Significance
Herbivory is one of the most important processes in the biosphere. When plants germinated underneath the soil or fallen leaves undergo skotomorphogenesis, they are especially prone to a vast range of seed predators and herbivorous arthropods. How greening plants protect themselves against these foes was thus far largely unknown. Here, we describe a mechanism how etiolated seedlings deter arthropod devourers. Our article thus contributes to the understanding of plant survival strategies in the natural environment.
Keywords: skotomorphogenesis, plant defense, herbivore deterrence, cysteine proteases, Kunitz protease inhibitor
Abstract
Water-soluble chlorophyll proteins (WSCPs) constitute a small family of unusual chlorophyll (Chl)-binding proteins that possess a Kunitz-type protease inhibitor domain. In Arabidopsis thaliana, a WSCP has been identified, named AtWSCP, that forms complexes with Chl and the Chl precursor chlorophyllide (Chlide) in vitro. AtWSCP exhibits a quite unexpected expression pattern for a Chl binding protein and accumulated to high levels in the apical hook of etiolated plants. AtWSCP expression was negatively light-regulated. Transgenic expression of AtWSCP fused to green fluorescent protein (GFP) revealed that AtWSCP is localized to cell walls/apoplastic spaces. Biochemical assays identified AtWSCP as interacting with RD21 (RESPONSIVE TO DESICCATION 21), a granulin domain-containing cysteine protease implicated in stress responses and defense. Reconstitution experiments showed tight interactions between RD21 and WSCP that were relieved upon Chlide binding. Laboratory feeding experiments with two herbivorous isopod crustaceans, Porcellio scaber (woodlouse) and Armadillidium vulgare (pillbug), identified the apical hook as Achilles’ heel of etiolated plants and that this was protected by RD21 during greening. Because Chlide is formed in the apical hook during seedling emergence from the soil, our data suggest an unprecedented mechanism of herbivore resistance activation that is triggered by light and involves AtWSCP.
Higher plants can pass through two different developmental programs after seed germination termed skotomorphogenesis and photomorphogenesis. Dark-grown (etiolated) seedlings undergoing skotomorphogenesis have long hypocotyls, an apical hook with closely apposed, unexpanded cotyledons, and a pale-yellow leaf color. By contrast, light-grown seedlings undergoing photomorphogenesis have short hypocotyls, open and fully expanded cotyledons, and are green. Because of their unique morphology, especially the presence of the apical hook, etiolated seedlings are enabled to grow through the soil without mechanical damage (1).
At the cellular level, etioplasts are the predominant plastid form of dark-grown plants, whereas chloroplasts are found in light-grown seedlings. Etioplasts contain a paracristalline internal membrane system termed the prolamellar body (1). In prolamellar bodies of barley and Arabidopsis thaliana etioplasts, two closely related isoforms of NADPH:protochlorophyllide (Pchlide) oxidoreductase, dubbed PORA and PORB, bind Pchlide and NADPH (2). Upon light exposure, Pchlide is converted to chlorophyllide (Chlide), which is subsequently esterified to chlorophyll (Chl) (2). Because the expression of the major light-harvesting Chl a/b-binding proteins (LHCs) is light-induced and because these proteins are not present during the early hours of greening (3), other proteins were suggested to bind the bulk of freshly formed Chlide (4). Early light-induced proteins (ELIPs) are candidates for this function because they are transiently expressed during the developmental switch from dark to light growth (5). However, we were able to show that a distant ortholog of water-soluble chlorophyll proteins (WSCPs) of Brassicaceae exists in barley, which is capable of binding Chlide during the transition of etioplasts to chloroplasts (6).
WSCPs constitute a small family of proteins that have been found in only a limited number of plant species belonging to Amaranthaceae, Brassicaceae, Chenopodiaceae, and Polygonaceae. WSCPs differ from the classical Chl-binding proteins of photosystems I and II in several aspects: (i) they are not constitutively expressed in green leaves, (ii) they are not membrane-bound, (iii) they bind less Chl and Chl a to Chl b at a different ratio, (iv) they do not interact with carotenoids, and (v) they contain a Kunitz protease inhibitor motif in their NH2-terminal parts not present in LHCs (7–10).
Despite considerable progress made over the last few years, definite answers on the actual role of WSCPs in planta are still missing (7–9). In the present work, we used reverse genetics to define the role of WSCP in Arabidopsis thaliana. Our data provide evidence for an unprecedented, organ-specific, and developmentally programmed WSCP function in herbivore deterrence.
Results
AtWSCP Belongs to a Small Family of Kunitz Protease Inhibitors.
WSCPs are characterized by the presence of the chlorophyll(ide)-binding signature PFCPLGI (10) and Kunitz trypsin inhibitor signature [LIVM]-x-D-x-[EDNTY]-[DG]-[RKHDENQ]-x-[LIVM]-x (5)-Y-x-[LIVM]) (7). Based on the occurrence of these motifs, a WSCP-related protein was identified in Arabidopsis thaliana, named AtWSCP, which is encoded by At1g72290. Sequence comparison revealed that AtWSCP belongs to a subfamily of Kunitz protease inhibitors in Arabidopsis (Fig. S1A). Interestingly, the protein encoded by At1g72290 lacks a predictable transit sequence for import into chloroplasts (www.cbs.dtu.dk/services/TargetP and www.cbs.dtu.dk/services/SignalP/) but contains a predictable signal peptide typical for proteins entering the secretory pathway (Fig. S1B) (7).
AtWSCP Is Expressed in Etiolated Seedlings.
