Significance
Double-stranded DNA is one of the stiffest polymers in biology, resisting both bending and twisting over hundreds of base pairs. However, tightly bent DNA loops are formed by proteins that turn off (repress) genes in bacteria. It has been shown that “architectural” proteins capable of kinking any DNA molecule without sequence preference facilitate this kind of gene repression. The mechanism of this effect is unknown for DNA loops involving the well-known Escherichia coli lac repressor. Here we adapt high-resolution protein-mapping techniques to show that an architectural protein directly binds tightly looped DNA to facilitate gene repression by the lac repressor.
Keywords: DNA looping, lac, architectural protein, Nhp6A, E. coli
Abstract
Double-stranded DNA is a locally inflexible polymer that resists bending and twisting over hundreds of base pairs. Despite this, tight DNA bending is biologically important for DNA packaging in eukaryotic chromatin and tight DNA looping is important for gene repression in prokaryotes. We and others have previously shown that sequence nonspecific DNA kinking proteins, such as Escherichia coli heat unstable and Saccharomyces cerevisiae non-histone chromosomal protein 6A (Nhp6A), facilitate lac repressor (LacI) repression loops in E. coli. It has been unknown if this facilitation involves direct protein binding to the tightly bent DNA loop or an indirect effect promoting global negative supercoiling of DNA. Here we adapt two high-resolution in vivo protein-mapping techniques to demonstrate direct binding of the heterologous Nhp6A protein at a LacI repression loop in living E. coli cells.
The local inflexibility of double-stranded DNA limits its bending and twisting over hundreds of base pairs, lengths relevant to DNA biological functions, and interactions with proteins (1, 2). In vitro cyclization kinetics experiments show that the length of DNA most likely to form a circle is ∼450 bp, with the probability of smaller circles dropping exponentially with length, as predicted by the worm-like chain polymer model (1). The bending and twisting persistence lengths of DNA (distances over which an initial trajectory is lost because of thermal energy) are both on the order of 150 bp (1, 2).
Although DNA is locally stiff, worm-like chain theory predicts that millimeter-length bacterial genomic DNAs spontaneously collapse to coils with volumes of a few hundred micrometers cubed. However, DNA packaging into nucleoids, nuclei, and viruses requires at least 400-fold additional compaction by DNA bending and looping beyond what is achieved by thermal energy (3). Eukaryotic nucleosome formation involves wrapping ∼150-bp DNA segments almost twice around histone octamer cores, and DNA segments shorter than one persistence length are also bent and twisted into bacterial repression loops, such as those regulating the lac and gal operons (1, 2, 4–6). Components of the lac operon switch can be reassembled to study DNA looping in vivo, where the β-galactosidase (lacZ) gene is controlled by simultaneous binding of the tetrameric lac repressor (LacI) to two operator sequences flanking a promoter. It has been shown that the resulting tight DNA loop inhibits promoter recognition by RNA polymerase (1, 4, 7) (Fig. 1 A and B). Thus, understanding the deformation of stiff DNA molecules is important in biology.
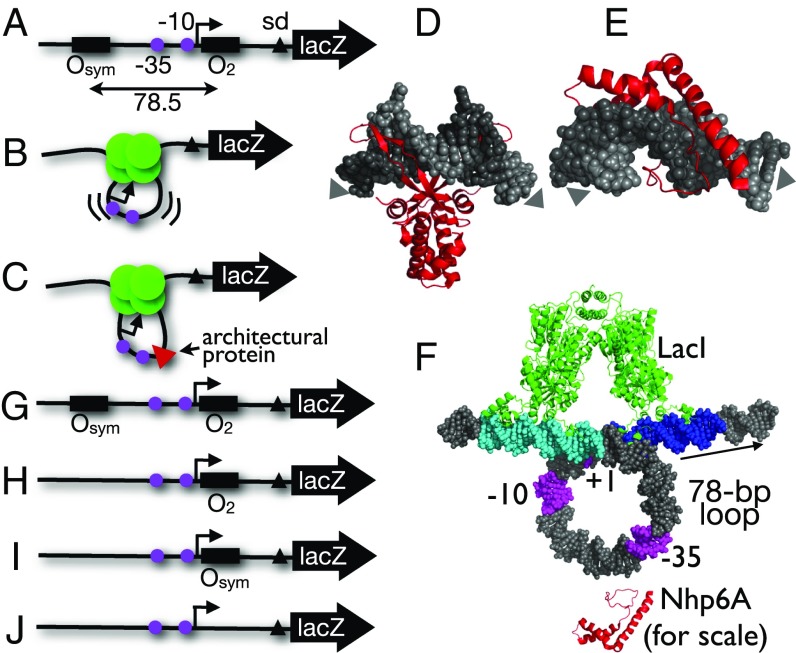
Fig. 1.
