Abstract
During germ cell development, epigenetic modifications undergo extensive remodeling. Abnormal epigenetic modifications usually result in germ cell loss and reproductive defect. Prmt5 (Protein arginine methyltransferase 5) encodes a protein arginine methyltransferase which has been demonstrated to play important roles in germ cell development during embryonic stages. In the present study, we found that Prmt5 was also abundantly expressed in male germ cells after birth. Inactivation of this gene by crossing with Stra8-Cre transgenic mice resulted in germ cell loss during spermatogenesis. Further study revealed that the germ cell development was grossly normal before P10. However, most of the germ cells in Prmt5Δ/f; Stra8-Cre mice were blocked at meiotic stage. The expression of meiosis associated genes was reduced in Prmt5Δ/f; Stra8-Cre testes compared to control testes at P10. γH2AX was detected in sex body of control germ cells at P12, whereas multiple foci were observed in Prmt5-deficient germ cells. Further study revealed that H4R3me2s was virtually absent in germ cells after Prmt5 inactivation. The results of this study indicate that Prmt5 also plays important roles in germ cell development during spermatogenesis.
Epigenetic modification is one of the important mechanisms regulating the gene expression, which is involved in a number of biological processes. Epigenetic processes include DNA methylation, histone modifications, and chromatin remodeling1. In mammals, germ cell is a special cell type which is different from other cell types that constitute the animal body. During germ cell development, both genetic and epigenetic mechanisms are involved2,3,4. In mice, primordial germ cells (PGCs) first emerge inside the extra-embryonic mesoderm at around E7.255,6. The somatic gene expression program needs to be suppressed in the PGC precursors, and epigenetic modifications might be important for this process. After arriving at the genital ridge by E11.5, PGCs will undergo extensive epigenetic reprogramming. The parental imprints are erased and the gender-specific new imprints are re-established at later developmental stages7. Epigenetic modifications also play important roles in later stage of germ cell development, including meiosis initiation and maturation of gametes. In male germ cells, proper regulation of epigenetic processes not only ensure proper sperm function, but also important for proper embryonic development. It has been demonstrated that aberrant epigenetic modification in spermatogenesis has a profound effect on both male fertility and embryonic development8.
Post-translational histone modifications include methylation, acetylation, phosphorylation, ubiquitylation and sumoylation. Methylation is one of the most prevalent histone modifications monitored by histone methyltransferases9. Arginine methylation is catalyzed by protein arginine methyltransferases (PRMTs)10. PRMT family members play pivotal roles in the regulation of diverse cellular processes ranging from transcription and RNA processing to signaling transduction, cell differentiation, apoptosis and tumorigenesis11,12. Prmt5 belongs to the PRMT family and is responsible for the formation of symmetric dimethylarginine (SMDA) in arginine-rich protein motifs13. It has been reported that Prmt5 is essential for maintaining the pluripotency of mouse embryonic stem cells (ES). Deletion of Prmt5 results in the down-regulation of pluripotency transcription factors and causes embryonic lethality before implantation14. However, Prmt5 is not required to maintain pluripotency in human ES cells15. Prmt5 is also expressed in primordial germ cells (PGCs) and directs histone arginine methylation in mouse germ cells16, recent studies found that inactivation of Prmt5 in PGCs using Blimp1-Cre resulted in germ cells death before E12.517,18, suggesting that Prmt5 plays essential roles in PGCs survival.
In this study, we found that Prmt5 was also abundantly expressed in germ cells of adult testis, suggesting that histone methylation probably also plays roles in spermatogenesis. To investigate the functions of Prmt5 in later stage of germ cell development, it was specifically inactivated in male germ cells by crossing Prmt5flox mice with Stra8-Cre transgenic mice. We found that the germ cells were gradually lost after day 12 and very few germ cells were survived in adult testes. The results of this study indicate that Prmt5 is required for male germ cell survival during spermatogenesis.
