Abstract
Recently, bacterial infection causing periodontal disease has attracted considerable attention as a risk factor for pancreatic cancer. Fusobacterium species is an oral bacterial group of the human microbiome. Some evidence suggests that Fusobacterium species promote colorectal cancer development; however, no previous studies have reported the association between Fusobacterium species and pancreatic cancer. Therefore, we examined whether Fusobacterium species exist in pancreatic cancer tissue. Using a database of 283 patients with pancreatic ductal adenocarcinoma (PDAC), we tested cancer tissue specimens for Fusobacterium species. We also tested the specimens for KRAS, NRAS, BRAF and PIK3CA mutations and measured microRNA-21 and microRNA-31. In addition, we assessed epigenetic alterations, including CpG island methylator phenotype (CIMP). Our data showed an 8.8% detection rate of Fusobacterium species in pancreatic cancers; however, tumor Fusobacterium status was not associated with any clinical and molecular features. In contrast, in multivariate Cox regression analysis, compared with the Fusobacterium species-negative group, we observed significantly higher cancer-specific mortality rates in the positive group (p = 0.023). In conclusion, Fusobacterium species were detected in pancreatic cancer tissue. Tumor Fusobacterium species status is independently associated with a worse prognosis of pancreatic cancer, suggesting that Fusobacterium species may be a prognostic biomarker of pancreatic cancer.
Keywords: Fusobacterium, microbiota, pancreas, miR-31, survival
INTRODUCTION
Pancreatic cancer is a highly aggressive malignancy, with < 50% patients surviving past 6 months after the diagnosis. Because chemotherapeutic options only marginally prolong life, the current mortality of patients with pancreatic cancer is nearly identical to its incidence [1–13]. Therefore, future studies are expected to elucidate the pathogenesis of pancreatic cancer and explore new possibilities for diagnostic and therapeutic approaches to the disease.
Recently, bacterial infection causing periodontal disease has attracted considerable attention as a risk factor for pancreatic cancer [3, 6, 7, 10, 14]. Individuals with periodontal disease have increased levels of markers of systemic inflammation, and oral bacteria can spread to the bloodstream [3, 6, 7, 15], gastrointestinal tract [16–19], liver [20, 21], or pancreas [22] and can even reach the brain [23]. Thus, oral bacteria may reach the pancreas too through the circulation or transductal transmission from the biliary tract and can contribute to pancreatic carcinogenesis by acting jointly with other pancreatic cancer risk factors that modulate inflammation and the immune response, e.g., obesity, smoking, and the ABO genetic variant [4, 6, 7].
Fusobacterium species (a group of non-spore-forming, anaerobic gram-negative bacteria) is an oral bacteria group of the human microbiome. The members of the Fusobacterium species are highly heterogeneous, and some of them have been recognized as opportunistic pathogens implicated not only in periodontitis [6, 7, 24] but also in inflammatory bowel diseases (IBD) [17–19], pancreatic abscess [22, 25], and hepatic abscess [20, 21, 25]. With regard to the association of Fusobacterium species with the pathogenesis of gastrointestinal cancer, metagenomic analyses involving whole-genome sequencing, transcriptome sequencing and 16S ribosomal RNA gene DNA sequencing have demonstrated enrichment of Fusobacterium species in colorectal cancer tissues [26–30]. Moreover, increased levels of Fusobacterium species are related to patient survival [29] and to specific molecular subsets of colorectal cancers [28].
Thus, accumulating evidence indicates that Fusobacterium species may play a role in gastrointestinal cancer; however, no previous studies have reported an association of Fusobacterium species with pancreatic cancer. Therefore, we examined whether Fusobacterium species exist in pancreatic cancer tissue and play a role in disease progression. To identify the role in pancreatic cancer, we also analyzed the association of tumor Fusobacterium species status with molecular characteristics, including epigenetic alterations and microRNA expression levels, and patient prognosis, using a database of 283 patients.
RESULTS
Detection of Fusobacterium species in cancer tissue specimens of pancreatic cancer
We assessed 302 formalin-fixed, paraffin-embedded (FFPE) tissue specimens of pancreatic cancers using the TaqMan Gene Expression Assay for Fusobacterium species and obtained 283 (94%) positive results. Fusobacterium species were detected in 8.8% (25/283) of the pancreatic cancer tissue specimens. Figure 1 shows DNA amounts (2−ΔCt) of the 25 Fusobacterium species-positive cases.
Figure 1. Distribution of Fusobacterium species-positive cases (n = 25) among pancreatic cancer tissue specimens.
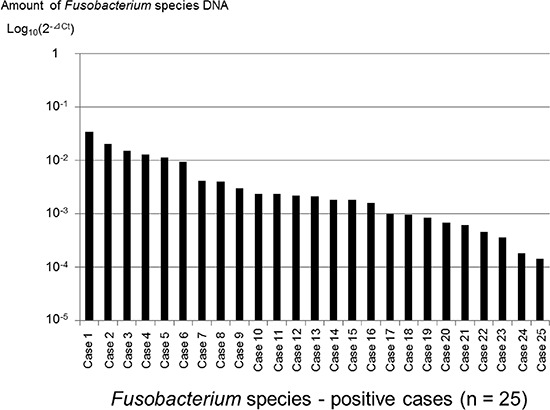
The Fusobacterium species-positive cases were ranked according to the amount of Fusobacterium species DNA.
