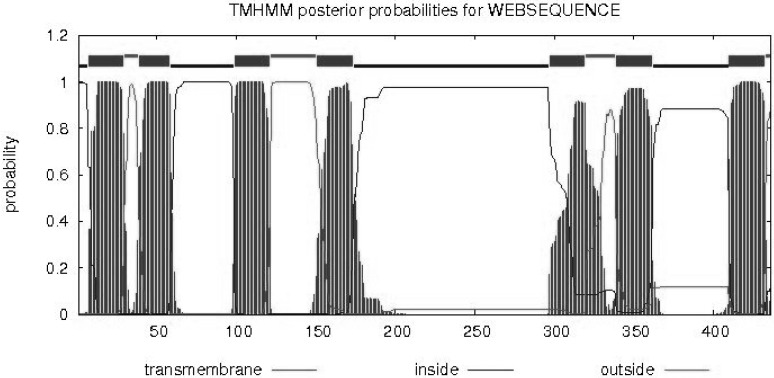
cDNA was obtained by reverse transcription and was used as a template for the PCR, and degenerate primers were designed. We obtained a 200-bp fragment that was connected with the pGEM-T easy vector. The vector was transformed into E. coli DH5α, and 10 colonies were sequenced. According to the BLAST homology search, the target sequence belongs to Lepidoptera olfactory receptor genes. The primers for 3' RACE and 5' RACE were designed based on the fragment sequence. The entire OrCo sequence was 1532 bp in length and was named LdisOrCo (GenBank accession number KF482409). The open reading frame was 1311 bp (436 amino acids; Fig. 1). The transmembrane structure indicated that the amino acids have seven putative transmembrane regions with α helices, similar to a typical G-protein coupled receptor (Fig. 2).
Figure 1.
Nucleotide and amino acid sequences of the OrCo gene in Lymantria dispar.
Figure 2.
Analysis of transmembrane region of the OrCo gene in Lymantria dispar.
Previous studies have shown that the OrCo receptor is highly conserved in insect evolution, and therefore known Lepidopteran relatives were selected for alignment, including bollworm (Helicoverpa armigera Or83b), tobacco budworm (Helicoverpa assulta Or83b), and cutworm (Spodoptera litura Or83b). Sequences of representative insects from Coleoptera, Diptera, and Hymenoptera were also chosen for the alignment. Our results showed that the LdisOrCo gene was 89% homologous with the related species (Spodoptera littoralis Or83b), 64% homologous with Taiwanese black beetles (Holotrichia plumbea Or83b), and >60% homologous with other insects (Fig. 3). The C-terminal was highly conserved.
Figure 3.
Alignment of amino acid sequences of LdisOrCo to selected OrCo proteins of various insect species. Amino acids that were identical in all sequences are shown in consensus lines. GenBank accession numbers for olfactory receptors are as follows: Lymantria dispar (Ldis,OrCo,mRNA,KF482409); Helicoverpa armigera (Harm,Or83b,mRNA,HQ186284); Helicoverpa assulta (Hass1,Or83b,mRNA,HQ186285); Helicoverpa assulta (Hass2,Or83b,mRNA,EU057178); Helicoverpa zea (Hzea,Or83b,mR-NA,AY843204); Heliothis viriplaca (Hvir,Or83b,mRNA,JQ394904); Spodoptera litura (Slit,Or83b,mRNA,JQ811935); Plutella xylostella (Pxyl,Or83b,mRNA,GQ923610); Bactrocera dorsalis (Bdor,Or83b,mRNA,EU621792); Bactrocera cucurbitae (Bcuc,Or83b,mRNA,HM745934); Ceratitis capitata (Ccap,Or83b,mRNA,AY843206); Haematobia irritans (Hirr,Or83b, mRNA,EU622915); Stomoxys calcitrans (Scal,Or83b,mRNA,EU622914); Drosophila melanogaster (Dmel,Or83b,mRNA,AY567998); Anopheles gambiae (Agam,Or83b,mRNA,AY843205); Aedes aegypti (Aaeg,Or83b,mRNA,XM_001651376); Holotrichia plumbea (Hplu,Or83b,mRNA,HQ110087); Holotrichia oblita (Hobl,Or83b,mRNA,JF718662); Holotrichia parallela (Hpar,Or83b,mRNA,JF826514).