Results
Prmt5 was expressed in the germ cells of testes during spermatogenesis
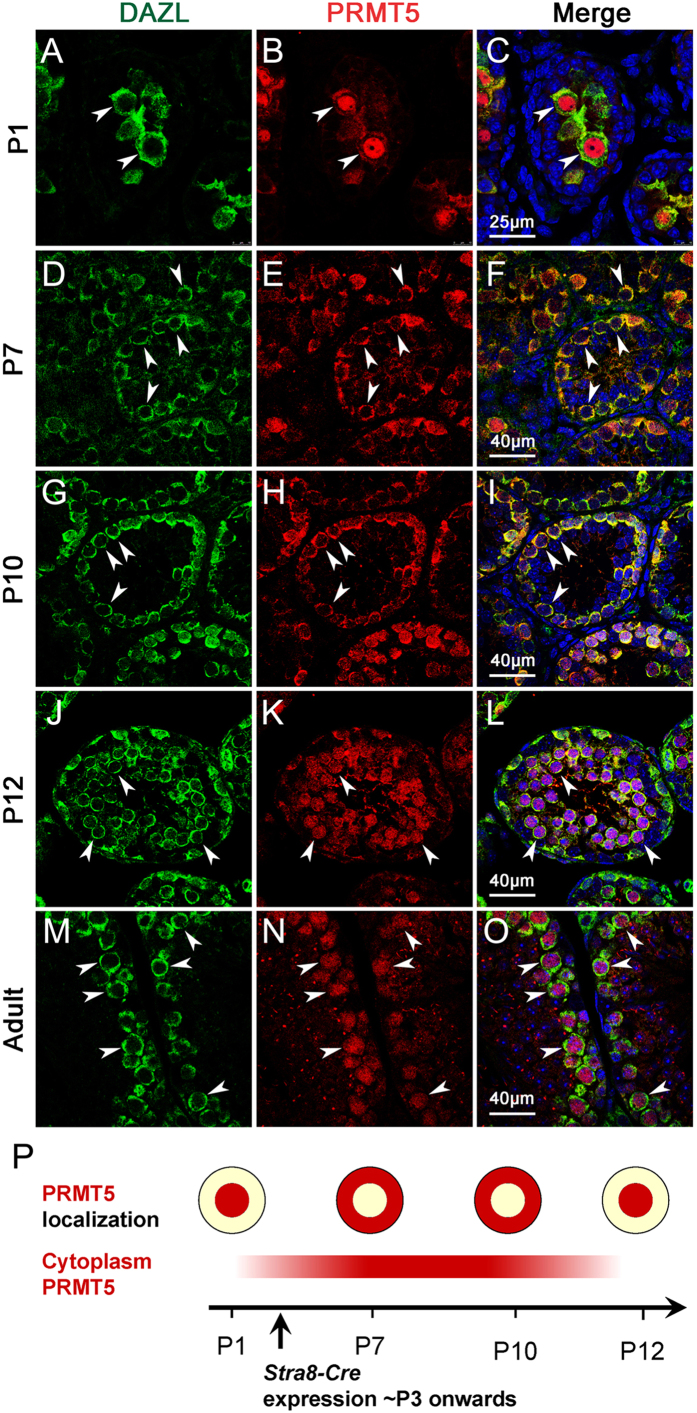
It has been reported that Prmt5 is abundantly expressed in the germ cells during embryonic stages, and inactivation of Prmt5 in PGCs results in germ cell loss in both male and female gonads16,17,18. In this study, the expression of Prmt5 in testes after birth was examined by immunofluorescence. As shown in Fig. 1, Prmt5 protein was localized in the nucleus of Dazl-posotive germ cells at P1 (B, C, white arrowheads). It was continually expressed in germ cells at P7 (E, F, white arrowheads) and P10 (H, I, white arrowheads), whereas the signal was mainly detected in cytoplasm at these stages. Interestingly, Prmt5 protein was translocated from cytoplasm to the nucleus of germ cells at P12 (K, L, white arrowheads). In adult testes, Prmt5 protein was abundantly expressed in the nucleus of spermatocytes (N, O, white arrowheads). The dynamics of nuclear-cytoplasm translocation of PRMT5 in germ cells postnatally was illustrated with the schematic diagram (Fig. 1P). These results indicate that Prmt5 is continually expressed in male germ cells postnatally, and its location is dynamic along with germ cell development.
Figure 1. The expression of Prmt5 in postnatal male germ cells was dynamic.
Immunofluorescence staining for PRMT5 (red) in testes at P1(B, C), P7(E, F), P10(H, I), P12(K, L) and adult(N, O). Germ cells were labeled with anti-DAZL antibody (A, D, G, J, M, green, white arrowheads). The nucleus were stained with DAPI (blue). P. Schematic diagram of nuclear-cytoplasm translocation of PRMT5 in male germ cells.
Inactivation of Prmt5 in germ cells resulted in germ cell loss and the defect of spermatogenesis
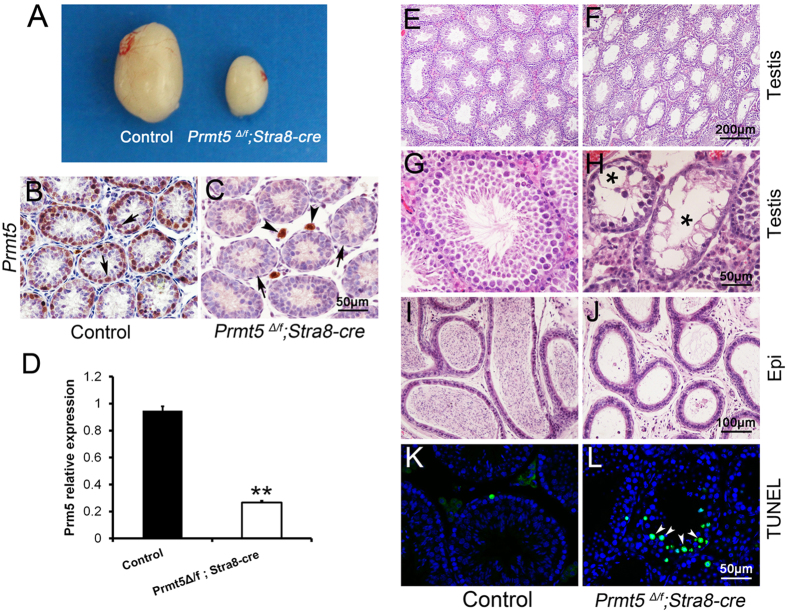
To examine the functions of Prmt5 in germ cells at later developmental stages, Prmt5flox mice were crossed with Stra8-Cre transgenic mice in which Cre is activated in germ cells of testes at approximately 3 days after birth19. Prmt5Δ/f; Stra8-Cre mice were grossly normal, no developmental defects were observed until 4 month of age. However, the size of testes from adult Prmt5Δ/f; Stra8-Cre males (Fig. 2A, right) was significantly smaller than that of control littermates (Fig. 2A , left) and most of seminiferous tubules were atrophic (Fig. 2F). High magnification images showed that very few germ cells were survived in the atrophic tubules (Fig. 2H, asterisks). In control mice, the caudal epididymis was filled with mature sperm (Fig. 2I), whereas no sperm was observed in epididymis from Prmt5Δ/f; Stra8-Cre males (Fig. 2J). A large number of TUNEL-positive apoptotic cells were noted in the seminiferous tubules of Prmt5Δ/f; Stra8-Cre mice (Fig. 2L, white arrowheads), but not in the control testes (Fig. 2K). These results indicate that Prmt5 plays important roles in spermatogenesis, and inactivation of this gene results in germ cell death.