Using the tumor Fusobacterium species-positive group, we also tested paired specimens of normal pancreatic mucosal tissues for Fusobacterium species. Our data demonstrated that Fusobacterium species were detected in 28% (7/25) of the paired specimens of normal tissues.
The association between tumor Fusobacterium species and clinical, pathological and molecular characteristics in pancreatic cancers
Table 1 summarizes clinical, pathological and molecular features of the 283 patients with pancreatic cancer, according to tumor Fusobacterium species status (positive versus negative). Tumor Fusobacterium species status was slightly but insignificantly associated with the female gender and high microRNA-21 (miR-21) expression levels. Neither microRNA-31 (miR-31) nor microRNA-143 (miR-143) correlate with tumor Fusobacterium species status.
Table 1. Clinical or molecular features of pancreatic cancers according to tumor Fusobacterium status.
| Clinical, pathological or molecular feature | Total | Tumor Fusobacterium species status | P | |
|---|---|---|---|---|
| Negative | Positive | |||
| All cases | 283 | 258 | 25 | |
| Gender | ||||
| Male | 161 (57%) | 151 (59%) | 10 (40%) | 0.074 |
| Female | 122 (43%) | 107 (42%) | 15 (60%) | |
| Age (mean ± SD) | 67.1 ± 9.1 | 67.3 ± 9.1 | 65.2 ± 8.4 | 0.25 |
| Tumor location in pancreas | ||||
| Head | 203 (72%) | 187 (73%) | 16 (64%) | 0.12 |
| Body | 62 (22%) | 57 (22%) | 5 (20%) | |
| Tail | 18 (6.4%) | 14 (5.4%) | 4(16%) | |
| Tumor size (cm) | ||||
| <2 | 44 (16%) | 41 (16%) | 3 (12%) | 0.94 |
| 2–4 | 191 (68%) | 174 (67%) | 17 (68%) | |
| 4–6 | 40 (14%) | 36 (14%) | 4 (16%) | |
| ≥6 | 8 (2.8%) | 7 (2.7%) | 1 (4.0%) | |
| Lymph node invasion | ||||
| Negative | 99 (35%) | 89 (35%) | 10 (40%) | 0.58 |
| Positive | 184 (65%) | 169 (66%) | 15 (60%) | |
| Disease Stage(UICC classification) | ||||
| I | 20 (7.1%) | 18 (7.0%) | 2 (8.0%) | 0.10 |
| II | 248 (88%) | 228 (88%) | 20 (80%) | |
| III | 10 (3.5%) | 9 (3.5%) | 1 (4.0%) | |
| IV | 5 (1.8%) | 3 (1.2%) | 2 (8.0%) | |
| Year of diagnosis | ||||
| Prior to 2009 | 157 (55%) | 139 (54%) | 18 (72%) | 0.076 |
| 2010–2013 | 126 (45%) | 119 (46%) | 7 (28%) | |
| KRAS gene | ||||
| Wild-type | 88 (31%) | 82 (32%) | 6 (24%) | 0.72 |
| Codon 12/13 mutated | 174 (61%) | 157 (61%) | 17 (68%) | |
| Codon 61/146 mutated | 21 (7.4%) | 19 (7.4%) | 2 (8.0%) | |
| MicroRNA-21 | ||||
| Low expression | 213 (75%) | 198 (77%) | 15 (60%) | 0.064 |
| High expression | 70 (25%) | 60 (23%) | 10 (40%) | |
| MicroRNA-31 | ||||
| Low expression | 213 (75%) | 196 (76%) | 17 (68%) | 0.38 |
| High expression | 70 (25%) | 62 (24%) | 8 (32%) | |
| MicroRNA-143 | ||||
| Low expression | 70 (25%) | 64 (25%) | 6 (24%) | 0.93 |
| High expression | 213 (75%) | 194 (75%) | 19 (76%) | |
| MLH1 status | ||||
| Unmethylated | 272 (96%) | 248 (96%) | 24 (96%) | 0.98 |
| Methylated | 11 (3.9%) | 10 (3.9%) | 1 (4%) | |
| CIMP status | ||||
| CIMP-low/zero | 249 (88%) | 226 (88%) | 23 (92%) | 0.52 |
| CIMP-high | 34 (12%) | 32 (12%) | 2 (8.0%) | |
Percentages (%) indicate the proportion of cases with a specific clinical, pathological, or molecular feature within a given dichotomous category of tumor Fusobacterium species status. The p values were calculated using t-test for age and χ2 test for gender; tumor location in pancreas; tumor size; lymph node invasion; year of diagnosis; KRAS mutation; expressions of microRNA-21, microRNA-31, and microRNA-143; MLH1 methylation and CIMP status or Fisher's exact test for disease stage.