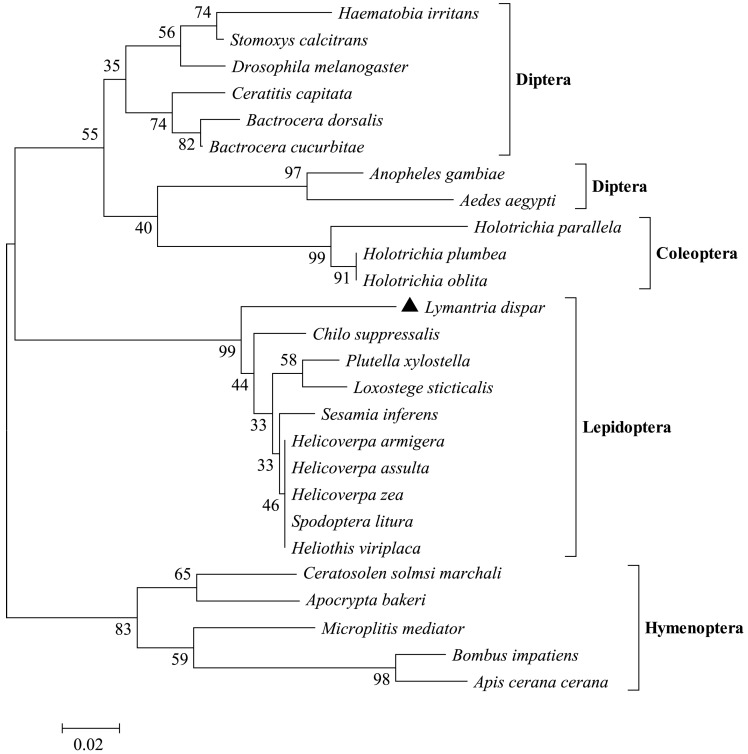
Using Mega 5.1, we performed phylogenetic analysis of odorant receptors in insects from the orders Diptera, Coleoptera, Hymenoptera, and Lepidoptera. After 1500 iterations, the evolutionary relationships between these insect orders became apparent (Fig. 4). The evolutionary tree showed that the 26 odorant receptor genes formed three major branches, with Coleoptera and Diptera belonging to one large branch, and Lepidoptera and Hymenoptera each occupying one branch. The closer evolutionary relationship between Coleoptera and Diptera was reflected in conservative characteristics for the genes of these orders. LdisOrCo had a close genetic relationship with Orco of other Lepidopteran insects.
Figure 4.
Neighbor-joining phylogenetic tree of the OrCo gene encoding olfactory receptors in various insects.
RNAi and electrophysiological assay
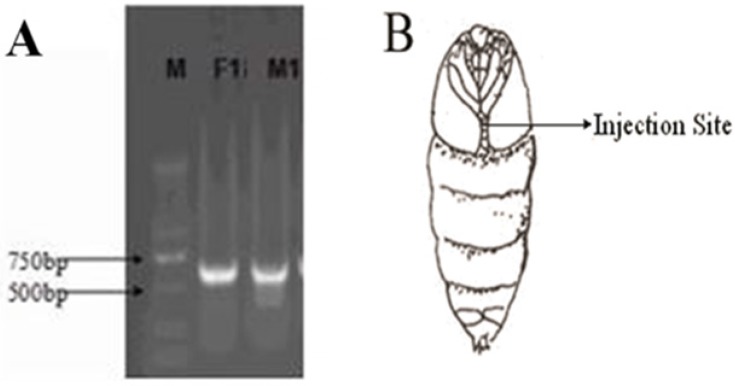
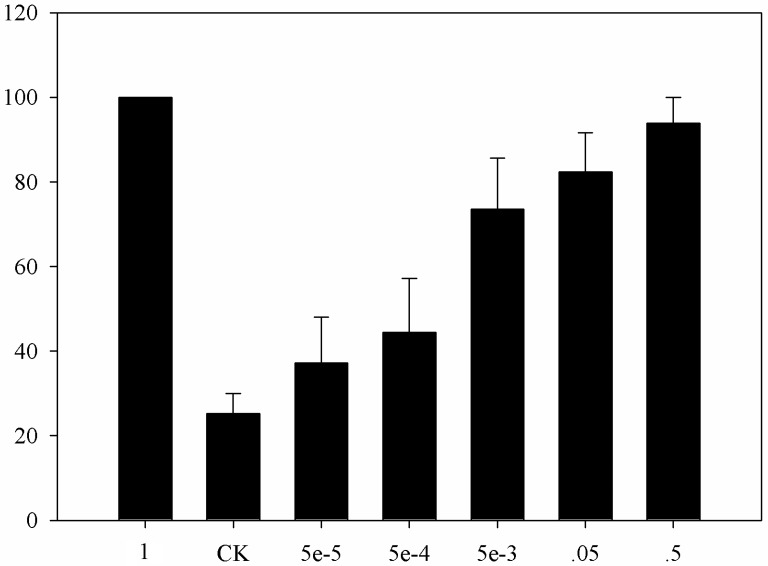
The dsRNA of each gene was detected by agarose gel electrophoresis and was approximately 600 bp (Fig. 5A), which was suitable for performing qPCR. After measuring the concentrations, 400 ng dsRNA was injected as illustrated in Figure 5B. The optimal concentration of pheromone to excite wild-type moths was screened (Fig. 6) and defined as 0.5 mg of sex pheromone.