Figure 2. Deletion of Prmt5 in the testes by Stra8-Cre resulted in massive germ cell loss.
The size of testes from adult Prmt5Δ/f; Stra8-Cre mice (A, right) was significantly smaller than that of control littermates (A, left). The protein (C) and mRNA (D) levels of Prmt5 was dramatically reduced in Prmt5Δ/f; Stra8-Cre at P10. Most of seminiferous tubules were atrophic in adult Prmt5Δ/f; Stra8-Cre testes (F, H, asterisks). No mature sperm were observed in the epididymis of Prmt5-deficient males (J). The number of apoptotic cells was significantly increased in Prmt5Δ/f; Stra8-Cre testes (L, white arrowheads). **p < 0.01.
To examine the efficiency of Stra8-Cre mediated deletion of Prmt5 gene, the expression of Prmt5 was examined by real-time PCR and immunohistochemistry. As shown in Fig. 1, Prmt5 was abundantly expressed in the germ cells of control testes at P10 (B, black arrows), whereas no PRMT5 was detected in the germ cells from Prmt5Δ/f; Stra8-Cre mice (C, black arrows) and only Leydig cells (C, black arrowheads) were labeled with anti-PRMT5 antibody. The results of immunofluorescence also showed that Prmt5 protein was absent in the germ cells of Prmt5Δ/f; Stra8-Cre testes at P7 (Fig. S1, E, F, white arrowheads). The mRNA level of Prmt5 was reduced approximately 70% in Prmt5Δ/f; Stra8-Cre testes at P10 (Fig. 2D), indicating that Prmt5 is deleted in germ cells with high efficiency.
The germ cells were gradually lost in Prmt5 Δ/f ; Stra8-Cre testes from P12 onwards
To further explore the underlying mechanisms which cause the defects of spermatogenesis in Prmt5Δ/f; Stra8-Cre mice, the histology of testes was examined by H&E staining and immunohistochemistry at early developmental stages. We found that the testes from Prmt5Δ/f; Stra8-Cre mice (Fig. S2B) was grossly normal at P10 compared to control testes (Fig. S2A). One layer of MVH-positive germ cells were observed at the peripheral region of seminiferous tubules in both control (Fig. 3A, black arrows) and Prmt5Δ/f; Stra8-Cre testes (Fig. 3B, black arrows) at this stage. Aberrant seminiferous tubules were first noted in Prmt5Δ/f; Stra8-Cre testes (Fig. S2D, asterisks) at P12, and atrophic tubules (asterisks) were observed in Prmt5-deficient testes at P14 (Fig. S2F, asterisks) and P21 (Fig. S2H, asterisks). Immunohistochemical results showed that the germ cells were localized in the lumen of seminiferous tubules from P12 to adult stage, and the number was gradually increased (Fig. 3C,E,G). By contrast, a single layer of germ cells were still located at the peripheral region in most of seminiferous tubules (Fig. 3D, asterisks) of Prmt5Δ/f; Stra8-Cre testes at P12. The number of germ cells was dramatically reduced at P21 (Fig. 3F, black arrowheads) and only a few germ cells were observed in the seminiferous tubules in testes of adult Prmt5Δ/f; Stra8-Cre males (Fig. 3H, black arrowheads). These results indicate that the germ cells were gradually lost from P12 after Prmt5 inactivation which causes the defect of spermatogenesis in Prmt5Δ/f; Stra8-Cre mice.
Figure 3. The germ cells were gradually lost in Prmt5Δ/f; Stra8-Cre testes from P12 onwards.