CIMP, CpG island methylator phenotype; SD, standard deviation; UICC, Unio Internationalis Contra Cancrum
Fusobacterium species status in pancreatic cancer and patient survival
During the follow-up of the 283 patients with pancreatic cancer, 109 patients died (all deaths were confirmed to be attributable to pancreatic cancer). The median follow-up period for cancer-specific survival was 30.3 months. In the Kaplan–Meier analysis, significantly shorter survival was observed in the Fusobacterium species-positive group [median cancer-specific survival (months): 17.2 versus 32.5; log-rank p = 0.021; Figure 2].
Figure 2. Kaplan–Meier curves of cancer-specific survival of patients with pancreatic cancer according to tumor Fusobacterium species status.
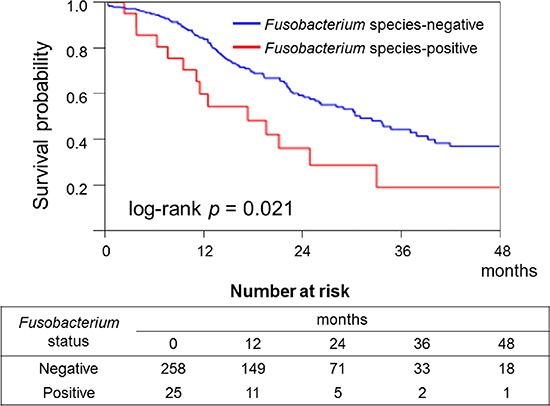
In the Kaplan–Meier analysis, significantly shorter survival was observed in the Fusobacterium species-positive group (log-rank p = 0.021).
The association of molecular alterations with patient survival in pancreatic cancer
KRAS mutation (codon 12, 13, 61, or 146) was detected in 69% of the 283 patients with pancreatic cancer (Table 1). BRAF (V600E), NRAS (codon 12, 13, or 61), and PIK3CA (exon 9 or 20) mutations were not found in these 283 cases. CpG island methylator phenotype (CIMP)-high status [three/four or more methylated promoters among CACNA1G, CDKN2A, IGF2, and RUNX3] was observed in 12% (34/283) of the pancreatic cancer cases (Table 1). Microsatellite instability (MSI)-high status was not detected among the 283 cases.
In the Kaplan–Meier analysis, high expression levels of miR-21 (log-rank p = 0.0025; Figure 3A), miR-31 (log-rank p = 0.0003; Figure 3B), and CIMP-high status (log-rank p = 0.024; Figure 4) significantly correlated with shorter survival. In contrast, neither KRAS mutations nor miR-143 was associated with patient survival (data not shown).
Figure 3. Kaplan–Meier curves of cancer-specific survival of patients with pancreatic cancer according to the amount of microRNA-21 (A) or microRNA-31 (B).
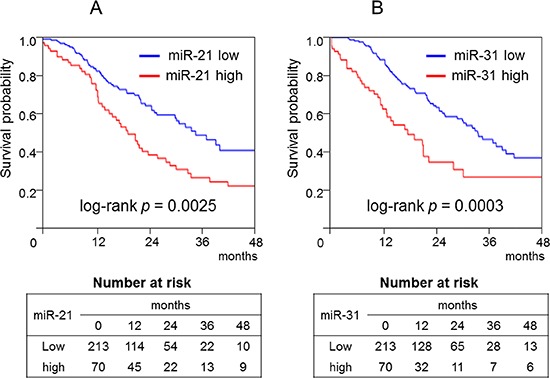
In the Kaplan–Meier analysis, high microRNA-21 (log-rank p = 0.0025) and microRNA-31 expression levels (log-rank p = 0.0003) significantly correlated with shorter survival.
Figure 4. Kaplan–Meier curves of cancer-specific survival of patients with pancreatic cancer according to CpG island methylator phenotype (CIMP) status.
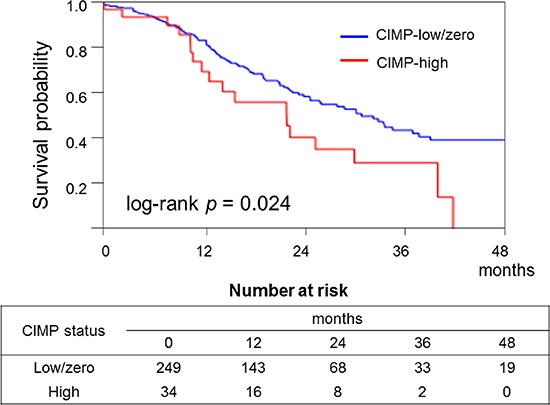
In the Kaplan–Meier analysis, CIMP-high status significantly correlated with shorter survival (log-rank p = 0.024).