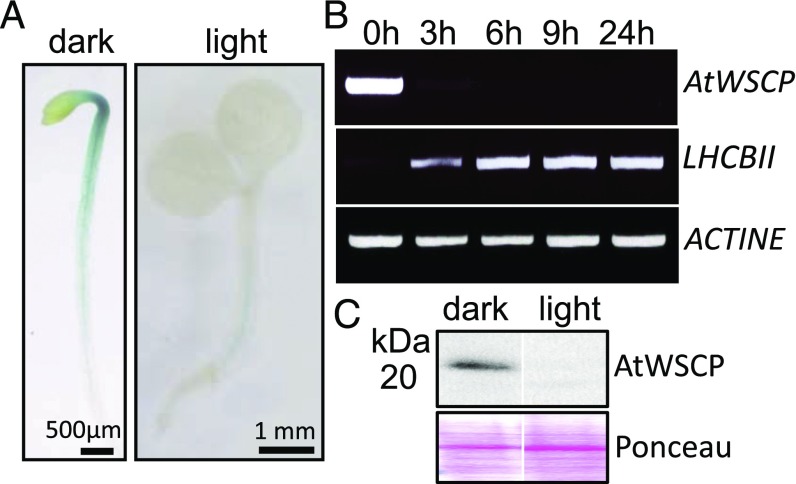
Database searches using the Arabidopsis eFP Browser at the Bio-Array Resource platform of the University of Toronto (11) revealed AtWSCP transcript accumulation in dark-grown, but not in light-grown, seedlings. To substantiate this point, transgenic lines were generated expressing bacterial β-glucuronidase (GUS) under the control of the AtWSCP promoter (SI Materials and Methods). When expression of GUS was analyzed, almost no AtWSCP promoter activity was detectable in the cotyledons of dark-grown seedlings. Instead, strong AtWSCP promoter activity was found around the vascular tissues and especially in those of the apical hook. In light-grown seedlings of the same age, no AtWSCP promoter activity was detectable at all (Fig. 1A).
Fig. 1.
AtWSCP gene expression in dark- and light-grown seedlings. (A) Promoter–β-glucuronidase (GUS) analysis in 3-d-old etiolated and light-grown seedlings of a transgenic line expressing GUS under the control of the AtWSCP promoter (AtWSCP promoter::GUS). (B) AtWSCP transcript levels analyzed by semiquantitative RT-PCR in 3-d-old etiolated seedlings before (0 h) and after 3, 6, 9, and 24 h of white light exposure. Controls show data for Actin as constitutively expressed gene and LHCBII as light-induced gene. (C) AtWSCP protein contents in etiolated and light-grown seedlings, respectively. For comparison, Ponceau-stained blots are shown each containing 50 µg of loaded protein.
Semiquantitative PCR was used to quantify AtWSCP transcript accumulations during greening. Consistent with the AtWSCP promoter::GUS studies, AtWSCP transcripts were detectable only in etiolated seedlings (Fig. 1B). In fact, levels of AtWSCP mRNA rapidly declined during greening (Fig. 1B). By contrast, abundance of LHCBII transcripts encoding the major light-harvesting Chl a/b-binding protein of photosystem II increased steadily (Fig. 1B). Protein gel blot analyses with polyclonal antibodies against bacterially expressed AtWSCP revealed the presence of a single band the size of which matched that of the mature AtWSCP lacking the predicted signal peptide (Fig. 1C). The amount of this AtWSCP protein rapidly declined during greening (Fig. 1C).
Knockout in the AtWSCP Gene Does Not Affect Greening.
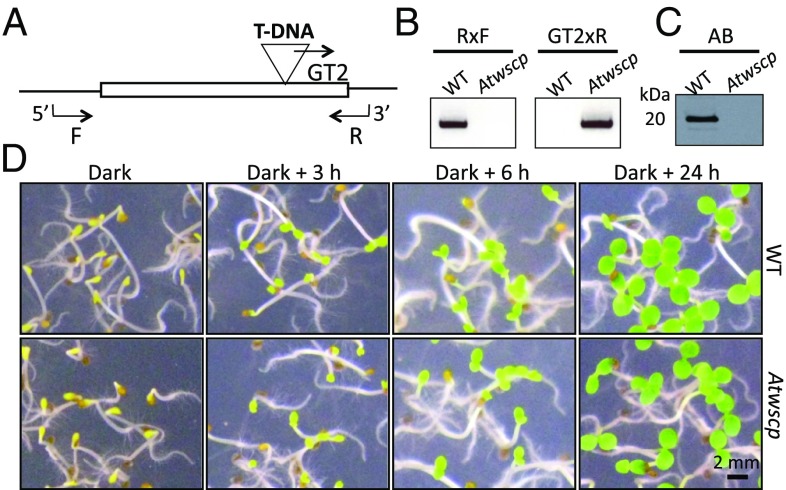
A knockout mutant line (SALK_009681; termed Atwscp) with a T-DNA insertion in the single exon of the AtWSCP gene (Fig. 2A) was obtained from the Salk Institute Genomic Analysis Laboratory collection (12) and used for performing tests to define the role of AtWSCP in planta. Genotyping confirmed the presence of homozygous plants (Fig. 2B). In Western blot analyses, no AtWSCP protein was detectable in etiolated Atwscp versus wild-type seedlings (Fig. 2C). When grown in the dark for 3 d and subsequently exposed to light for 3, 6, and 24 h, respectively, etiolated wild-type and Atwscp seedlings looked indistinguishable and greened with similar kinetics (Fig. 2D).
Fig. 2.
Phenotype of etiolated A. thaliana wild-type and Atwscp knockout seedlings. (A) Schematic presentation of the AtWSCP gene including the 5′ and 3′ untranslated regions, position of the T-DNA insertion in the single exon (triangle), and primers used to prove the presence of the T-DNA (R, F, and GT2). (B) Genotyping of the Atwscp mutant with the indicated gene-specific primers. (C) Western blot with the polyclonal WSCP antibody. (D) Greening of 3-d-old etiolated wild-type and Atwscp mutant seedlings after 3, 6, or 24 h of white light exposure.
AtWSCP Is Not Localized to Plastids.
Quite astonishing for a protein implicated in Chl binding, the structure predictions described before suggested AtWSCP to enter the secretory pathway (Fig. S1B). To gain insights into the localization of AtWSCP in planta, transgenic plants were generated constitutively expressing an AtWSCP::green fluorescent (GFP) fusion protein under the control of the 35S cauliflower mosaic virus (CaMV) promoter. For comparison, transgenic lines were established expressing fusions of GFP with WSCP from Lepidium virginicum (LvWSCP) and precursor ferredoxin (pFD) from Silene pratensis, respectively. pFD is a well-known example of a plastid protein with a typical chloroplast transit peptide directing the protein into the stroma. Similar to AtWSCP, LvWSCP is synthesized with a predictable signal peptide for entering the secretory pathway but nevertheless has been detected in chloroplasts from which it could be isolated and characterized (13).