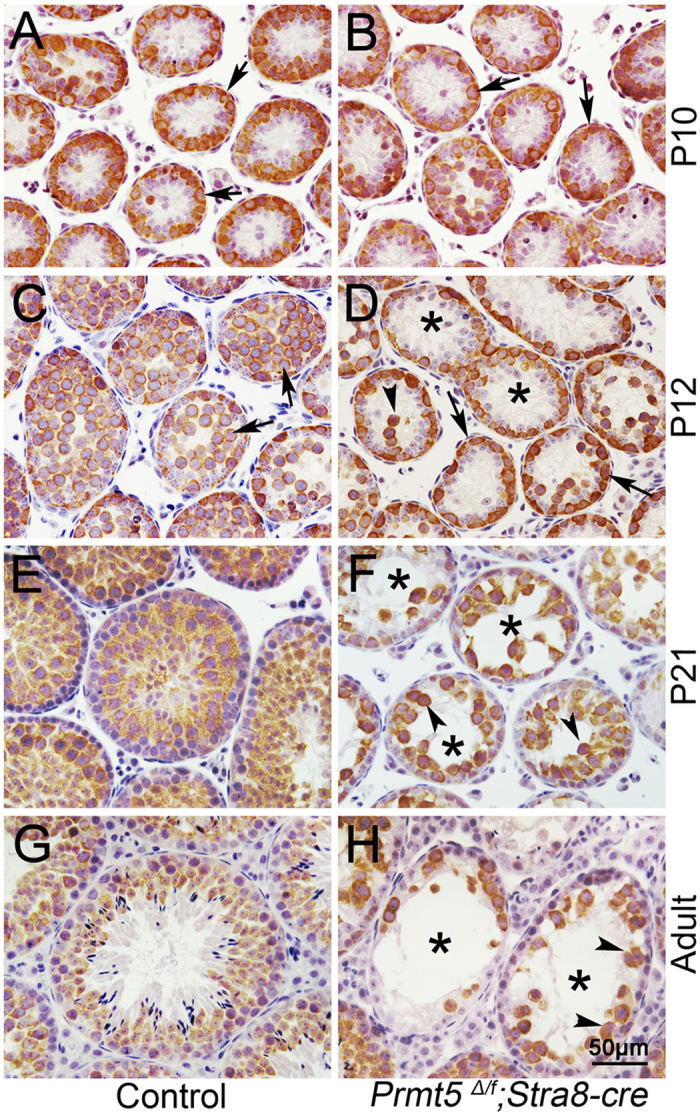
Germ cells were labeled with antibody against MVH (brown, black arrow). MVH-positive germ cells were localized at the peripheral region of seminiferous tubules in both control (A, black arrows) and Prmt5Δ/f; Stra8-Cre mice (B, black arrows) at P10. The germ cells were observed in the lumen of seminiferous tubules from P12 to adult stage in control mice (C, E, G). By contrast, single layer of germ cells were observed at the peripheral region in most of seminiferous tubules (D, asterisks) of Prmt5Δ/f; Stra8-Cre testes at P12. The number of germ cells was dramatically reduced at P21 (F, black arrowheads) and only a few germ cells were observed in the seminiferous tubules in adult Prmt5Δ/f; Stra8-Cre testes (H, black arrowheads).
Aberrant meiosis was observed in Prmt5-deficient germ cells
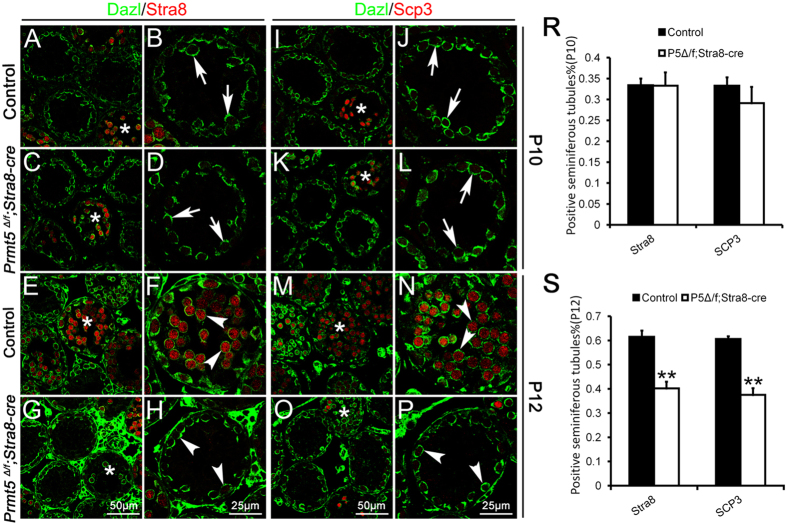
To examine whether the germ cell meiosis is affect after Prmt5 inactivation, the expression of meiosis-associated genes was analyzed by immunofluorescence and real-time PCR analysis. Stra8-positive and Scp3-positive germ cells were detected in both control (Fig. 4A,I, asterisks) and Prmt5Δ/f; Stra8-Cre testes (Fig. 4C,K, asterisks) at P10, and the percentage of seminiferous tubules containing meiotic germ cells was not changed between control and Prmt5Δ/f; Stra8-Cre mice (Fig. 4R). Stra8-positive (Fig. 4E, asterisks; Fig. 4F, white arrowheads) and Scp3-positive (Fig. 4M, asterisks; Fig. 4N, white arrowheads) germ cells were detected in most of seminiferous tubules of control testes at P12. By contrast, the number of tubules with Stra8-positive (Fig. 4G, asterisks) and Scp3-positive (Fig. 4O, asterisks) germ cells was significantly reduced in Prmt5Δ/f; Stra8-Cre testes (Fig. 4S). The expression of Dmc1 and γH2AX was also examined by immunofluorescence. In control testes, Dmc1 protein (Fig. S3A, red) was detected in most of germ cells at P12, and co-localized with Scp3 (Fig. S3A, green). γH2AX (green) was mainly observed in sex body (Fig. S3D) of control germ cells. Dmc1 protein (Fig. S3 B, red) was also detected in a small number of germ cells in Prmt5-deficient germ cells, and co-localized with Scp3 (Fig. S3B, green). A small portion of germ cells in Prmt5Δ/f; Stra8-Cre testes were also γH2AX (green) positive, whereas multiple foci were noted (Fig. S3E). Dmc1 (Fig. S3C) and γH2AX (Fig. S3F) protein was absent in most of germ cells from Prmt5Δ/f; Stra8-Cre testes.