Multivariate Cox regression analysis in patients with pancreas cancer
In the univariate Cox regression analysis, compared to Fusobacterium species-negative cases, significantly higher mortality rates were observed among Fusobacterium species-positive cases [hazard ratio (HR): 1.91; 95% confidence interval (CI): 1.04–3.24; p = 0.037; Table 2].
Table 2. The association between tumor Fusobacterium species status and mortality of patients with pancreatic cancer.
| Fusobacterium species status | Total N | Fusobacterium species status | |
|---|---|---|---|
| UnivariateHR (95% CI) | Multivariatestage-stratifiedHR (95% CI) | ||
| Negative | 258 | 1 (referent) | 1 (referent) |
| Positive | 25 | 1.91 (1.04–3.24) | 2.16 (1.12–3.91) |
| P | 0.037 | 0.023 | |
The multivariate, stage-stratified Cox model included the Fusobacterium species variable adjusted by gender; age at diagnosis; tumor size; tumor location; year of diagnosis; KRAS mutation status; CIMP status and expression levels of microRNA-21, microRNA-31, and microRNA-143. A backward stepwise elimination with a threshold of p = 0.10 was used to select variables in the final model. Of the variables, Fusobacterium species, CIMP status, and expression level of microRNA-31 were included in the final model.
CI, confidence interval; CIMP, CpG island methylator phenotype; HR, hazard ratio
A multivariate model initially included gender, age at diagnosis, tumor size, tumor location, year of diagnosis, KRAS mutation, CIMP status and miR-21, miR-31, and miR-143 expression levels. Similarly, compared to Fusobacterium species-negative cases, an independent association with poorer prognosis was observed in Fusobacterium species-positive cases in the multivariate analyses of cancer-specific survival (HR: 2.16; 95% CI: 1.12–3.91; p = 0.023; Table 2).
DISCUSSION
In the present study on patients with pancreatic cancer who underwent surgical treatment, we tested whether Fusobacterium species exist in pancreatic cancer tissue. The detection rate of Fusobacterium species in pancreatic cancer tissue specimens in the present study was 8.8%. Moreover, we found that Fusobacterium species status of pancreatic cancer tissue specimens was independently associated with worse prognosis. Therefore, our data suggest that Fusobacterium species exist in pancreatic cancer tissue and may be related to the malignant potential. Although our data need to be confirmed in other (independent) data sets, tumor Fusobacterium species status may serve as a prognostic biomarker of pancreatic cancer.
At present, the pathogenesis of pancreatic cancer is poorly understood. With respect to the association of bacterial infections with pancreatic cancer, another study showed that a history of periodontal disease is associated with an increased risk of pancreatic cancer in a prospective cohort study [10]. Among the oral bacteria, Porphyromonas gingivalis (P. gingivalis), a pathogen responsible for periodontal disease, is associated with pancreatic carcinogenesis [6]. In addition, a high level of antibodies to P. gingivalis in serum correlates with a lower risk of pancreatic cancer [7]. According to culture methods, the microbiota isolated from the pancreas has similarities to the oral microbiota, particularly in cases of pancreatitis [6, 22]. Moreover, the spread of oral bacteria to the pancreas via dissemination has been documented for both animal models and human subjects [7, 15, 31].
Aside from periodontal disease, the genus Fusobacterium species is associated with inflammatory disorders of the gastrointestinal tract [16–19], with pancreatic abscesses [22, 25], hepatic abscesses [20, 21], cerebral abscesses [23] and Lemierre's syndrome [32]. These data suggest that oral Fusobacterium species may reach the pancreas through the circulation (i.e., blood or lymphatic vessels). Furthermore, multiple studies have shown that the oral microbiota overlaps with the gastrointestinal tract microbiota; these data suggest that there are multiple avenues for the dissemination of oral bacteria within the body [7, 33, 34]. Transductal transmission from the biliary tract (either ascending or descending) has also been considered [31, 35]. Further research is needed to identify the route of Fusobacterium species' spread to the pancreas.
Recent studies showed that Fusobacterium species are detected in gastrointestinal tract cancers, such as gastric cancer [34] and colorectal cancer [26, 29, 30]; however, published studies have not examined the relationship of Fusobacterium species with the cancer outcome in a large sample (n > 100) of such patients. In the present study, we found that tumor Fusobacterium species status is independently associated with shorter survival in patients with pancreatic cancer. To the best of our knowledge, this is the first report to demonstrate an association of tumor presence of Fusobacterium species with the outcome of pancreatic cancer. Although no significant association was found, Fusobacterium species were highly detected in pancreatic tail cancer (4/18; 22%) than in head (16/203; 7.9%) or body cancer (5/62; 8.0%). The high prevalence of Fusobacterium species in pancreatic tail cancers remains uncertain. One possible explanation for this result is the difference of the vascular supply between the pancreatic tail and head or body. Moreover, in the tumor Fusobacterium species-positive cases, Fusobacterium species were detectable in 28% of the paired specimens of normal tissues, suggesting that these bacteria may play a role in the pathogenesis of pancreatic cancer. Our findings should help to elucidate the details of pancreatic carcinogenesis and could lead to the development of a new diagnostic or therapeutic method (i.e., eradication) for patients with pancreatic cancer.