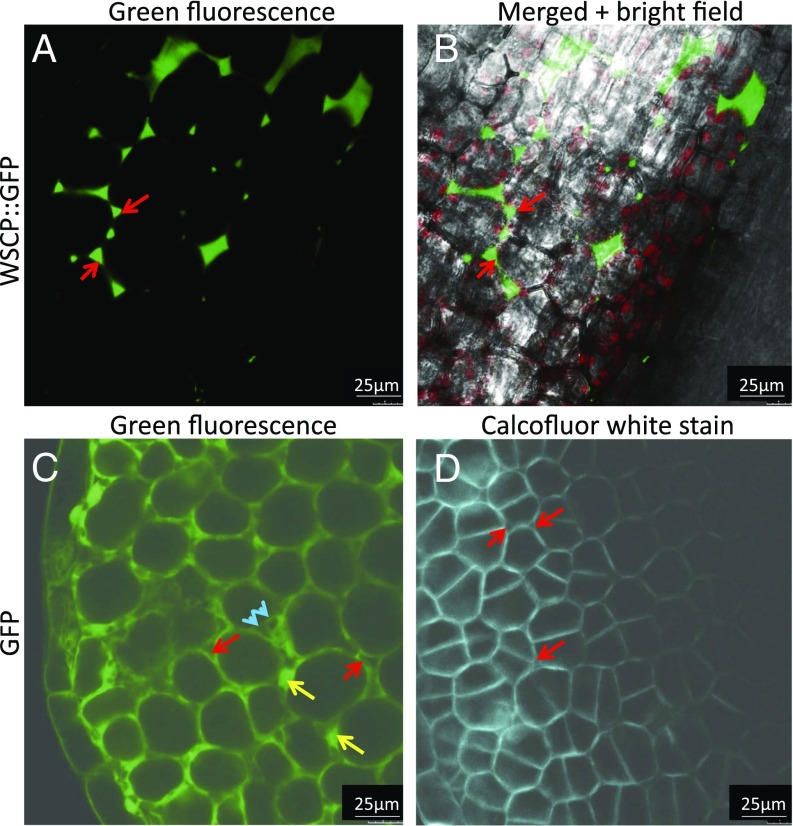
Agrobacterium-mediated transformation of A. thaliana wild-type plants was used to obtain transgenic plants expressing AtWSCP::GFP, LvWSCP::GFP, pFD::GFP, or GFP alone. Etiolated seedlings of the T3 generations of the different transgenic lines were analyzed by confocal laser scanning microscopy. Obviously, AtWSCP::GFP signals were obtained in both the cotyledons and the apical hook. In the cotyledons, AtWSCP::GFP was not localized to the plastids in all transgenic lines examined (replicates of nine plants from five independent transgenic lines). Instead, AtWSCP::GFP fluorescence appeared in the outermost parts of the cell, either representing cell walls/apoplastic spaces or the tonoplast surrounding the vacuole (Fig. 3 A and B; see also Fig. S2 A–C). Often, huge vacuoles are present in plant cells and occupy large areas. In the microscopic images analyzed, the lack of intense “background” fluorescence covering uniformly the cell at first glance seems to disprove that AtWSCP::GFP accumulated in the soluble part of the vacuole. We cannot exclude, however, that AtWSCP::GFP was present in the tonoplast, tightly adhering/being tightly appressed to the cell wall. Such localization would be consistent with the structure predictions identifying a vacuolar localization signal in AtWSCP (Fig. S1).
Fig. 3.
Localization of AtWSCP::GFP in planta. (A and B) Fluorescence analysis of AtWSCP::GFP accumulation in the cotyledons of etiolated seedlings of respective transgenic plants of the T3 generation. (C) Localization of GFP in T3 plants expressing GFP without AtWSCP attached to it. (D) Calcofluor white stain to visualize cell walls in AtWSCP::GFP plants. Red flashes mark apoplastic spaces, yellow flashes mark GFP fluorescence in cytoplasm and nuclei, whereas blue arrowheads indicate sparing of GFP fluorescence by plastids.
For GFP alone without AtWSCP attached to it, a cytoplasmic localization was evident (Fig. 3C; see also Fig. S2 D–F). Counterstaining with Calcofluor white confirmed the presence of thin primary cell walls typical for growing cells and reinforced the idea of AtWSCP::GFP localization in cell walls/apoplastic spaces and/or the tonoplast (Fig. 3D). In transgenic lines constitutively overexpressing a fusion protein consisting of the predicted signal peptide of AtWSCP and GFP (SP::GFP), fluorescence was confined to similar regions of the cell representing cell walls/apoplastic spaces and/or vacuoles (Fig. S3). Because the chimeric protein comprised only the first 30 amino acids of AtWSCP and did not contain the putative vacuolar targeting signal (Fig. S1), we assume that this reporter protein was localized in cell walls/apoplastic spaces, while not necessarily precluding a localization of the full-length AtWSCP in the vacuole. For LvWSCP::GFP, a completely different localization in plastids was found in all transgenic lines examined (Fig. S4 A–C), as was seen for pFd::GFP used as control (Fig. S4 D–F).
AtWSCP::GFP fluorescence distribution in the apical hook was similar to that in the cotyledons and revealed the presence of the protein in cell walls/apoplastic spaces (Fig. S5 A–C). In fact, no fluorescence signal was present in plastids (Fig. S5 A–C), whereas LvWSCP::GFP (Fig. S5 D–F) and pFd::GFP (Fig. S5 G–I) gave rise to strong plastid signals.
AtWSCP Does Not Operate in Vascular Tissue Formation and Hypocotyl Elongation.