Figure 4. Aberrant germ cell meiosis was observed in Prmt5Δ/f; Stra8-Cre mice.
The germ cells were labeled with Dazl (green) and meiotic germ cells were labeled with Stra8 (red) and Scp3 (red). In control testes, Stra8-positive (A, asterisks) and Scp3-positive (I, asterisks) germ cells were only detected in a small portion of seminiferous tubules at P10. Stra8-positive (C, asterisks) and Scp3-positive (K, asterisks) germ cells were also observed in seminiferous tubules of Prmt5Δ/f; Stra8-Cre mice at P10, and no statistical difference was noted compared to control mice (R). Stra8-positive (E, asterisks; F, white arrowheads) and Scp3-positive (M, asterisks; N, white arrowheads) germ cells were detected in most of seminiferous tubules of control testes at P12. The number of tubules with Stra8-positive (G) and Scp3-positive (O) germ cells was significantly reduced in Prmt5Δ/f; Stra8-Cre testes at P12 (S).
The differentially expressed genes in Prmt5 Δ/f ; Stra8-Cre testes
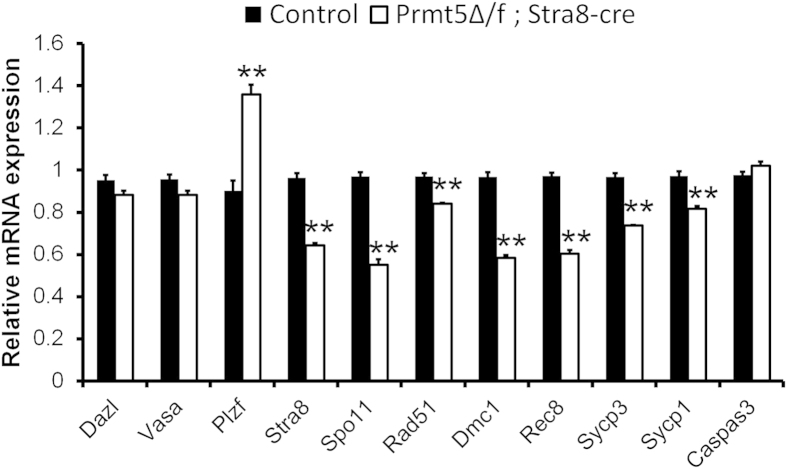
The differentially expressed genes between control and Prmt5Δ/f; Stra8-Cre testes at P10 was examined by real-time PCR analysis. As shown in Fig. 5, the mRNA levels of meiosis associated genes (e.g. Stra8, Spo11, Rad51, Scp1, Scp3, Dmc1, Rec8) were significantly reduced in testes from Prmt5Δ/f; Stra8-Cre males. By contrast, the expression of germ cell specific genes, Dazl and Vasa, were not decreased in Prmt5-deficient germ cells. Interestingly, the mRNA level of Plzf was significantly increased in Prmt5Δ/f; Stra8-Cre testes. It has been reported that the expression of transposable elements in germ cells was repressed by PRMT5 during embryonic stages17. However, the results of real-time PCR showed that the mRNA level of both IAP-LTR and L1-ORF2 was not increased in Prmt5Δ/f; Stra8-Cre testes (Fig. S4)
Figure 5. The expression of meiosis-associated genes was decreased in Prmt5Δ/f; Stra8-Cre testes at P10.
The expression of germ cell specific and meiosis-associated genes was examined by real-time PCR analysis. The mRNA level of Dazl and Vasa was not changed between control and Prmt5-deficient testes, whereas the expression of meiosis-associated genes Stra8, Spo11, Dmc1, and Rec8 was significantly reduced in testes of Prmt5Δ/f; Stra8-Cre testes compared to control testes. **p < 0.01.