Our study has some limitations because of its cross-sectional (observational) design and the risk that an unknown bias (e.g., the selection bias) may confound the results. Furthermore, our analysis does not include treatment data; thus, another unknown bias, e.g., differential treatment assignment, may skew the results. In addition, we excluded cases without available tumor tissue: another possible bias. Nevertheless, the results of our regression analyses are adjusted for potential confounders, including disease stage, year of diagnosis, CIMP status, and miR-21 and miR-31 expression levels.
The role of Fusobacterium species in pancreatic carcinogenesis remains unknown. Recent studies showed that Fusobacterium species increase production of reactive oxygen species (ROS) and inflammatory cytokines (e.g., IL-6 and TNF) in colorectal cancer [30]. Inflammation and ROS can cause epigenetic silencing of the mismatch repair protein MLH1 [36]. Accordingly, we assessed here the possible association of tumor Fusobacterium species status with epigenetic alterations, such as MLH1 methylation and CpG island methylator phenotype (CIMP) in pancreatic cancer.
The term “CIMP” has been repeatedly used over the past decade to describe CpG island promoter methylation in various human malignancies, including pancreatic cancer [12, 28, 37–39]. In contrast to colorectal cancer research, in the field of pancreatic research, CIMP is an unfamiliar term. One study showed that CIMP-high status is observed in five of 36 (14%) pancreatic cancers; however, there is no significant association between the CIMP status and the prognosis [12]. In contrast, our present data show that CIMP-high status (according to methylated promoters that are widely used in colorectal cancer research), and MLH1 methylation are detected in 12% and 3.9% of pancreatic cancers. Although neither CIMP status nor MLH1 methylation correlate with tumor Fusobacterium species status, our results show that the CIMP-high status is associated with an unfavorable prognosis of pancreatic cancer. CIMP is believed to be useful not only for molecular characterization but also for the assessment of a response to treatment in a variety of human cancers [37]. Therefore, further research is needed to clarify the role of epigenetic alterations in pancreatic cancer.
MicroRNAs constitute a class of small non-coding RNA molecules that function as post-transcriptional gene regulators and have been increasingly recognised as biomarkers of various human cancers [2, 5, 8, 9, 40–42]. Certain microRNAs are induced during the macrophage inflammatory response and have the ability to regulate host-cell responses to pathogens [43]. In addition, pathogens themselves may regulate microRNA expression [44]. MicroRNAs influence networks that control innate and adaptive immunity and apoptosis by regulating signaling pathways [43]. Previous studies suggest that cells can secrete microRNAs that can be delivered into recipient cells where they can change gene expression [45, 46]. Accordingly, it is possible that immune system-related microRNAs can travel to the pancreas, even if the pathogens are located elsewhere. Nevertheless, no previous studies have reported an association between microbiota and microRNA expression in gastrointestinal cancer.
With regard to microRNA expression levels in pancreatic cancer, a recent study demonstrated that miR-21, miR-31 [2, 9, 11], and miR-143 expression [47] levels are significantly associated with higher stage or worse survival. Therefore, we analyzed the association between tumor Fusobacterium species status and miR-21, miR-31, or miR-143 expression levels in pancreatic cancer. No significant associations were found. In line with previous studies [2, 9, 11], our present data also show that miR-21 and miR-31 expression levels are related to shorter survival among patients with pancreatic cancer. Thus, these microRNAs could be diagnostic biomarkers or therapeutic targets in patients with pancreatic cancer.
In conclusion, Fusobacterium species were detected in pancreatic cancer tissue. Although no significant association was found between Fusobacterium species status and molecular alterations of pancreatic cancers, the tumor Fusobacterium species status was independently associated with worse prognosis of pancreatic cancer. Thus, our data suggest that tumor Fusobacterium species is a promising biomarker of pancreatic cancer and could open up exciting opportunities to improve our understanding of the pathogenesis of this fatal disease.
MATERIALS AND METHODS
Patients and cancer tissue specimens
We collected FFPE tissue specimens of 302 pancreatic cancers [pancreatic ductal adenocarcinomas (PDACs)] of patients who underwent surgical treatment at Sapporo Medical University Hospital, Teine Keijinkai Hospital or Otaru Municipal Hospital between 2003 and 2013. To avoid the selection bias as much as possible, we collected the consecutive FFPE cancer tissue specimens. To clarify the association between Fusobacterium and survival in patients with pancreatic cancer, we limited the patients who received adjuvant chemotherapy to those treated with gemcitabine or 5-FU (including S-1). Patients who were treated with chemotherapy or radiotherapy prior to resection were excluded.