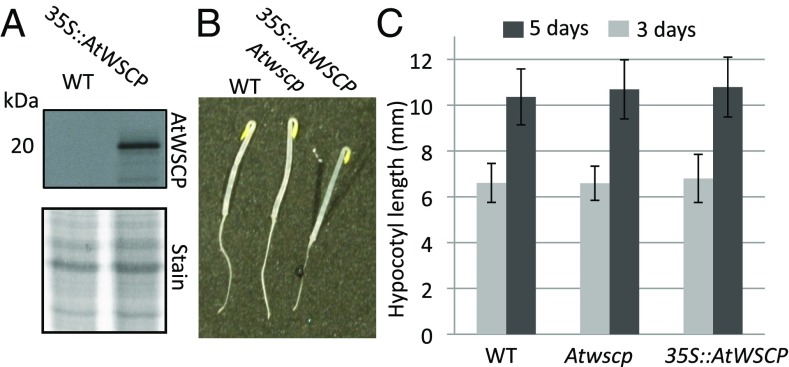
One Kunitz trypsin inhibitor in chickpea, termed TPI-1, has been implicated in regulating hypocotyl growth and apical hook formation (14–17). If AtWSCP were to accomplish a similar role, changing its amount should have severe anatomical (phenotypic) effects. To test this hypothesis, we compared seedling morphology for wild-type and Atwscp mutant seedlings with that of transgenic lines constitutively overexpressing AtWSCP (35S::AtWSCP) under the control of the 35S promoter. Several independent 35S::AtWSCP lines were obtained that overexpressed AtWSCP (one is presented in Fig. 4A). However, in none of these AtWSCP overexpressors could differences in hypocotyl length, light-triggered apical hook straightening, and overall seedling morphology be observed (Fig. 4B). When hypocotyl length was measured after 3 and 5d, respectively, of growth in the dark provoking skotomophogenesis, no difference became apparent for wild-type, Atwscp mutant, and 35S::AtWSCP overexpressing plants (Fig. 4C).
Fig. 4.
Phenotype of etiolated A. thaliana wild-type and Atwscp mutant plants versus transgenic plants overexpressing AtWSCP (35S::AtWSCP). (A) AtWSCP protein levels in leaves of 3-wk-old wild-type and 35S::AtWSCP plants, as assessed by Western blotting using the AtWSCP antiserum (Upper) and Coomassie staining of a replicate SDS/PAGE gel (Lower). (B) Morphology of 3-d-old etiolated wild-type, Atwscp and 35S::AtWSCP seedlings. (C) Hypocotyl length of 3-d-old etiolated wild-type, Atwscp and 35S::AtWSCP seedlings. The data represent the mean of three independent replicates (n = 40; ± defines the SE).
AtWSCP Interacts with the Granulin Domain-Containing Cysteine Proteases RD21.
Halls and coworkers (8) identified AtWSCP as a potent inhibitor of the recombinant proaleurain maturation protease (At3g19390) and papain. There is a small family of granulin domain-containing proteases in Arabidopsis encoded by At3g19390, At5g43060, At4g34460 (XYLEM BARK CYSTEINE PEPTIDASE 3, XBCP3), and At1g47120 (RESPONSIVE TO DESICCATION 21, RD21). RD21 is most interesting because it has been implicated in various reactions conferring resistance to biotic and abiotic foes (18–21). Like many other cysteine endoproteases, RD21 is encoded as a preproprotein having an NH2-terminal propeptide with autoinhibitory activity and a COOH-terminal granulin-domain containing propeptide with unknown function (Fig. S6). The NH2-terminal propeptide is cleaved by a yet-unknown mechanism either requiring an autocatalytic processing under low pH or activity of a processing enzyme (22).
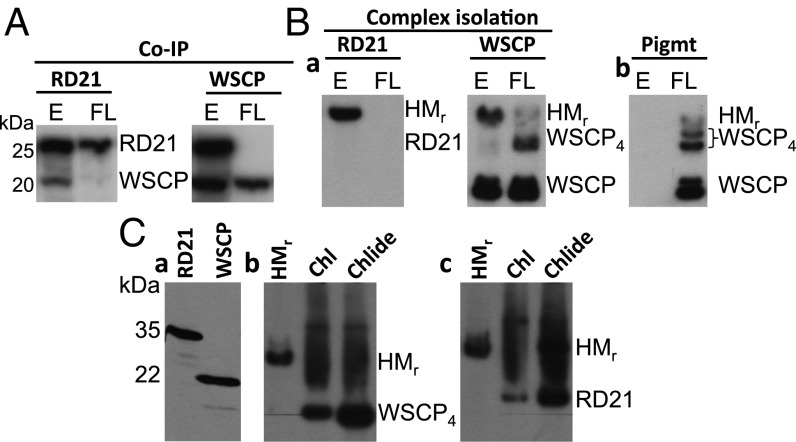
We asked whether RD21 may interact with AtWSCP in etiolated plants. To test for such interactions, pull-down assays were conducted on protein extracts that had been prepared from the apical hook of 4.5-d-old etiolated wild-type plants. Interestingly, both antibodies used, the one against RD21 and the one against AtWSCP, coprecipitated both proteins from the tissue extract of etiolated wild-type plants and, thus, confirmed the interaction of RD21 and AtWSCP in the apical hook. With protein extracts from flashed seedlings, however, the RD21 antibody precipitated only RD21 and the AtWSCP antibody precipitated only AtWSCP (Fig. 5A). This result showed that RD21 and AtWSCP no longer interacted in flash-illuminated seedlings.
Fig. 5.
Interaction of RD21 with WSCP. (A) Pull-down assay to detect RD21–WSCP interactions in plant extracts prepared from the apical hook region of 4.5-d-old etiolated seedlings and seedlings that had been flash-illuminated and kept in the dark for another 2 h. (B) Isolation of high molecular mass (HMr) complexes containing RD21 and WSCP from transgenic plants expressing AtWSCP-(His)6. Protein extracts were prepared from 4.5-d-old etiolated seedlings and flashed seedlings as before and used for nondenaturing PAGE plus Western blotting with the indicated RD21 and WSCP antisera (a) as well as pigment fluorescence analysis (b). (C) In vitro reconstitution of RD21–WSCP complexes and their Chl- and Chlide-dependent dissociation (b and c), as assessed by nondenaturing PAGE. C, a shows 35S-RD21-Flag and 35S-WSCP-(His)6 that had been produced from respective cDNA clones by coupled in vitro transcription/translation and analyzed on a denaturing SDS/PAGE gel.
Next, we isolated WSCP-containing complexes from the apical hook of transgenic plants expressing AtWSCP-(His)6 constitutively. The protein complexes in turn were subjected to nondenaturing PAGE and detected by (i) Western blotting using the WSCP and RD21 antisera and (ii) scoring red light-induced pigment autofluorescence on X-ray films in case the protein would be complexed with Chl and/or Chlide. As shown in Fig. 5B, both assays revealed the presence and light-induced dissociation of higher molecular mass (HMr) complexes containing both WSCP and RD21 in the apical hook of etiolated plants. Several different WSCP bands of descent size were obtained, which most likely represent dissociation intermediates. Interestingly, a pool of monomeric WSCP was present already in the dark but gave rise to an autofluorescing band containing pigment only during greening (Fig. 5B, compare a and b).