Figure 5.
A: Electrophoretic diagram of purified dsRNA of the OrCo gene in male and female gypsy moths. M, DNA marker; F1, female OrCo; M1, male OrCo. B: dsRNA injection site.
Figure 6.
Electrophysiological recording of adult male response to different amounts of pheromone. CK indicates solvent. One milligram of sex pheromone was regarded as the reference with which the other values were compared.
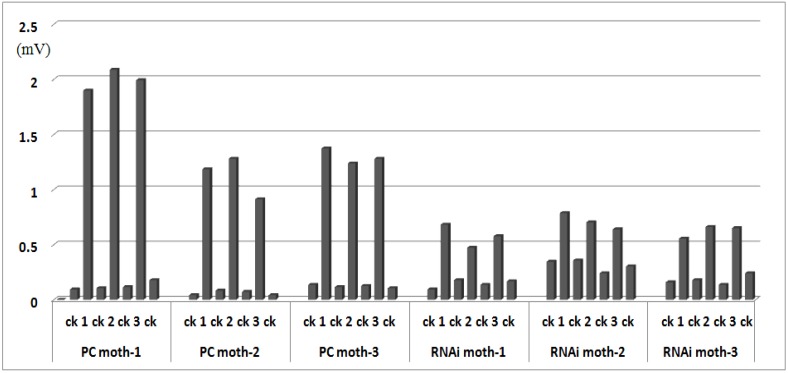
Adults (2- or 3-day-old) were analyzed by electrophysiological assay; the reaction values are illustrated in Fig.7. The average response values of positive controls injected with water were 0.101 mV± 0.003 to n-hexane and 1.472 mV±0.046 to sex pheromone, and RNAi moths injected with dsRNA were 0.212 mV±0.007 to n-hexane, 0.636 mV±0.01 to sex pheromone, decreasing 57% compared with the response of positive control moth to sex pheromone. The t-tests showed that the response to sex pheromone was significantly reduced (P < 0.05) after injection with Orco dsRNA, and the response to n-hexane was not significantly different between the positive control and RNAi moths. Because the female control did not respond to the sex pheromone, the reaction values were recorded for male antennae only.
Figure 7.
Electrophysiological response in adult males. PC indicates positive control moths injected with water; RNAi indicates treated moths injected with Orco dsRNA. Each treatment included three moths. Antennae were stimulated three times (labeled 1, 2, and 3) with 0.5 mg sex pheromone dissolved in n-hexane (ck), and ck was used to stimulate the antennae before and after pheromone stimulation.
Real-time qPCR
We conducted general PCR amplification using the primers shown in Table 3. The target OrCo fragments and actin genes were ~200 bp after sequencing. The qPCR data were further processed using the 2-ΔΔCT method to analyze differences in expression between target and reference genes (Table 5).
Table 5.
Differential gene expression in adult female and male Lymantria dispar.
| dsRNA combination | Target | Reference | Mean CT of target | Mean CT of reference | ΔCT | ΔΔCT | 2-ΔΔCT |
|---|---|---|---|---|---|---|---|
| F-Orco | Orco | actin | 32.074 | 31.284 | 0.790 | 0.711 | 0.77 ± 0.23 |
| F-CK | Orco | actin | 33.520 | 33.440 | 0.079 | 0 | 1 |
| M-Orco | Orco | actin | 30.440 | 27.724 | 2.716 | 2.316 | 0.26 ± 0.09 |
| M-CK | Orco | actin | 33.567 | 33.167 | 0.340 | 0 | 1 |
CK, Positive control; CT, Cycle threshold; ΔCT = Mean CT of target - mean CT of reference; ΔΔCT = ΔCT (Orco) - ΔCT (CK); 2-ΔΔCT shows the expression difference between Orco and CK (± SE).