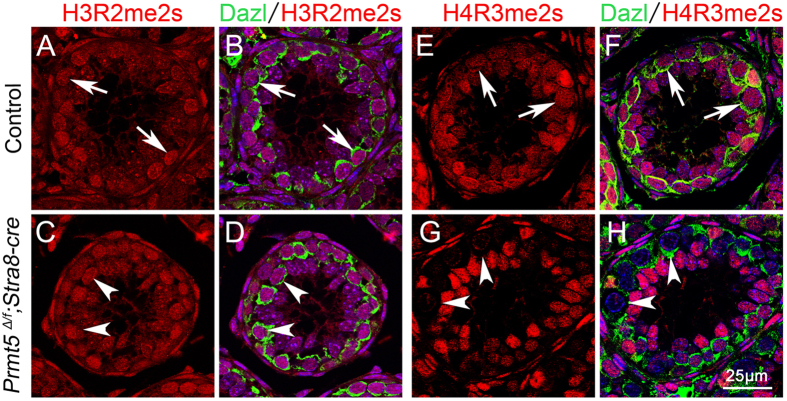
Symmetrical dimethylation on arginine-3 of histone H4 (H4R3me2s) was dramatically reduced in germ cells of Prmt5 Δ/f ; Stra8-Cre testes
As a protein arginine methyltransferase, PRMT5 is responsible for the symmetrical dimethylation of H4R3 in PGCs, and H4R3me2s is virtually absent in PGCs after Prmt5 inactivation in mouse model17,18. H3R2me2s is another major form of histone methylation, which has been reported to be catalyzed by PRMT520. To examine whether H4R3me2s and H3R2me2s are also catalyzed by PRMT5 in germ cells during spermatogenesis, immunofluorescence experiments were performed. As shown in Fig. 6, H3R2me2s was detected in germ cells of both control (A,B, white arrows) and Prmt5Δ/f; Stra8-Cre testes (C,D, white arrowheads) at P10. Robust signal of H4R3me2s was detected in germ cells of control testes (E,F, white arrows), whereas no signal was observed in the germ cells of Prmt5Δ/f; Stra8-Cre testes (G,H, white arrowheads) at this stage.
Figure 6. H4R3me2s was dramatically reduced in Prmt5-deficient germ cells.
The histone methylation in control and Prmt5Δ/f; Stra8-Cre testes was examined by immunofluroresence at P10 . H3R2me2s was detected in germ cells of both control (A, B, white arrows) and Prmt5Δ/f; Stra8-Cre testes (C, D, white arrowheads). H4R3me2s was detected in germ cells of control testes (E, F, white arrows). By contrast, H4R3me2s signal was completely absent in the germ cells of Prmt5Δ/f; Stra8-Cre testes (G, H, white arrowheads).
Discussion
The functions of epigenetic modification in germ cell development have been reported previously. DNA methylation plays critical roles in retrotransposon silencing and germ cell development, deletion of Dnmt3L in mouse model up-regulates the transcription of LINE and IAP retrotransposons in spermatogonia and spermatocytes. The mutant mice display meiotic failure with widespread non-homologous chromosome synapsis and progressive loss of germ cells by the mid-pachytene stage21,22,23. The precise timing of establishment and removal of histone methylation marks is also critical for spermatogenesis. Double knockout of the H3K9 trimethyltransferase genes Suv39h1 and Suv39h2 causes defects in meiosis in male germ cells24. Prmd9 (PR domain-containing 9) is an H3K4 trimethyltransferase that is specifically expressed in early meiotic germ cells both in the testis and ovary. Analysis of Prdm9-null mice indicates that it is involved in germ cell meiosis by regulating synapsis formation and recombination of homologous chromosomes during meiotic prophase25. Reduction of the histone methyltransferase MII2 activity results in a dramatic decrease of the number of spermatocytes by an apoptotic process26. Homozygous mutation of Ehmt2 (euchromatic histone-lysine N-methyltransferase 2) in germ cells causes the arrest of meiosis at the early pachytene stage in both testis and ovary, indicating that H3K9 mono- and dimethylation is also essential for early meiotic progression25.
Prmt5 encodes a protein arginine methyltransferase which catalyzes the symmetric dimethylarginine (SMDA) in both glycine and arginine-rich protein motifs. Previous studies have demonstrated that Prmt5 is expressed in PGCs during embryonic stages. Specifically deletion of this gene in PGCs by crossing with Blimp1-Cre causes germ cell loss before sex determination. Kim et al. found that Prmt5 functions in PGCs development by repressing transposable elements and that inactivation of Prmt5 results in the up-regulation of IAP and LINE117, whereas Li’s study revealed that Prmt5 is required for germ cell survival by enabling the correct splicing of primary RNA transcripts that function in the DNA damage response pathway18.