Histological data on pancreatic cancer tissue specimens were evaluated by pathologists (T.S. and T.H.) who were blinded to the clinical and molecular information. Cancer-specific survival was defined as the period from the surgical treatment of pancreatic cancer to death or last follow up. The patients were followed until death or until July 2014, whichever came first. Informed consent was obtained from all the patients before specimen collection. This study was approved by the institutional review boards of the three participating institutions and complied with the tenets of the Helsinki Declaration. Our analysis of the pancreatic cancer tissue specimens is fully compliant with the REMARK guidelines [48].
DNA extraction and quantitative PCR for Fusobacterium species
Genomic DNA was extracted from FFPE tissue specimens using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). We used custom-made TaqMan primer/probe sets (Applied Biosystems, Foster City, CA, USA) specific for Fusobacterium species (the sequences are available upon request). The cycle threshold (Ct) values for Fusobacterium species were normalized to prostaglandin transporter (PGT) in each reaction as previously described [27, 49]. The PCR mix consisted of 4 μl of genomic DNA (20 ng/μl), 10 μl of 1× TaqMan Environmental Master Mix 2.0 (Applied Biosystems), 1.0 μl of Fusobacterium species-specific 20× TaqMan primer and probe sets, 1.0 μl of the PGT primer and probe sets and 4 μl of microbial-DNA-free water (Qiagen). The conditions for quantitative PCR were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The SDS v1.4 software (Applied Biosystems) was used for comparative analysis of the cycle thresholds (ΔCt).
Pyrosequencing of KRAS, NRAS, BRAF, and PIK3CA and analysis of MSI
Genomic DNA was used for PCR and targeted pyrosequencing of KRAS (codon 12, 13, 61, or 146), NRAS (codon 12, 13, or 61), BRAF (V600E), and PIK3CA (exon 9 or 20) as described previously [38, 50]. MSI analysis was performed using two markers (BAT25 and BAT26) as described previously [50].
Sodium bisulfite treatment and real-time PCR (MethyLight) to assess promoter methylation of CACNA1G, CDKN2A (p16), IGF2, MLH1, and RUNX3
Bisulfite modification of genomic DNA was performed using the BisulFlash™ DNA Modification Kit (Epigentek, Brooklyn, NY, USA) [39]. DNA methylation was quantified in five promoters [CACNA1G, CDKN2A (p16), IGF2, MLH1, and RUNX3] using Real-Time PCR (MethyLight). CIMP-high status was defined as the presence of three/four or more methylated promoters [CACNA1G, CDKN2A (p16), IGF2, and RUNX3], and CIMP-low/zero was defined as the presence of zero/four to two/four methylated promoters as described previously [39].
RNA extraction and quantitative reverse transcription-PCR (RT-PCR) of miR-21, miRNA-31, and miRNA-143
Total RNA was extracted from FFPE tissues using the miRNeasy FFPE Kit (Qiagen). MiR-21-5p, miR-31-5p, and miR-143-3p expression levels were analyzed by quantitative RT-PCR using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and TaqMan microRNA Assays (Applied Biosystems) as previously described [50]. U6 small nuclear RNA (snRNA; RNU6B; Applied Biosystems) served as an endogenous control. We defined high expression level groups of miR-21 and miR-31 as the fourth level (Q4) in a quartile as described previously [50]. In contrast, low expression level group of miR-143 was defined as the first level (Q1) in a quartile.
Statistical analysis
The JMP (version 10) and SAS (version 9) software applications were used for all calculations (SAS Institute, Cary, NC, USA). To assess associations among the clinical, pathological and molecular characteristics, we have used either the χ2 test for gender; tumor location in pancreas; tumor size; lymph node invasion; year of diagnosis; KRAS mutation; expressions of miR-21, miR-31, and miR-143; MLH1 methylation and CIMP status or Fisher's exact test for disease stage (UICC classification). To compare mean patient ages, t-test was used.
The Kaplan–Meier method and log-rank test were performed to assess the association between Fusobacterium species status of pancreatic cancer and patient mortality. In the analysis of cancer-specific mortality, deaths from causes other than pancreatic cancer were censored. To adjust the results for confounders, we used multivariate Cox proportional hazards regression models to calculate HR according to tumor Fusobacterium species status of pancreatic cancer. We included disease stages (I, II, III, IV, or unknown) as a stratifying variable using the “strata” option in the SAS “proc phreg” command. The multivariate model initially included gender (male versus female), age at diagnosis (continuous variable), tumor size (<2, 2–4, 4–6, or ≥6 cm), tumor location in the pancreas (head, body or tail), year of diagnosis (continuous variable), KRAS mutation status (in codon 12/13/61/146; present versus absent), CIMP status (CIMP-high versus CIMP-low/zero), and miR-21, miR-31, and miR-143 expression levels (high versus low expression levels). To avoid overfitting, variables in the final model were selected using backward stepwise elimination with a threshold of p = 0.10.
Acknowledgments
We deeply thank the pathology departments of Sapporo Medical University Hospital, Teine Keijinkai Hospital, and Otaru municipal Hospital for providing us with tissue specimens.