In a final experiment, cDNA-encoded Flag-tagged RD21 containing the granulin domain and His-tagged mature AtWSCP lacking its targeting signal were produced by coupled in vitro transcription/translation of respective cDNA clones, purified, and reconstituted into larger complexes. These complexes in turn were supplemented with Chl or Chlide and complex dissociation monitored by nondenaturing PAGE as described before. Fig. 5C depicts HMr complexes containing RD21 and WSCP and shows their Chlide-dependent dissociation. Results shown in the supporting information proved the specificity of the WSCP-pigment interaction and that the dissociation of the HMr complexes led to an activation of RD21’s proteolytic activity. This dissociation is illustrated in Fig. S7, showing the decline of both WSCP and RD21 to vanishingly low levels after prolonged incubations in white light.
Reduced Herbivory on Atwscp Seedlings.
The results presented so far suggested a light-dependent, Chlide- and WSCP-mediated mechanism of RD21 activation in the apical hook of etiolated plants during greening. We asked whether such mechanism could contribute to plant protection against herbivorous arthropods that prey on seeds and young seedlings (23). Examples for such herbivorous arthropods are provided by Porcellio scaber (woodlouse) and Armadillidium vulgare (pillbug), two largely nocturnal isopod crustaceans that are generally considered to represent seed predators and detritivores but can also live as facultative herbivores (23). If deprived for nutrients, woodlice climbed light-grown plants and severed inflorescence stems and petioles (23). Both isopod species also attacked etiolated seedlings and selected the apical hook as primary target. Quantitative assays revealed that approximately 50% of the etiolated seedlings analyzed in three replicate experiments were severed by the isopods, breaking the apical hook and dropping the cotyledons (Fig. 6). The dropped cotyledons in turn were consumed, beginning at the mid ribs and outer leaf edges, but this feeding activity was delayed and occurred only 24 h after the first attack breaking the apical hook.
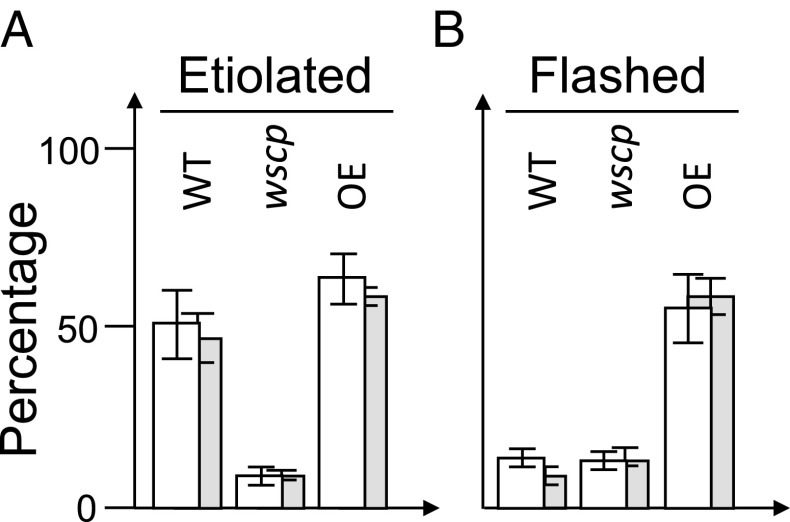
Fig. 6.
Apical hook damage by P. scaber (woodlice, white columns) and A. vulgare (pillbug, gray columns) of etiolated wild-type (WT), Atwscp mutant, and 35S::AtWSCP overexpressor (OE) seedlings. Percentages refer to 4.5-d-old seedlings that had been illuminated for 30 min with white light shifted back to darkness for 2 h (B) or kept in the dark (A). The number of seedlings with damaged apical hooks was counted and is expressed as percentage of the total number of seedlings analyzed in three independent experiments comprising each 120 plantlets and 3 isopods. Error bars are indicated.
Remarkably, the primary attack at the apical hook, but not subsequent feeding activity on the cotyledons, was significantly reduced in the Atwscp mutant containing free RD21 and, in fact, accounted to only 10% of that seen on etiolated wild-type seedlings (Fig. 6). Interestingly, this percentage was similar to the value obtained for wild-type seedlings that had been flashed and subsequently transferred to darkness for 2 h. In marked contrast to these results, no such light effect on the primary and secondary feeding activity of woodlice and pillbugs was observed for seedlings constitutively overexpressing AtWSCP (Fig. 6).
Discussion
AtWSCP Does Not Operate in the Establishment of the Photosynthetic Apparatus.
Previously, a role of WSCPs as Chlide carrier and putative protease inhibitor was proposed (9, 13). Both possibilities are not mutually exclusive. Here, we add more information on AtWSCP’s temporal and spatial expression, its intracellular localization, and its putative function during seedling development and show the following. First, AtWSCP accumulation is not restricted to the transmitting tract tissue of developing flowers, as reported in ref. 9. Substantial amounts of AtWSCP were also found in etiolated plants. Nevertheless, the expression pattern of AtWSCP during light-induced greening was not consistent with a role as transient Chlide carrier during the establishment of the photosynthetic apparatus. For example, AtWSCP promoter activity and AtWSCP transcript accumulation were confined to etiolated plants and were not detected in light-grown seedlings. Moreover, maximum promoter activity was restricted to the apical hook of etiolated seedlings, but no activity was detected in the cotyledons where the bulk of Chl synthesis occurs. Second, AtWSCP was not found in plastids (etioplasts) as should be expected for a protein binding Chl or one of its precursors, but accumulated in cell walls/apoplastic spaces.
AtWSCP and LvWSCP both do not possess predictable chloroplast transit sequences, but contain predictable signal peptides for proteins entering the secretory pathway. Interestingly, a class of chloroplast proteins was identified in Arabidopsis thaliana that are targeted to their final destination through the endoplasmic reticulum (24–26). LvWSCP could be a member of this unique family of N-glycosylated chloroplast proteins (13). Because also AtWSCP appears to enter the secretory pathway but is not targeted to chloroplasts, differences in the structure of the respective signal peptides and/or N-glycosylation sites could be responsible for the differential targeting of AtWSCP and LvWSCP in planta that need to be explored in future work.