In this study, we found that Prmt5 was continually expressed in the germ cells of adult testis. Deletion of this gene after birth caused massive germ cell loss, and only a few germ cells were noted in adult testes. These results indicate that Prmt5 is also required for postnatal germ cell survival in testis which is consistent with the functions during embryonic stages17,18. Stra8-Cre is reported to be activated in germ cells of testis at approximately 3 days after birth19, and we also found that both the mRNA and protein level of Prmt5 were virtually absent in germ cells at P7. However, the development of germ cells was grossly normal in Prmt5Δ/f; Stra8-Cre testes at P10, Stra8-positive and Scp3-positive germ cells were observed in both control and Prmt5-deficient testes, indicating that the initiation of meiosis is not affected in Prmt5-deficient germ cells. The defect of germ cell development was first noted at P12 in Prmt5Δ/f; Stra8-Cre testes. In control mice, the number of germ cells was significantly increased at P12, and Stra8-positive and Scp3-positive germ cells were noted in the lumen of most seminiferous tubules. By contrast, most of the germ cells were still localized at the peripheral region of seminiferous tubules in Prmt5-deficient testes at this stage. A small number of Stra8-positive and Scp3-positive germ cells were detected in Prmt5-deficient testes. However, multiple foci of γH2AX were noted in Prmt5-deficient germ cells. In control germ cell, γH2AX was only detected in sex bodies. These results suggest that the DNA double strain breaks during meiosis are not properly repaired in Prmt5-deficient germ cells. Immunofluorescence results also showed that Prmt5 protein was mainly expressed in spermatocytes, not in germ cells at other developmental stages. Based on these results, we speculate that Prmt5 is probably not involved in the development of germ cells before meiosis, but it is required for the process of meiosis. The death of Prmt5-deficient germ cells is probably due to the defects of meiosis. However, we could not exclude the possibility that the blockage of meiosis is a deleterious effect after Prmt5 inactivation.
Previous studies found that both H4R3me2s and H3R2me2s are catalyzed by PRMT516,20. In this study, we found that H4R3me2s was virtually absent in germ cells after Prmt5 inactivation, whereas H3R2me2s was not changed in Prmt5-deficient germ cells, suggesting that PRMT5 is responsible for H4R3me2s in germ cell during later developmental stages. The functions of Prmt5 in germ cell development are probably mediated by H4R3me2s which are consistent with other histone modifiers24,25,26. However, whether Prmt5 is involved in germ cell development mediated by histone methylation or indirectly catalyzed the methylation of other proteins is unclear which need further investigation.
It has been reported that Prmt5 is required for germ survival during embryonic stage by repressing the transcription of retrotransposon17. In this study, we found that the transcription level of IAP and LINE1 was not increased in Prmt5-deficient germ cells, suggesting that Prmt5 is required for germ cell survival at different developmental stages but the underlying mechanisms are different.
Taken together, in the present study, we found that Prmt5 was also expressed in male germ cells after birth which also plays important roles in spermatogenesis. Deletion of this gene results in aberrant spermatogenesis and most of germ cells die in adult testes. The results suggest that Prmt5 is probably required for germ cell meiosis and the death of Prmt5-deficient germ cells is likely due to the defects of meiosis. However, we could not exclude that the defect of meiosis is a deleterious effects after Prmt5 inactivation.
Material and methods
Mice
All animal works were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Institute of Zoology, Chinese Academy of Sciences (CAS). All the mice were maintained in a C57BL/6;129/SvEv mixed background. Prmt5flox mice strain was obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM; Prmt5tm2a(EUCOMM)Wtsi)27, Prmt5Δ/+mice were obtained by crossing with ZP3-Cre mice. Prmt5Δ/f; Stra8-Cre mice were obtained by crossing Prmt5Δ/+; Stra8-Cre males with Prmt5flox/flox females. Genotyping was performed by PCR as described previously using DNA isolated from tail tips28,29.
Tissue collection and histological analysis
Testes were dissected from Prmt5Δ/f; Stra8-Cre and control mice immediately after euthanasia, fixed in 4% paraformaldehyde for up to 24 hrs, stored in 70% ethanol, and embedded in paraffin. Five-micrometer-thick sections were cut and mounted on glass slides. After deparaffinization, sections were processed for immunohistochemistry and immunofluorescence analysis.
Immunohistochemistry analysis
IHC analysis of tissues from at least three mice for each genotype was performed using a Vectastain ABC (avidin–biotin–peroxidase) kit (Vector Laboratories, Burlingame, CA) as recommended and using antibodies to MVH (1:500, Abcam, ab13840), PRMT5 (1:200, Millipore, 07-405). The IHC procedure was performed as described previously28. After staining, the sections were examined with a Nikon Microscope, and images were captured with Nikon DS-Ri1 CCD camera.
Immunofluorescence analysis
After rehydration and antigen retrieval, the 5 μm sections were incubated with 5% donkey serum in 0.3% triton X-100 for 1 hr. Then the sections were incubated with the primary antibodies for 1.5 hrs, and the corresponding TRITC-conjugated donkey anti-rabbit IgG (1:150, Jackson) and FITC-conjugated donkey anti-mouse IgG (1:150, Jackson) for 1.5 hrs at room temperature. The following dilutions of primary antibodies were used: PRMT5 (1:200, Millipore, 07-405), DAZL (1:100, AbD Serotec, MCA2336), STRA8 (1:200, Abcam, ab49405), SCP3 (1:200, Abcam, ab15093), γH2AX (1:400, Millipore, 05-636), DMC1 (1:100, Santa Cruz, sc-22768 ), H3R2me2s (1:50, Millipore, ABE460), H4R3me2s (1:100, Abcam, ab5823). After three times wash in PBS, the nuclei was stained with DAPI. The sections were examined with confocal laser scanning microscope (Carl Zeiss Inc., Thornwood, NY).
