Abstract
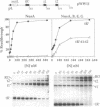
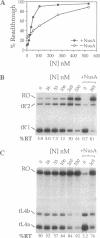
During transcription of phage lambda early operons, the N gene product alters host RNA polymerase (RNAP) so that transcription proceeds through multiple stop signals. Here, we reproduce the essence of N activity with purified components in synthetic transcription units that contain lambda pL promoter and the N-recognition site, nutL, followed by a variety of intrinsic terminators. We show that three host factors (NusA, NusE, and NusG) are essential for N to allow appreciable transcription through multiple terminators and that this persistent antitermination is stimulated by a fourth factor, NusB. Remarkably, in the absence of all four factors, N suppresses various terminators placed near the nut site. This basal antitermination activity of N is enhanced by NusA and is diminished by high salt and temperature. We postulate that N interacts with RNAP directly, inducing the termination-resistant state. While NusA facilitates this interaction, the other factors strengthen it sufficiently over time and distance so that RNAP bypasses multiple terminators. The dispensability of NusB for persistent antitermination in vitro, but not in vivo, raises the possibility that NusB performs two functions: it increases the stability of N antitermination complex and also counteracts an inhibitory factor in the cell.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barik S., Ghosh B., Whalen W., Lazinski D., Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987 Sep 11;50(6):885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Das A., Ghosh B., Barik S., Wolska K. Evidence that ribosomal protein S10 itself is a cellular component necessary for transcription antitermination by phage lambda N protein. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4070–4074. doi: 10.1073/pnas.82.12.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992 Nov;174(21):6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Wolska K. Transcription antitermination in vitro by lambda N gene product: requirement for a phage nut site and the products of host nusA, nusB, and nusE genes. Cell. 1984 Aug;38(1):165–173. doi: 10.1016/0092-8674(84)90537-3. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baron L. S. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974 Mar;58(1):141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baumann M., Baron L. S. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976 Aug;73(1):119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984 Dec;48(4):299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Johnson L. L., Alessi D., Craven M. G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990 Dec;4(12A):2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Schauer A. T., Baumann M. R., Baron L. S., Adhya S. L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Wilgus G. S., Mural R. J. Gene N regulator function of phage lambda immun21: evidence that a site of N action differs from a site of N recognition. J Mol Biol. 1973 Dec 25;81(4):505–516. doi: 10.1016/0022-2836(73)90519-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B., Das A. nusB: a protein factor necessary for transcription antitermination in vitro by phage lambda N gene product. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6305–6309. doi: 10.1073/pnas.81.20.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh B., Grzadzielska E., Bhattacharya P., Peralta E., DeVito J., Das A. Specificity of antitermination mechanisms. Suppression of the terminator cluster T1-T2 of Escherichia coli ribosomal RNA operon, rrnB, by phage lambda antiterminators. J Mol Biol. 1991 Nov 5;222(1):59–66. doi: 10.1016/0022-2836(91)90737-q. [DOI] [PubMed] [Google Scholar]
- Ghysen A., Pironio M. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J Mol Biol. 1972 Mar 28;65(2):259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980 Jun 15;140(1):57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985 Aug;42(1):259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981 May;24(2):421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981 Mar 25;147(1):11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Houman F., Diaz-Torres M. R., Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990 Sep 21;62(6):1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Cashel M., Friedman D. I., Nakamura Y., Walter W. A., Gross C. A. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J Mol Biol. 1988 Nov 20;204(2):247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Leirmo S., Harrison C., Cayley D. S., Burgess R. R., Record M. T., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987 Apr 21;26(8):2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Li J., Mason S. W., Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993 Jan;7(1):161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Limberger R. J., Campbell A. M. Functional elements of DNA upstream from the integrase operon that are conserved in bacteriophages 434 and lambda. Gene. 1987;61(2):135–144. doi: 10.1016/0378-1119(87)90108-9. [DOI] [PubMed] [Google Scholar]
- Linderoth N. A., Calendar R. L. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J Bacteriol. 1991 Nov;173(21):6722–6731. doi: 10.1128/jb.173.21.6722-6731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S. W., Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991 Aug;5(8):1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- Mason S. W., Li J., Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J Mol Biol. 1992 Jan 5;223(1):55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- Mason S. W., Li J., Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J Biol Chem. 1992 Sep 25;267(27):19418–19426. [PubMed] [Google Scholar]
- Nodwell J. R., Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993 Jan 29;72(2):261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- Nodwell J. R., Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991 Nov;5(11):2141–2151. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- Patterson T. A., Zhang Z., Baker T., Johnson L. L., Friedman D. I., Court D. L. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J Mol Biol. 1994 Feb 11;236(1):217–228. doi: 10.1006/jmbi.1994.1131. [DOI] [PubMed] [Google Scholar]
- Reynolds R., Bermúdez-Cruz R. M., Chamberlin M. J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992 Mar 5;224(1):31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978 Sep 5;124(1):195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- Sparkowski J., Das A. Simultaneous gain and loss of functions caused by a single amino acid substitution in the beta subunit of Escherichia coli RNA polymerase: suppression of nusA and rho mutations and conditional lethality. Genetics. 1992 Mar;130(3):411–428. doi: 10.1093/genetics/130.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C. L., Greenblatt J., Li J., Condon C., Squires C. L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S. L., Gottesman M. E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992 Mar 6;68(5):989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Sullivan S. L., Ward D. F., Gottesman M. E. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J Bacteriol. 1992 Feb;174(4):1339–1344. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen W. A., Das A. Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product. New Biol. 1990 Nov;2(11):975–991. [PubMed] [Google Scholar]
- Whalen W., Ghosh B., Das A. NusA protein is necessary and sufficient in vitro for phage lambda N gene product to suppress a rho-independent terminator placed downstream of nutL. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2494–2498. doi: 10.1073/pnas.85.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell W. S., Roberts J. W. The phage lambda gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell. 1992 Jun 26;69(7):1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]