Experimental design. (A) lac promoter construct showing cis elements (−35, −10 elements as magenta circles, Shine–Dalgarno element as black triangle). (B) Repression by LacI tetramer (green circles) via a strained DNA loop in cells lacking the E. coli HU architectural protein. (C) Hypothetical facilitation of DNA looping by yeast sequence-nonspecific architectural protein Nhp6A (red triangle). (D) DNA kinking by Anabaena HU [PDB ID code 1P51 (54)]. (E) DNA kinking by S. cerevisiae Nhp6A [PDB ID code 1J5N (25)]. Arrows indicate DNA helix axis trajectory. (F) Model of lac promoter (−10, −35, +1 elements in magenta) captured in a repression loop anchored by LacI tetramer (green) simultaneously binding to upstream (cyan) and proximal (blue) operators. Arrow shows direction of transcription. An Nhp6A architectural protein (red) is indicated near the loop to illustrate scale. (G–J) Experimental lac promoter constructs evaluated here.
Classic (8–13) and more recent (3, 7, 14–20) experiments have manipulated components of the lac operon in vivo to characterize the biophysics of this switch. Changing the relative spacing and DNA affinities of lac operators, and the concentration of LacI allow modeling of the thermodynamic properties of the switch and the elasticities of the polymer components. One of the mysteries resulting from these analyses is the apparent “softness” of DNA in vivo relative to expectations based on in vitro observations (1). Apparent bend-and-twist flexibilities have been estimated to be two- to sevenfold higher in vivo (8, 9, 21). We are interested in understanding the origin of this apparent DNA softening.
A plausible explanation for DNA softening in cells is the presence of abundant sequence-nonspecific “architectural” proteins with the ability to kink DNA, potentially relieving bending strain (Fig. 1C) (22). Architectural proteins include the bacterial histone-like U93 (HU) protein (Fig. 1D) (23, 24) and the eukaryotic high-mobility group B (HMGB) proteins (Fig. 1E) (25–27). Because they bind and kink DNA (28), such proteins reduce the persistence length of DNA in vitro (29–32) and in simulations (33). Architectural DNA bending proteins may facilitate formation of tight repression loops.
Prior studies have explored the role of architectural proteins in the biophysics of bacterial DNA loops at the lac and gal operons. The Adhya laboratory showed that the bacterial HU protein facilitates gal repression by direct binding to kink the looped DNA (34). Such an effect has never been directly shown for loops anchored by LacI. However, we and others have shown that lac repression is substantially weakened in bacteria lacking HU (14, 20) and we demonstrated that heterologous eukaryotic architectural DNA binding proteins can complement this defect (16). It has recently been shown that the presence of HU proteins can buffer sequence-dependent looping effects in vitro and in vivo (20) and Monte Carlo simulations predict how decoration of tightly looped DNA by HU will occur to minimize DNA distortion in the resulting complexes (35). Thus, tight DNA looping might be facilitated by direct binding of architectural DNA binding proteins within the DNA loop.