Nucleic acid isolation and quantitative reverse transcription PCR
The testes were dissected at P10, and stored at −80 °C after snap frozen with liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy kit in accordance with the manufacturer’s instructions. Two micrograms of total RNA was used to synthesize first-strand cDNA. To quantify gene expression, real-time SYBR Green assay was performed with the isolated RNA. Gapdh was used as an endogenous control. All gene expression was quantified relative to Gapdh expression. The relative concentration of the candidate gene expression was calculated using the formula 2−ΔΔCT as described in the SYBR Green user manual. Primers used for real-time PCR were listed in Table S1.
TUNEL assay
TUNEL assays were conducted with the In Situ Cell Death Detection Kit, Fluorescein (Promega BioSciences, San Luis Obispo, CA, USA), as recommended. The images were captured by Nikon DS-Ri1 CCD camera.
Statistical analysis
Experiments were repeated at least three times. Three to five control or Prmt5-deficient gonads at each time point were used for immunostaining. The quantitative results are presented as mean ± SEM. The data were evaluated for statistical differences using student T-test. P-value < 0.05 was considered as significant.
Additional Information
How to cite this article: Wang, Y. et al. Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 5, 11031; doi: 10.1038/srep11031 (2015).
Supplementary Material
Acknowledgments
This work was supported by the Major Research Plan (2011CB944303 and 2013CB945001), and the National Natural Science Foundation of China (31471348,31071271, 31171373). Collaborative Innovation Center for Cardiovascular Disorders.
Footnotes
Author Contributions F.G., S.L.B. conceived the project; Y.B.W. performed the experiments and analyzed the data, T.X.Z., Q.L.L., C.Y.L., F.H., M.C., L.J.Z., X.H.C. and Q.Y. helped to analyze the data, F.G. and Y.B.W. wrote the manuscript. All authors read and approved the final manuscript.
References
- Anway M. D., Cupp A. S., Uzumcu M. & Skinner M. K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M. A., Hayashi K. & Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 (2007). [DOI] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W. & Reik W. Epigenetic reprogramming in mammals. Human molecular genetics 14 Spec No 1, R47–58 (2005). [DOI] [PubMed] [Google Scholar]
- Kimmins S. & Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature 434, 583–589 (2005). [DOI] [PubMed] [Google Scholar]
- Ginsburg M., Snow M. H. & McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development 110, 521–528 (1990). [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S. C. & Surani M. A. A molecular programme for the specification of germ cell fate in mice. Nature 418, 293–300 (2002). [DOI] [PubMed] [Google Scholar]
- Hajkova P. et al. Epigenetic reprogramming in mouse primordial germ cells. Mechanisms of development 117, 15–23 (2002). [DOI] [PubMed] [Google Scholar]
- Kishigami S. et al. Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Developmental biology 289, 195–205 (2006). [DOI] [PubMed] [Google Scholar]
- Aletta J. M., Cimato T. R. & Ettinger M. J. Protein methylation: a signal event in post-translational modification. Trends in biochemical sciences 23, 89–91 (1998). [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Boue S. & Izpisua Belmonte J. C. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nature reviews. Genetics 12, 231–242 (2011). [DOI] [PubMed] [Google Scholar]
- Bedford M. T. Arginine methylation at a glance. Journal of cell science 120, 4243–4246 (2007). [DOI] [PubMed] [Google Scholar]
- Pal S. & Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. Journal of cellular physiology 213, 306–315 (2007). [DOI] [PubMed] [Google Scholar]
- Najbauer J., Johnson B. A., Young A. L. & Aswad D. W. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. The Journal of biological chemistry 268, 10501–10509 (1993). [PubMed] [Google Scholar]
- Tee W. W. et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes & development 24, 2772–2777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Li Z., Chin C. J., Lee S. A. & Clark A. T. PRMT5 is required for human embryonic stem cell proliferation but not pluripotency. Stem cell reviews 10, 230–239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K. et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nature cell biology 8, 623–630 (2006). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. PRMT5 Protects Genomic Integrity during Global DNA Demethylation in Primordial Germ Cells and Preimplantation Embryos. Molecular cell 56, 564–579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. et al. The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. The EMBO journal (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou P. I., Payne C. J., Dearth A. T. & Braun R. E. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori V. et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nature structural & molecular biology 19, 136–144 (2012). [DOI] [PubMed] [Google Scholar]
- Bourc'his D. & Bestor T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). [DOI] [PubMed] [Google Scholar]
- Webster K. E. et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America 102, 4068–4073 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K., Kusumi M., Yokomine T., Li E. & Sasaki H. Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Molecular reproduction and development 73, 116–122 (2006). [DOI] [PubMed] [Google Scholar]
- Peters A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- Tachibana M., Nozaki M., Takeda N. & Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. The EMBO journal 26, 3346–3359 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S. et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics & chromatin 2, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi M. et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes & development 27, 1903–1916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. et al. The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proceedings of the National Academy of Sciences of the United States of America 103, 11987–11992 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N. et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. The EMBO journal 18, 5931–5942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.