Although antennae from females did not respond to sex pheromones, Orco expression was reduced and the value of 2-ΔΔCT was 0.77, 23% less than that of the control. Orco expression in males was significantly decreased, and the value of 2-ΔΔCT was 0.26, 74% lower than the control. The magnitude of the decline was slightly higher than that of the EAG value.
Discussion
In this study, we identified genes encoding OrCo proteins in Lymantria dispar. This is the first olfactory receptor gene identified in this economic pest to date. Sequence alignment and homology analysis demonstrated significant conservation of OrCo among different insects, even among different orders. As shown in Figure 3, the C-terminal domains were especially highly conserved among all aligned sequences, which indicated their relevance to the function of OrCo. OrCo is broadly expressed in most olfactory sensory neurons as the critical, constant subunit of the heteromultimeric insect Or that forms a receptor complex 13, 15, 21-24. In addition, the transmembrane topology of OrCo differs from that of conventional Ors, with the intracellular location of the N-terminus and the extracellular location of the C-terminus 19, 25. The C-terminus was presumed to strongly influence this Or-OrCo complex, and the complex was regarded as a ligand-gated ion channel 18, 19, 26.
EAG showed that female L. dispar did not respond to the sex pheromone. We suggest a two-part explanation for this finding. First, Or-based recognition of the sex pheromone are not expressed in females, and OrCo and Or cannot form the complex that is required for this function. Second, olfactory receptor neurons in female moths may only be capable of responding to general odors. qPCR showed that OrCo expression declined to different degrees in male and female moths, which could suggest that knockdown of the OrCo gene influences the response to pheromone. Similar results were reported for RNAi of Orco in Tribolium castaneum and Phyllotreta striolata 27, 28. Results showed that gypsy moths showed severe olfactory defects when the expression of OrCo was reduced, which demonstrated the crucial role of this gene in this species' olfactory sensory mechanism. EAG showed 57% decrease and qPCR 74% decrease, and the difference of decrement indicated that not all the Ors form the Orco-Or complex to recognize the chemical signals. However, this experiments made cannot distinguish between the different hypotheses suggested for the physiological function of Orco. The function is also related to temporal and spatial gene expression; Orco is expressed in antennae and in maxillary palps of larval, pupal, and adult Anopheles gambiae, and in all developmental stages of male Culex pipiens pallens 14, 29. In Lepidoptera, OrCo is specifically expressed in antenna throughout the life cycle, except for the embryonic period 30. The developmental characteristics of OrCo function in gypsy moths require further clarification.
Olfactory signaling is a target for pest management, with attempts to interfere with and thus disrupt odorant- or pheromone-driven behaviors. Many investigations of pheromones have been performed in recent decades; it has been assumed that no single, general design can be applied in developing pheromones that are analogous for all species 31. However, OrCo could be a key to controlling various insect pests. Agonistic and antagonistic effects have been investigated in insects based on the conserved Orco, such as the stimulant VUAA1 32, 33, which provides a foundation for pest control. The sequence and function of OrCo identified here in gypsy moths can inform further research on olfactory-based pest-management strategies and can be applied to improve and protect forest health.
Acknowledgments
This work was supported by the Fundamental Research Funds of the Chinese Academy of Inspection and Quarantine (Grant No. 2013JK004, 2014JK018 ), the Beijing Nova Program (Z131105000413048) and the National Science and technology support program (2015BAD08B01).
Author contributions
WL and YY designed and performed the experiments. JZ, PZ and LD performed the RNAi experiment and analyzed the qPCR and EAG data. HC and SZ wrote the manuscript.
Abbreviations
- cDNA
complementary DNA
- CT
cycle threshold
- dsRNA
double-stranded RNA
- EAG
electroantennography
- IPTG
isopropyl thio-β-D-galactoside
- OrCo
olfactory receptor co-receptor
- Ors
olfactory receptors
- qPCR
quantitative PCR
- RACE
rapid amplification of cDNA ends
- RNAi
RNA-interference
- RT-PCR
reverse transcription-polymerase chain reaction
- X-gal
5-bromo-4-chloro-3-indolyl-β-D-galactoside.