Although this direct binding model is intuitive and supported for gal, other possible indirect mechanisms exist for architectural protein facilitation of DNA looping. One possibility is related to DNA supercoiling. It has been shown that DNA looping can be stabilized by the unrestrained negative supercoiling typical of bacterial cells (36–39). Supercoiling compacts DNA, raising the local concentration of all DNA sites. Furthermore, DNA supercoiling generates plectonemes where the cost of tight DNA looping is paid by superhelical strain (3, 7, 40). We have shown that deletion of genes encoding various nucleoid proteins, including HU, can change the global superhelical density in Escherichia coli (15). Thus, it is possible that architectural proteins act indirectly to stabilize tight DNA loops by promoting processes that increase global supercoiling.
Here we test the hypothesis that architectural proteins facilitate LacI DNA looping by direct binding to the looped DNA. The model is summarized in Fig. 1F. We complement the looping defect of an HU-deficient E. coli strain by ectopic expression of the Saccharomyces cerevisiae non-histone chromosomal protein 6A (Nhp6A) tagged with a Myc epitope or fused to micrococcal nuclease (MNase). We then adapt two high-resolution methods for mapping protein binding to DNA in living E. coli cells. For three different DNA loop sizes we detect binding of the Nhp6A architectural protein at a single sequence in the lac promoter. Nhp6A binding is not observed in unlooped DNA or when this preferred sequence is missing.
Results and Discussion
Experimental Design.
This study involves mapping DNA binding by the heterologous S. cerevisiae Nhp6A protein complementing the lac repression looping defect of an E. coli strain deleted for the hupA and hupB genes encoding both subunits of the nucleoid HU protein (16). We confirmed and extended our previous results showing that yeast Nhp6A can be expressed in bacteria as an epitope-tagged monomer with or without fusion to MNase, a sequence-nonspecific nuclease that can be activated by Ca2+ ions (Fig. S1A). Importantly, both forms of Nhp6A functionally complement the lac looping defect in ΔHU cells (Fig. S1B). Expression of these Nhp6A proteins allows mapping of Nhp6A binding to four DNA test constructs (Fig. 1 G–J) integrated into the large F′ episome of E. coli. DNA looping is only expected in the Osym/O2 construct (Fig. 1G) (14), where a pair of lac operators is present. Operators are spaced by ∼78 bp [an integral number of DNA helical turns, given our consistent observation of 11 bp per turn for this region in vivo (3)] to allow formation of an untwisted loop.
High-resolution mapping of Nhp6A binding was achieved by two methods adapted for the current project. A chromatin immunoprecipitation exonuclease ligation-mediated PCR (ChIP-exo-LMPCR) method to map protein binding sites at high resolution on a single E. coli promotor was adapted from a published genome-wide eukaryotic protocol (41, 42). The method is outlined in Fig. S2. Briefly, formaldehyde cross-linking and immunoprecipitation of endogenous epitope-tagged protein–DNA complexes is followed by DNA fragmentation and phage λ exonuclease treatment. Cross-linked proteins are detected as obstacles to processive exonuclease digestion, leaving DNA termini adjacent to the complexes. After cross-link reversal, extension of a gene-specific primer, ligation-mediated PCR, and Southern blotting, detection of the immunoprecipitated protein binding sites is achieved at base pair resolution in sequencing gels.
The chromatin endogenous cleavage LMPCR (ChEC-LMPCR) method was adapted for E. coli analysis by modifying protocols also previously implemented in eukaryotes (43, 44). The method is outlined in Fig. S2. Briefly, formaldehyde cross-linking of an endogenously expressed DNA binding protein fused to MNase is followed by transient Ca2+ activation of the nuclease to induce site-specific affinity cleavage of DNA at the site of the bound protein. After reversal of cross-links, capping of nonspecific nicks, polishing of DNA termini, extension of a gene-specific primer, ligation-mediated PCR, and Southern blotting, detection of the MNase fusion protein binding sites is achieved at base pair resolution.
ChIP-exo-LMPCR Mapping of Nhp6A at a LacI Repression Loop.