AtWSCP Interacts with RD21 in the Apical Hook of Etiolated Seedlings.
AtWSCP interacts with RD21, a papain-like cysteine proteinase involved in stress responses and defense (18–21). Localization studies using the eFP browser (11) identified both AtWSCP and RD21 to have overlapping spatial expression patterns and to accumulate in endoplasmic reticulum (ER) bodies, lytic vacuoles, and cell walls/apoplastic spaces (Fig. S8). Like AtWSCP, RD21 is synthesized as a preproprotein bearing an NH2-terminal signal peptide (predomain) directing the protein to the secretory pathway (22). RD21 additionally contains a prodomain exhibiting autoinhibitory activity and has with the protease domain, proline-rich domain, and granulin domain three supplementary protein modules (ref. 22; compare Fig. S6). The granulin domain shares homology to granulins/epithelins in animals, which are growth hormones that are released upon wounding (27). Activation of RD21 involves removal of the prodomain, giving rise to an intermediate form (iRD21). Then, iRD21 undergoes further processing/maturation that is associated with the removal of the granulin domain (Fig. S6). In planta, RD21 isoforms containing or lacking the granulin domain have been detected (28–30).
The propeptide regulates RD21 activity by autoinhibition during seed development, but on germination when pH drops below 5, intramolecular conformational changes take place that lead to the destabilization and cleavage of the propeptide. Thus, to avoid uncontrolled RD21 activity in both time and space, another layer of regulation exists that consists of protease inhibitors. AtWSCP is such an inhibitor that expresses in etiolated seedlings and controls RD21 activity throughout the skotomorphogenic phase of seedling growth.
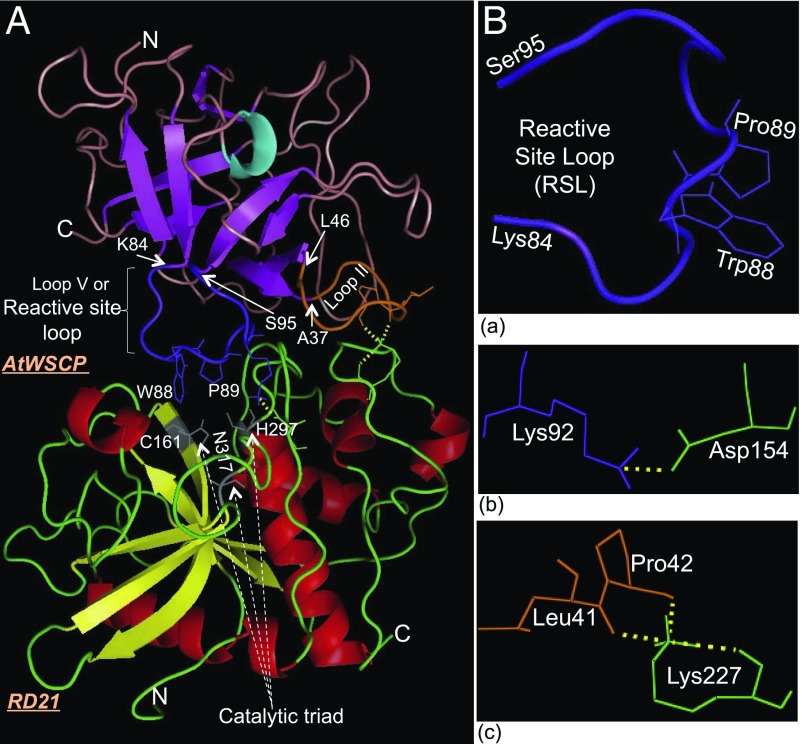
Molecular modeling and docking analyses were carried out to suggest a scenario of how AtWSCP and RD21 may interact. Using established methods (refs. 31–33; SI Materials and Methods), individual 3D models were first built for AtWSCP and RD21. The 3D structure for AtWSCP most closely resembled that of soybean Kunitz-type trypsin inhibitor and tamarind Kunitz inhibitor (TKI), with an α-turn and 10 antiparallel β-strands that form a barrel-like structure (Fig. S9A) (34, 35). Similarly, the molecular modeling of RD21 revealed a typical papain-like structure, with two almost equally sized lobes dubbed R (right) and L (left), divided by an active site cleft (Fig. S9B) (36). The R domain of RD21 is predominantly comprised of an extended NH2-terminal loop and four antiparallel β-strands, whereas the L domain is primarily composed of α-helices and the COOH-terminal end (Fig. S9B).
Based on the 3D models, we next sought to understand the AtWSCP–RD21 interactions. A clue to this end was provided by studies on oryzacystatin-I and papain-like proteases and on TKI and its interactions with factor Xa and trypsin (34, 35, 37). Specifically, the second loop (Ala37-Leu46, orange) of AtWSCP, which spans between β-strands 2 and 3, encompasses the LHCII signature sequence and the fifth loop (Lys84-Ser95, purple), which connects β-strands 5 and 6 is proposed to be the reactive-site loop (RSL) (Fig. 7A and Fig. S9A). In our interaction model, Try88 and Pro89 in the RSL of AtWSCP are predicted to intrude into the active site region of RD21 containing Cys161 and His297 and, thereby, to block its proteolytic activity (Fig. 7 A and B). Moreover, one amino acid residue, Lys92 in the RSL, and two amino acid residues, Leu41 and Pro42 in the LHCII signature sequence, are predicted to form hydrogen bonds with amino acid residues Asp154 and Lys227, respectively, in RD21 (Fig. 7B; see also Fig. S9 C and D). Together, these hydrogen bonds may stabilize the observed AtWSCP–RD21 interaction. However, the presence and close physical proximity of the LHCII signature of AtWSCP to the catalytic triad of RD21 could explain the observed light-triggered, Chlide-dependent dissociation of the AtWSCP–RD21 complex in vitro and in planta.
Fig. 7.