References
- 1.Whittle A, Lenhart S, White KA. Optimal control of gypsy moth populations. Bull Math Biol. 2008;70(2):398–411. doi: 10.1007/s11538-007-9260-7. [DOI] [PubMed] [Google Scholar]
- 2.Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25(8):2364–73. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- 3.Horst R, Damberger F, Luginbuhl P, Guntert P, Peng G, Nikonova L. et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci USA. 2001;98(25):14374–9. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal WS. Pheromone Reception. In: Schulz S, editor. Topics in Current Chemistry. Berlin: Springer Berlin Heidelberg; 2005. pp. 1–36. [Google Scholar]
- 5.Leal WS, Ishida Y, Pelletier J, Xu W, Rayo J, Xu X. et al. Olfactory proteins mediating chemical communication in the navel orangeworm moth, Amyelois transitella. PLoS One. 2009;4(9):e7235. doi: 10.1371/journal.pone.0007235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura N, Nakagawa T, Touhara K, Ishikawa Y. Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem Molec. 2010;40(1):64–73. doi: 10.1016/j.ibmb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307(5715):1638–42. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 8.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446(7135):542–6. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 9.Olafson PU. Molecular characterization and immunolocalization of the olfactory co-receptor Orco from two blood-feeding muscid flies, the stable fly (Stomoxys calcitrans, L.) and the horn fly (Haematobia irritans, L.) Insect Mol Biol. 2013;22(2):131–42. doi: 10.1111/imb.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patch HM, Velarde RA, Walden K, Robertson HM. A Candidate Pheromone Receptor and Two Odorant Receptors of the Hawkmoth Manduca sexta. Chem Senses. 2009;34(4):305–16. doi: 10.1093/chemse/bjp002. [DOI] [PubMed] [Google Scholar]
- 11.Vosshall LB, Hansson BS. A Unified Nomenclature System for the Insect Olfactory Coreceptor. Chem Senses. 2011;36(6):497–8. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- 12.Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S. et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci USA. 2004;101(47):16653–8. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger J, Klink O, Mohl C, Raming K, Breer H. A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189(7):519–26. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 14.Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101(14):5058–63. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–14. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Benton R. On the ORigin of smell: odorant receptors in insects. Cell Mol Life Sci. 2006;63(14):1579–85. doi: 10.1007/s00018-006-6130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stengl M, Funk NW. The role of the coreceptor Orco in insect olfactory transduction. J Comp Physiol A. 2013;199(11SI):897–909. doi: 10.1007/s00359-013-0837-3. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 19.Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452(7190):1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundin C, Kall L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR. et al. Membrane topology of the Drosophila OR83b odorant receptor. Febs Lett. 2007;581(29):5601–4. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller R, Tu Z. Odorant receptor c-terminal motifs in divergent insect species. J Insect Sci. 2008;8:53. [Google Scholar]
- 23.Krieger M, Ross KG. Identification of a major gene regulating complex social behavior. Science. 2002;295(5553):328–32. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
- 24.Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–32. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- 25.Wistrand M, Kall L, Sonnhammer EL. A general model of G protein-coupled receptor sequences and its application to detect remote homologs. Protein Sci. 2006;15(3):509–21. doi: 10.1110/ps.051745906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa T, Pellegrino M, Sato K, Vosshall LB, Touhara K. Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. Plos ONE. 2012;7(3):e32372. doi: 10.1371/journal.pone.0032372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engsontia P, Sanderson AP, Cobb M, Walden KK, Robertson HM, Brown S. The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol. 2008;38(4):387–97. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhao YY, Liu F, Yang G, You MS. PsOr1, a potential target for RNA interference-based pest management. Insect Mol Biol. 2011;20(1):97–104. doi: 10.1111/j.1365-2583.2010.01049.x. [DOI] [PubMed] [Google Scholar]
- 29.Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect Biochem Mol Biol. 2006;36(3):169–76. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Zhong G, Hu M, Yi X, Zhao H, Wang H. Molecular cloning and functional identification of an insect odorant receptor gene in Spodoptera litura (F.) for the botanical insecticide rhodojaponin III. J Insect Physiol. 2013;59(1):26–32. doi: 10.1016/j.jinsphys.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Renou M, Guerrero A. Insect parapheromones in olfaction research and semiochemical-based pest control strategies. Annu Rev Entomol. 2000;45:605–30. doi: 10.1146/annurev.ento.45.1.605. [DOI] [PubMed] [Google Scholar]
- 32.Jones PL, Pask GM, Rinker DC, Zwiebel LJ. Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci USA. 2011;108(21):8821–5. doi: 10.1073/pnas.1102425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones PL, Pask GM, Romaine IM, Taylor RW, Reid PR, Waterson AG. et al. Allosteric antagonism of insect odorant receptor ion channels. PLoS One. 2012;7(1):e30304. doi: 10.1371/journal.pone.0030304. [DOI] [PMC free article] [PubMed] [Google Scholar]