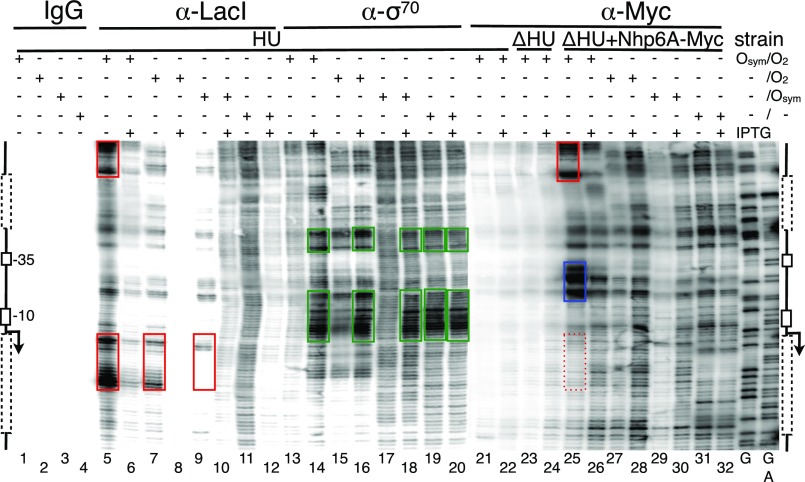
ChIP-exo-LMPCR mapping was applied to four lac constructs (Fig. 1 G–J) to map binding sites of endogenous LacI, the σ70 subunit of E. coli RNA polymerase, and heterologous Nhp6A tagged with a Myc epitope. Results were obtained in the presence and absence of the lac inducer isfopropyl-β-d-thiogalactopyranoside (IPTG) and are shown in Fig. 2. Banding patterns in the Southern blot of a representative sequencing gel can be interpreted relative to the flanking diagrams indicating positions of operators (when present, dotted lines in Fig. 2), the −10 and −35 promoter elements, and transcription start point (broken arrow). Maxam–Gilbert chemical DNA sequencing lanes (G, G+A) were used for reference. We first mapped LacI and the σ70 subunit of E. coli RNA polymerase as positive controls before applying the technique to map Nhp6A.
Fig. 2.
High-resolution in vivo mapping of proteins bound to the lac promoter region of the F′ episome using ChIP-exo-LMPCR. Bacterial cultures were grown to log phase in the presence or absence of 2 mM IPTG, as indicated. Immunoprecipitation of the indicated four formaldehyde cross-linked bacterial lysates was then performed using the indicated antibodies (IgG, α-LacI, α-RNAP σ70, and α-Myc), followed by λ exonuclease digestion of DNA to mark protein complexes, and LMPCR processing. Samples were resolved on 6% (wt/vol) denaturing polyacrylamide gels and imaged following Southern blotting as described in Methods. Promoter region schematic illustrations are shown. Dotted boxes indicate the location of proximal and distal (if present) operators. The position of the transcription start site is indicated by the broken arrow, with the −35 and −10 boxes outlined. Sequencing ladders (G and G+A) were created by standard Maxam and Gilbert chemical modifications of genomic DNA in vitro. Red and green boxes indicate exonuclease terminations associated with LacI and σ70 binding, respectively. Blue box indicates Nhp6A binding. Data are representative of at least three replicates.
In the absence of specific immunoprecipitation, no signal is seen (Fig. 2, lanes 1–4). In contrast, immunoprecipitation of cross-linked LacI protein followed by exonuclease treatment led to strong banding patterns just upstream of occupied operators in the absence (Fig. 2, lane 5), but not in the presence (Fig. 2, lane 6), of IPTG. Interestingly, the position of exonuclease termination is consistently upstream of LacI bound to the strong Osym operator (Fig. 2, lanes 5 and 9), but largely within the binding site of LacI bound to the weaker O2 (Fig. 2, lane 5). This finding suggests that the technique detects subtle differences in protein affinity and DNA sequence-dependent cross-linking with formaldehyde. For cases with one or zero operators (Fig. 2, lanes 7–12), LacI binding is weaker, as expected in the absence of cooperative interactions, and there are no distinct exonuclease terminations in the absence of lac operators (Fig. 2, lanes 11–12). Exonuclease termination signals were not observed further upstream or downstream from the lac promoter. These results for LacI confirmed the sensitivity and specificity of the method.