Structural model for the AtWSCP–RD21 interaction, predicted by using ClusPro. (A) Ribbon diagram of the AtWSCP–RD21 complex (front view). The fifth or reactive site loop (RSL) that spans between the fifth and sixth β-strands of AtWSCP is shown in purple, and it’s beginning at Lys84 and end at Ser95 are marked by arrows. The Trp88 and Pro89 residue at the RSL of AtWSCP that intrudes between the catalytic triad, i.e., Cys161, His297, and Asn317 at the active site cleft of RD21 are shown by lines. The β-strands and α-helices of RD21 are shown respectively in yellow and red, whereas the β-strands of AtWSCP are depicted in magenta, an α-turn in cyan, and connecting loops are shown in deep salmon. (B) An enlarged view of the reactive site loop showing the intruding amino acid residues and their respective locations in AtWSCP (a). Specific amino acid residues from AtWSCP and RD21 that form hydrogen bonds are shown in purple or orange and green, respectively. These interactions involve the following: AtWSCP:Lys92-RD21:Asp154 (b), and AtWSCP:Leu41 and Pro42-RD21:Lys227 (c). Amino acid numbering for RD21 is based on their locations in the full-length preproprotein, and for AtWSCP is based on the mature protein after removal of the signal peptide.
AtWSCP Is Operative in Herbivore Resistance Activation During Greening.
As mentioned, the interaction between AtWSCP and RD21 in the apical hook appears to be part of a mechanism of keeping RD21 and, perhaps, also other papain-like cysteine proteinases in an inactive state as long as the seedling etiolates underneath the soil or fallen leaves. Once the seedling de-etiolates, the light-triggered switch to photomorphogenesis then would release RD21 from AtWSCP. Our biochemical studies suggest Chlide as trigger of this dissociation step. How Chlide is transported from the developing chloroplast to cell walls/apoplastic spaces is unknown but may involve stromules, providing recently discovered highways from plastids to the remainder of the cell (38). Superimpose on this very rapid effect is the light-induced depression of AtWSCP gene expression that depletes AtWSCP from etiolated plants and, thereby, increases the amount of free RD21 ready of counteracting proteases present in the arthropod gut. Presumably in concert with other proteinases, efficient protection is conferred onto the seedling’s Achilles’ heel, the apical hook, against arthropod devourers. What destiny the Chlide-complexed WSCP oligomers may have is unclear and should be studied in future work.
Materials and Methods
Plant Material and Growth Conditions.
Seeds of the following Arabidopsis thaliana lines were used for the experiments: ecotype Columbia (Col-0; referred to as wild-type), SALK_009681 (renamed Atwscp) that carries a T-DNA insertion in the gene At1g72290 encoding AtWSCP (12), transgenic lines expressing AtWSCP constitutively (wild-type transformed with the plasmid pB7WG2 containing the coding frame for AtWSCP; referred to as 35S::AtWSCP), lines carrying the promoter of AtWSCP in front of the β-glucuronidase coding sequence (AtWSCP::GUS), and transgenic lines expressing AtWSCP, LvWSCP, and pFD fused to green fluorescent protein (GFP) under the control of 35S promoter (35S::AtWSCP/LvWSCP/pFD::GFP). For comparison, transgenic seedlings expressing a fusion consisting of the signal peptide of AtWSCP and GFP (SP::GFP) were produced. Seeds were plated onto Murashige–Skoog agar medium and germinated in the dark or in white light for appropriate periods.
Whole-Plant Predation Assay.
Populations of 120 wild-type, Atwscp mutant, and 35S::AtWSCP overexpressing plants were grown for 4.5 d in darkness on Petri dishes, and the open dishes were then transferred to a large box, filled to a depth of 6 cm with soil, and containing P. scaber and A. vulgare. Cultivation of the isopods was performed essentially as described in ref. 23. For feeding experiments, P. scaber and A. vulgare were fed with pesticide-free Arabidopsis wild-type plants, then starved for 3 d and placed at low density of 1–3 per liter of soil into the nutrient chamber. At different times, feeding was measured by counting the number of plants with damaged apical hooks and/or dropped cotyledons.
Protein Analyses.
Protein extracts of etiolated seedlings were prepared according to Hurkman and Tanaka (39). Briefly, tissues were extracted with doubly concentrated SDS sample buffer (40), separated on 12% (wt/vol) SDS/PAGE gels, and blotted onto nitrocellulose membranes. Western blotting was carried out according to Towbin et al. (41) by using an alkaline phosphatase-based system with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium or enhanced chemiluminescence (ECL Western Blotting Analysis system; Amersham), respectively. Pull-down assays on protein extracts of the apical hook region were performed with antibodies raised against the bacterially expressed and purified AtWSCP (SI Materials and Methods) and antibodies against RD21, using standard procedures. Isolation of HMr AtWSCP complexes was achieved by Ni-NTA agarose chromatography of apical hook extracts that had been prepared from transgenic seedlings overexpressing a AtWSCP(His)6 protein. In vitro reconstitution experiments and pigment autofluorescence screens were carried out with cDNA-encoded, wheat germ-translated proteins containing or lacking [35S]methionine and isolated pigments (Chl, Chlide, Pchlide) (42).
cDNA Synthesis and Semiquantitative PCR.
Semiquantitative PCR was carried out on DNA templates that had been generated by first-strand cDNA synthesis, using appropriate primers, as described in the SI Materials and Methods.
Promoter Studies.
AtWSCP promoter activity was scored in transgenic plants expressing β-glucuronidase (GUS) by using standard procedures (SI Materials and Methods). Image acquisition was made with an Eclipse E-600 (Nikon) microscope or a SZX12 (Olympus) binocular and documented with an Olympus DP70 camera.
Supplementary Material
Acknowledgments
We thank Dr. Claudia Rossig for a gift of transgenic seeds expressing pFd::GFP fusion proteins and Ikuko Hara-Nishimura (Kyoto University) for a gift of the antibody directed against RD21 (see ref. 28). This work was supported by the Chaire d’Excellence Program of the French Ministry of National Education and Research (to C.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507714112/-/DCSupplemental.