Abbreviations
- CI
confidence interval
- FFPE
formalin-fixed paraffin-embedded
- CIMP
CpG island methylator phenotype
- HR
hazard ratio
- IBD
inflammatory bowel disease
- miR-21
microRNA-21
- miR-31
microRNA-31
- miR-143
microRNA-143
- MSI
microsatellite instability
- PDAC
pancreatic ductal adenocarcinoma
- RT-PCR
reverse transcription-PCR
- SD
standard deviation
- UICC
Unio Internationalis Contra Cancrum
Footnotes
CONFLICT OF INTEREST
No conflicts of interest.
Writing assistance
The authors would like to thank Enago (www.enago.jp) for the English language review.
GRANT SUPPORT
This work was supported by the Japan Society for the Promotion of Science (JSPS) Challenging Exploratory Research (grant number 25670371 to Y.S.), Ono Cancer Research Foundation (to K.N.), Takeda Science Foundation (to K.N.), The Suhara Memorial Foundation (to K.N.), Yuasa Memorial Foundation (to K.N.), Pancreas Research Foundation of Japan (to K.N.) and Daiwa Securities Health Foundation (to H.I.).
Statement of author contributions
Study concept and design; K.M., K.N., Y.S.
Acquisition of data; K.M., Y.M., H.K., H.I., M.I., S.K., T.N., M.T., Y.K., K.H.
Analysis and interpretation of data; K.M., K.N., T.S., T.H., Y.A.,
Drafting of the manuscript; K.M., K.N.
Critical revision of the manuscript for important intellectual content; K.M., K.N., Y.S., Y.M., R.M., H.M.
Statistical analysis; K.M., K.N., Y.S.
Material support; T.N., T.T., H.M., T.S., T.H., M.I., Y.K., K.H.
Study supervision; K.N., H.M., H.S., H.Y., K.H., K.I., Y.S.
Final approval of manuscript; All authors
REFERENCES
- 1.Vincent A, Hong SM, Hu C, Omura N, Young A, Kim H, Yu J, Knight S, Ayars M, Griffith M, Van Seuningen I, Maitra A, Goggins M. Epigenetic silencing of EYA2 in pancreatic adenocarcinomas promotes tumor growth. Oncotarget. 2014;5:2575–2587. doi: 10.18632/oncotarget.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma MZ, Kong X, Weng MZ, Cheng K, Gong W, Quan ZW, Peng CH. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risch HA. Pancreatic cancer: Helicobacter pylori colonization, N-nitrosamine exposures, and ABO blood group. Mol Carcinog. 2012;51:109–118. doi: 10.1002/mc.20826. [DOI] [PubMed] [Google Scholar]
- 5.Mace TA, Collins AL, Wojcik SE, Croce CM, Lesinski GB, Bloomston M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J Surg Res. 2013;184:855–860. doi: 10.1016/j.jss.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34:2193–2197. doi: 10.1093/carcin/bgt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjonneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764–1770. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frampton AE, Giovannetti E, Jamieson NB, Krell J, Gall TM, Stebbing J, Jiao LR, Castellano L. A microRNA meta-signature for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn. 2014;14:267–271. doi: 10.1586/14737159.2014.893192. [DOI] [PubMed] [Google Scholar]
- 10.Michaud DS, Joshipura K, Giovannucci E, Fuchs CS. A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J Natl Cancer Inst. 2007;99:171–175. doi: 10.1093/jnci/djk021. [DOI] [PubMed] [Google Scholar]
- 11.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 12.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 13.Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, Thompson PA, Jaggi M, Chauhan SC. MicroRNA-14 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget. 2014;5:7599–7609. doi: 10.18632/oncotarget.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi C, Gudino CV, Gibson FC, 3rd CA. Review: Pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol. 2010;25:305–316. doi: 10.1111/j.2041-1014.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swidsinski A, Dorffel Y, Loening-Baucke V, Theissig F, Ruckert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kuhn S, Schilling J, Dorffel WV. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 17.Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minami M, Ando T, Okamoto A, Sasaki N, Ohkura T, Torii K, Hasegawa T, Ohta M, Goto H. Seroprevalence of Fusobacterium varium in ulcerative colitis patients in Japan. FEMS Immunol Med Microbiol. 2009;56:67–72. doi: 10.1111/j.1574-695X.2009.00550.x. [DOI] [PubMed] [Google Scholar]
- 19.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 20.Yoneda M, Kato S, Mawatari H, Kirikoshi H, Imajo K, Fujita K, Endo H, Takahashi H, Inamori M, Kobayashi N, Kubota K, Saito S, Tohnai I, Watanuki K, Wada K, Maeda S, et al. Liver abscess caused by periodontal bacterial infection with Fusobacterium necrophorum. Hepatol Res. 2011;41:194–196. doi: 10.1111/j.1872-034X.2010.00748.x. [DOI] [PubMed] [Google Scholar]
- 21.Athavale NV, Leitch DG, Cowling P. Liver abscesses due to Fusobacterium spp that mimick malignant metastatic liver disease. Eur J Clin Microbiol Infect Dis. 2002;21:884–886. doi: 10.1007/s10096-002-0844-8. [DOI] [PubMed] [Google Scholar]