Results for ChIP-exo-LMPCR mapping of the σ70 subunit of E. coli RNA polymerase are shown in Fig. 2, lanes 13–20. The results confirm expectations: the exonuclease termination signals in the promoter are upstream of the −10 and −35 boxes and strongly IPTG-dependent for the tightly controlled promoter within a repression loop (Fig. 2, lanes 13–14), but less IPTG-dependent when the weak proximal O2 lacks an auxiliary operator (Fig. 2, lanes 15–16), more IPTG-dependent again when the stronger proximal Osym is present (Fig. 2, lanes 17–18), and essentially constitutive in the absence of operators (Fig. 2, lanes 19–20). The IPTG-dependent complementarity between promoter occupation signals because of LacI (Fig. 2, lanes 5–12) vs. σ70 (Fig. 2, lanes 13–20), together with the satisfying position-specificity of the signals, provide unprecedented insight into in vivo protein binding by these factors. These observations demonstrate that ChIP-exo-LMPCR is an effective tool for mapping protein binding. The method was therefore applied to map the binding of Nhp6A.
In the absence of Myc-tagged Nhp6A protein, only background exonuclease termination signals are detected (Fig. 2, lanes 21–24) in ChIP-exo-LMPCR, regardless of the HU status of the cells. In contrast, Myc-tagged Nhp6A creates a very strong pair of exonuclease termination signals just downstream from the −35 box of the lac promoter (Fig. 2, lane 25). We assign these signals to Nhp6A architectural protein bound within the lac loop at this position. This is, to our knowledge, the first such in vivo finding. Note that the strong exonuclease termination signals near the top of the image (Fig. 2, red box in lane 25) are also seen when Osym is occupied by LacI (Fig. 2, upper red box in lane 5). We therefore assign these signals not to Nhp6A, but to LacI cross-linked simultaneously with Nhp6A on the same DNA molecules, acting as a bystander source of exonuclease terminations when Nhp6A is immunoprecipitated. As expected, a corresponding Nhp6A bystander signal is observed in cells expressing Nhp6A when LacI is immunoprecipitated (Fig. S3). The presence of strong exonuclease termination signals attributed to Nhp6A binding within the repression loop correlates with loss of downstream exonuclease termination signals expected for coimmunoprecipitated LacI bound at O2 (Fig. 2, compare lower red box in lane 5 and dotted red box in lane 25). We interpret this suppression as evidence that a large fraction of the captured DNA molecules were cross-linked to LacI at Osym and Nhp6A within the promoter. This finding would explain why most captured DNAs terminated exonuclease cleavage upstream of O2. The strong exonuclease termination signals assigned to Nhp6A within the LacI loop are much attenuated upon gene induction by IPTG (Fig. 2, lane 26), and in the remaining lanes (Fig. 2, lanes 26–32), consistent with the absence of DNA looping in these cases. Thus, formation of the novel Nhp6A complex is strictly dependent on tightly looped DNA.
ChEC-LMPCR Mapping of Nhp6A at a LacI Repression Loop.