References
- 1.von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. Plant Cell. 1995;7(7):1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reinbothe C, et al. Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 2010;15(11):614–624. doi: 10.1016/j.tplants.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Apel K, Kloppstech K. The plastid membranes of barley (Hordeum vulgare). Light-induced appearance of mRNA coding for the apoprotein of the light-harvesting chlorophyll a/b protein. Eur J Biochem. 1978;85(2):581–588. doi: 10.1111/j.1432-1033.1978.tb12273.x. [DOI] [PubMed] [Google Scholar]
- 4.Reinbothe S, Reinbothe C. The regulation of enzymes involved in chlorophyll biosynthesis. Eur J Biochem. 1996;237(2):323–343. doi: 10.1111/j.1432-1033.1996.00323.x. [DOI] [PubMed] [Google Scholar]
- 5.Adamska I. 2001. The Elip family of stress proteins in the thylakoid membranes of pro- and eukaryota. Regulation of Photosynthesis, eds Aro E-M, Andersson B (Kluwer Academic Publishers, Dordrecht, The Netherlands), pp 487–505.
- 6.Reinbothe C, Satoh H, Alcaraz JP, Reinbothe S. A novel role of water-soluble chlorophyll proteins in the transitory storage of chlorophyllide. Plant Physiol. 2004;134(4):1355–1365. doi: 10.1104/pp.103.033613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh H, Uchida A, Nakayama K, Okada M. Water-soluble chlorophyll protein in Brassicaceae plants is a stress-induced chlorophyll-binding protein. Plant Cell Physiol. 2001;42(9):906–911. doi: 10.1093/pcp/pce117. [DOI] [PubMed] [Google Scholar]
- 8.Halls CE, et al. A Kunitz-type cysteine protease inhibitor from cauliflower and Arabidopsis. Plant Sci. 2006;170:1102–1110. [Google Scholar]
- 9.Bektas I, Fellenberg C, Paulsen H. Water-soluble chlorophyll protein (WSCP) of Arabidopsis is expressed in the gynoecium and developing silique. Planta. 2012;236(1):251–259. doi: 10.1007/s00425-012-1609-y. [DOI] [PubMed] [Google Scholar]
- 10.Green BR, Kühlbrandt W. Sequence conservation of light-harvesting and stress-response proteins in relation to the three-dimensional molecular structure of LHCII. Photosynth Res. 1995;44(1-2):139–148. doi: 10.1007/BF00018304. [DOI] [PubMed] [Google Scholar]
- 11.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi S, et al. Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll-binding protein (WSCP) from Virginia pepperweed (Lepidium virginicum), a unique WSCP that preferentially binds chlorophyll b in vitro. Planta. 2013;238(6):1065–1080. doi: 10.1007/s00425-013-1952-7. [DOI] [PubMed] [Google Scholar]
- 14.Jiménez T, Martín I, Labrador E, Dopico B. A chickpea Kunitz trypsin inhibitor is located in cell wall of elongating seedling organs and vascular tissue. Planta. 2007;226(1):45–55. doi: 10.1007/s00425-006-0465-z. [DOI] [PubMed] [Google Scholar]
- 15.Jiménez T, Martín I, Hernández-Nistal J, Labrador E, Dopico B. The accumulation of a Kunitz trypsin inhibitor from chickpea (TPI-2) located in cell walls is increased in wounded leaves and elongating epicotyls. Physiol Plant. 2008;132(3):306–317. doi: 10.1111/j.1399-3054.2007.01010.x. [DOI] [PubMed] [Google Scholar]
- 16.Hernández-Nistal J, Martín I, Jiménez T, Dopico B, Labrador E. Two cell wall Kunitz trypsin inhibitors in chickpea during seed germination and seedling growth. Plant Physiol Biochem. 2009;47(3):181–187. doi: 10.1016/j.plaphy.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Downing WL, et al. A Brassica napus transcript encoding a protein related to the Künitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J. 1992;2(5):685–693. [PubMed] [Google Scholar]
- 18.Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129(2):175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: Sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 1992;33(3):217–224. [Google Scholar]
- 20.Gepstein S, et al. Large-scale identification of leaf senescence-associated genes. Plant J. 2003;36(5):629–642. doi: 10.1046/j.1365-313x.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 21.Shindo T, Misas-Villamil JC, Hörger AC, Song J, van der Hoorn RA. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE. 2012;7(1):e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu C, et al. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS ONE. 2012;7(3):e32422. doi: 10.1371/journal.pone.0032422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farmer EE, Dubugnon L. Detritivorous crustaceans become herbivores on jasmonate-deficient plants. Proc Natl Acad Sci USA. 2009;106(3):935–940. doi: 10.1073/pnas.0812182106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanjo Y, et al. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;18(10):2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villarejo A, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2008;10(2):220–227. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 26.Faye L, Daniell H. Novel pathways for glycoprotein import into chloroplasts. Plant Biotechnol J. 2006;4(3):275–279. doi: 10.1111/j.1467-7652.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- 27.Bateman A, Bennett HPJ. The granulin gene family: From cancer to dementia. BioEssays. 2009;31(11):1245–1254. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 28.Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001;127(4):1626–1634. [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi Y, et al. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 2001;42(9):894–899. doi: 10.1093/pcp/pce144. [DOI] [PubMed] [Google Scholar]
- 30.Carter C, et al. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16(12):3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32(Web Server issue):W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song HK, Suh SW. Kunitz-type soybean trypsin inhibitor revisited: Refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J Mol Biol. 1998;275(2):347–363. doi: 10.1006/jmbi.1997.1469. [DOI] [PubMed] [Google Scholar]
- 35.Patil DN, Chaudhary A, Sharma AK, Tomar S, Kumar P. Structural basis for dual inhibitory role of tamarind Kunitz inhibitor (TKI) against factor Xa and trypsin. FEBS J. 2012;279(24):4547–4564. doi: 10.1111/febs.12042. [DOI] [PubMed] [Google Scholar]
- 36.Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem Biol. 2006;13(6):637–647. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Benchabane M, Schlüter U, Vorster J, Goulet M-C, Michaud D. Plant cystatins. Biochimie. 2010;92(11):1657–1666. doi: 10.1016/j.biochi.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Hanson MR, Sattarzadeh A. Trafficking of proteins through plastid stromules. Plant Cell. 2013;25(8):2774–2782. doi: 10.1105/tpc.113.112870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurkman WJ, Tanaka CK. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986;81(3):802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinbothe C, Buhr F, Pollmann S, Reinbothe S. In vitro reconstitution of light-harvesting POR-protochlorophyllide complex with protochlorophyllides a and b. J Biol Chem. 2003;278(2):807–815. doi: 10.1074/jbc.M209738200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.