- 22.Brook I, Frazier EH. Microbiological analysis of pancreatic abscess. Clin Infect Dis. 1996;22:384–385. doi: 10.1093/clinids/22.2.384. [DOI] [PubMed] [Google Scholar]
- 23.Han XY, Weinberg JS, Prabhu SS, Hassenbusch SJ, Fuller GN, Tarrand JJ, Kontoyiannis DP. Fusobacterial brain abscess: a review of five cases and an analysis of possible pathogenesis. Journal of neurosurgery. 2003;99:693–700. doi: 10.3171/jns.2003.99.4.0693. [DOI] [PubMed] [Google Scholar]
- 24.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 25.Shahani L, Khardori N. Fusobacterium necrophorum—beyond Lemierres syndrome. BMJ case reports. 2011 doi: 10.1136/bcr.07.2011.4527. doi:10.1136/bcr.07.2011.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estecio MR, Issa JP. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–90. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 30.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz S, Hackert T, Hartwig W, Rossmanith F, Strobel O, Schneider L, Will-Schweiger K, Kommerell M, Buchler MW, Werner J. Bacterial translocation and infected pancreatic necrosis in acute necrotizing pancreatitis derives from small bowel rather than from colon. Am J Surg. 2010;200:111–117. doi: 10.1016/j.amjsurg.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Zheng L, Giri B. Gastrointestinal Variant of Lemierre Syndrome: Fusobacterium nucleatum Bacteremia-Associated Hepatic Vein Thrombosis: a Case Report and Literature Review. Am J Ther. doi: 10.1097/MJT.0000000000000084. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Scientific reports. 2014;4:4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runkel NS, Rodriguez LF, Moody FG. Mechanisms of sepsis in acute pancreatitis in opossums. Am J Surg. 1995;169:227–232. doi: 10.1016/S0002-9610(99)80142-1. [DOI] [PubMed] [Google Scholar]
- 36.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes LA, Melotte V, de Schrijver J, de Maat M, Smit VT, Bovee JV, French PJ, van den Brandt PA, Schouten LJ, de Meyer T, van Criekinge W, Ahuja N, Herman JG, Weijenberg MP, van Engeland M. The CpG island methylator phenotype: what's in a name? Cancer Res. 2013;73:5858–5868. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 38.Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, Iafrate AJ, Fuchs CS, Haigis KM, Ogino S. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157–163. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, Takahashi T, Adachi Y, Nojima M, Sasaki Y, Tokino T, Baba Y, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 40.Dong J, Zhao YP, Zhou L, Zhang TP, Chen G. Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch Med Res. 2011;42:8–14. doi: 10.1016/j.arcmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu S, Hamada T, Fukuyama T, Nakano R, Uchiyama A, Kawamoto M, Yamaguchi K, Hashimoto H. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol. 2012;25:112–121. doi: 10.1038/modpathol.2011.142. [DOI] [PubMed] [Google Scholar]
- 42.Farrell JJ, Toste P, Wu N, Li L, Wong J, Malkhassian D, Tran LM, Wu X, Li X, Dawson D, Wu H, Donahue TR. Endoscopically Acquired Pancreatic Cyst Fluid MicroRNA 21 and 221 Are Associated With Invasive Cancer. Am J Gastroenterol. 2013;108:1352–1359. doi: 10.1038/ajg.2013.167. [DOI] [PubMed] [Google Scholar]
- 43.Virtue A, Wang H, Yang XF. MicroRNAs and toll-like receptor/interleukin-1 receptor signaling. J Hematol Oncol. 2012;5:66. doi: 10.1186/1756-8722-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honda T, Takahashi N, Miyauchi S, Yamazaki K. Porphyromonas gingivalis lipopolysaccharide induces miR-146a without altering the production of inflammatory cytokines. Biochem Biophys Res Commun. 2012;420:918–925. doi: 10.1016/j.bbrc.2012.03.102. [DOI] [PubMed] [Google Scholar]
- 45.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 46.Rechavi O, Erlich Y, Amram H, Flomenblit L, Karginov FV, Goldstein I, Hannon GJ, Kloog Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes & development. 2009;23:1971–1979. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavano F, di Mola FF, Piepoli A, Panza A, Copetti M, Burbaci FP, Latiano T, Pellegrini F, Maiello E, Andriulli A, di Sebastiano P. Changes in miR-143 and miR-21 expression and clinicopathological correlations in pancreatic cancers. Pancreas. 2012;41:1280–1284. doi: 10.1097/MPA.0b013e31824c11f4. [DOI] [PubMed] [Google Scholar]
- 48.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 49.Boutaga K, Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunology & Medical Microbiology. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, Yamamoto E, Kurokawa S, Adachi Y, Takahashi H, Okuda H, Kusumi T, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35:776–783. doi: 10.1093/carcin/bgt374. [DOI] [PubMed] [Google Scholar]