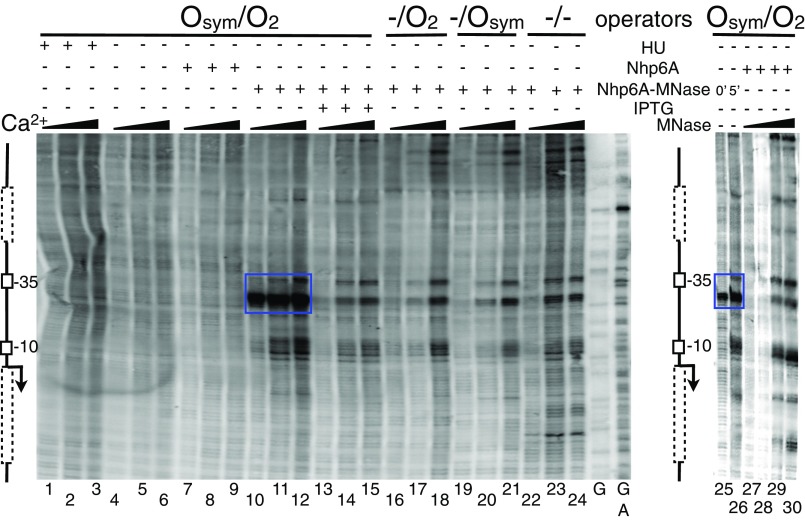
With the described ChIP-exo-LMPCR data showing evidence that the heterologous Nhp6A protein binds directly in LacI loops, we sought to corroborate this result with an independent in vivo protein mapping method. We therefore adapted a ChEC-LMPCR mapping method (43) and applied it to the same four lac constructs to map DNA binding by a Nhp6A-MNase fusion protein. Results are shown in Fig. 3. For the strongly looped Osym/O2 construct, only background signals are observed in the absence of the Nhp6A-MNase fusion (Fig. 3, lanes 1–9). In contrast, a very strong cleavage signal is detected just downstream of the −35 promoter element with increasing Ca2+ activation of the Nhp6A-MNase fusion (Fig. 3, lanes 10–12). This signal is greatly diminished upon IPTG induction (Fig. 3, lanes 13–15), in repressed promoters without DNA looping (Fig. 3, lanes 16–21), and in the constitutive promoter (Fig. 3, lanes 22–24). Three areas of weaker nuclease activity are seen in these cases (Fig. 3, lanes 13–24), corresponding to A/T sequences in the promoter. To interpret this background reactivity, Nhp6A-MNase reactivity on tightly looped DNA (Fig. 3, lanes 25–16) was compared with the same template in the absence of Nhp6A-MNase, treated instead with four increasing concentrations of exogenous MNase after formaldehyde cross-linking in the presence of Nhp6A. The results (Fig. 3, lanes 27–30) confirm that low nonspecific MNase reactivity at A/T promoter sequences is responsible for the background cleavage signal, whereas the dominant cleavage signal near the center of the lac loop in Fig. 3, lanes 10–12 and 25–26, is a result of direct binding of Nhp6A-MNase in the loop. Endogenous MNase fusion proteins create detectable DNA cleavage even before addition of Ca2+ (Fig. 3, lane 25). This is likely because of intracellular Ca2+ concentrations (reportedly near 100 nM) in E. coli (45). These ChEC-LMPCR data confirm the results of ChIP-exo-LMPCR (Fig. 2).
Fig. 3.
In vivo mapping of Nhp6A-MNase fusion protein bound to the lac promoter region of the F′ episome using ChEC-LMPCR. Bacterial cultures were grown to log phase in the presence or absence of 2 mM IPTG, as indicated. Formaldehyde cross-linked bacterial lysates were then analyzed. Flanking schematic diagrams are as in Fig. 2. Where indicated, cleavage by the endogenous MNase fusion protein was activated for 0, 5, or 10 min by addition of 10 mM Ca2+. As a control, increasing concentrations of commercial MNase were added to cross-linked lysates containing expressed Nhp6A (lanes 27–30). Sequencing ladders (G and G+A) were as described in Fig. 2. Blue box indicates Nhp6A binding. Data are representative of at least three replicates.
A Tightly Bent lac Promoter Sequence Recruits an Architectural DNA Binding Protein.
We wished to determine if Nhp6A is recruited to a specific sequence or to a preferred position within the tight repression loop. We therefore compared Nhp6A binding in stable lac repression loops of 67.5 bp, 78.5 bp, and 89.5 bp. Remarkably, Nhp6A binding is detected at the same sequence in all three loops, both by ChIP-exo-LMPCR and ChEC-LMPCR (Figs. S4 and S5). Because the relative position of this sequence is different in each of the loops, this result shows that architectural protein binding is determined by sequence in these tightly bent DNAs.
Summary and Prospects.
We previously showed that loss of the abundant E. coli HU architectural protein disables DNA looping by LacI in vivo (14), and that heterologous eukaryotic HMGB architectural proteins can complement this defect (16). Loss of HU reduces the global unrestrained negative superhelical density in E. coli (15), an effect that might indirectly reduce DNA looping by LacI (38). On the other hand, tight DNA looping in the gal operon has been shown to be facilitated by direct HU binding in the DNA loop, and fitting to a thermodynamic model of in vivo E. coli LacI looping data, as well as Monte Carlo simulations of LacI loops, also raise the possibility of loop facilitation by direct architectural protein binding. Our data do not rule out a role for global supercoiling effects. However, using two novel adaptations of high-resolution in vivo methods we provide clear evidence of direct DNA loop binding by Saccharomyces Nhp6A, an architectural protein complementing an HU defect in the test strains. The ChIP-exo-LMPCR and ChEC-LMPCR methods provide base pair-resolution data documenting signals because of λ exonuclease and tethered MNase, respectively. Mapping data for LacI, σ70, and Nhp6A are summarized in Fig. 4A. The complementing patterns of LacI and σ70 binding under repressed vs. induced conditions provide unprecedented insight into protein occupancy of this series of engineered looped and unlooped E. coli promoters and lac operators. The results are entirely consistent with expectations, and also reveal the potential sensitivity of ChIP-exo-LMPCR to subtle effects of operator sequence and affinity on patterns of formaldehyde cross-linking and exonuclease termination. This sensitivity can complicate mapping of precise protein binding sites (42). The method is also shown to detect cross-linked bystander proteins coimmunoprecipitated with local target proteins of interest.
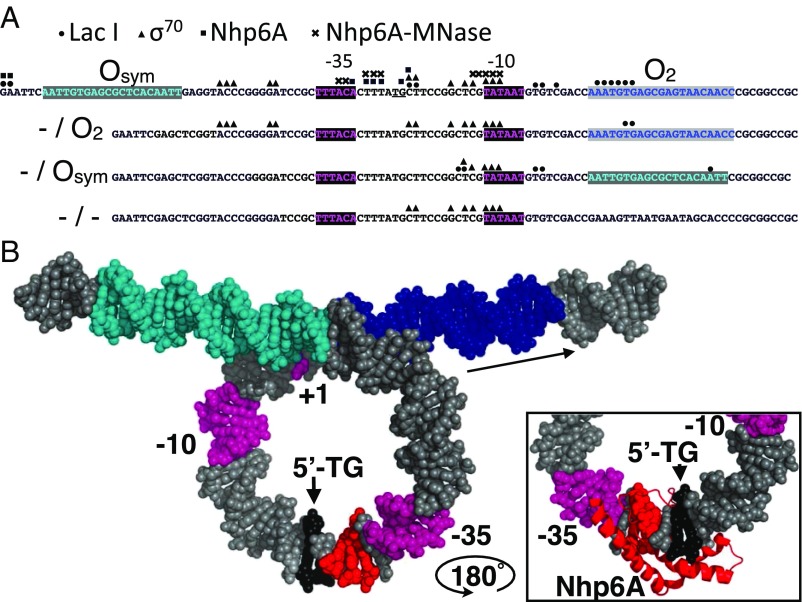
Fig. 4.
Summary of high-resolution protein binding data and model. (A) Four tested lac promoter constructs with the indicated lac operators and promoter elements showing protein binding sites identified by ChIP-exo-LMPCR (LacI, circles; σ70, triangles; Nhp6A, squares near TG/CA dinucleotide, underlined), and by ChEC-LMPCR (Nhp6A-MNase, crosses). (B) Model of LacI loop showing operators (cyan and blue), promoter elements (magenta), and Nhp6A cleavage sites (red) identified by ChEC-LMPCR near the TG/CA dinucleotide (black) proposed as the kinked binding site. (Inset) Illustration of plausible intercalation of Nhp6A methionine 29 at the TG/CA dinucleotide based on [PDB ID code 1J5N (25)] after rotation of the complex by 180° about a vertical axis.
Of greatest significance to testing the current hypothesis is the clear detection of the Nhp6A protein bound to a specific lac promoter sequence within the DNA looped by LacI (Fig. 4A). Exonuclease termination sites just downstream of the −35 box of the test promoter suggest a single Nhp6A binding site in the loop (Fig. 4B). Comparison of the apparent Nhp6A binding sequence with the reported high-resolution Nhp6A-DNA NMR structure (25) immediately suggests a model for this interaction (Fig. 4B and Fig. S6). Nhp6A binding appears to map to a 5′-TG/CA base pair step in the lac promoter, a sequence known to be readily kinked (46). Nhp6A kinking at such a dinucleotide involves intercalation of methionine 29. We propose that this kinkable site within the lac promoter plays a natural role in recruiting architectural protein stabilizers of the tight repression loop as has been suggested for HU (20). We show using ChIP-exo-LMPCR and ChEC-LMPCR that this is the preferred Nhp6A binding sequence in repression loops of three different sizes, and that a loop lacking this sequence does not recruit Nhp6A. Furthermore, Nhp6A binding is detected only in cases of tight DNA looping by LacI. Thus, we show that the sequence of the lac promoter encodes an architectural DNA binding protein site reminiscent of the architectural protein binding site at the gal loop.
Efforts are underway to extend the present methods to map binding of the endogenous heterodimeric HU protein where recent in vitro, in vivo, and simulation studies propose repression facilitation by direct loop binding (34, 35). In addition, the constructs studied here are based on components of the lac operon control switch, but differ in subtle sequence and spacing details relative to the wild-type lac promoter. We are now mapping protein binding sites on the wild-type lac operon promoter in vivo.
Our experimental results are directly relevant to the fascinating recent experimental and simulation work of the Phillips and coworkers (20) and Olson and coworkers (35). Boedicker et al. (20) confirm our prior in vivo result (14) that E. coli architectural protein HU facilitates gene repression by LacI, and extend the work in vitro using tethered particle motion experiments. The authors show that DNA looping sequence effects become latent in the presence of HU. Using data fitting to an insightful statistical mechanics model, Boedicker et al. (20) go on to propose that loop facilitation is because of two HU proteins binding directly within the lac loop. The absence of supercoiling effects in the tethered particle motion experiments tends to support this direct loop binding hypothesis. Wei et al. (35) use Monte Carlo simulations to argue that, at equilibrium, sequence-nonspecific architectural proteins, such as HU, will spontaneously decorate tight DNA loops at preferred positions due to thermodynamic effects (minimizing the free energy of the strained system). The authors simulate random uptake of HU proteins onto the 92-bp wild-type lacI loop and find one or two HU proteins bound in cases of successfully closed structures. In light of these predictions and the data presented here, it will be very interesting to apply the ChIP-exo-LMPCR and ChEC-LMPCR methods to experimentally map HU binding sites on the wild-type LacI loop in living bacteria. These experiments promise important new insights into the mystery of apparent DNA softening in vivo.
Methods
Bacterial Strains.
The four DNA promoter/operator looping DNA constructs (Fig. 1 G–J) used in this study were based on plasmid pJ992 (14) created by modification of pFW11-null (47). See SI Methods for full details.
Protein Expression Constructs.
Nhp6A and Nhp6A-MNase protein expression constructs were created by inserting purified PCR products into plasmid pJ1035, a modified version of pLX20 containing a promoter driving moderate levels of protein expression (14). Both full-length Nhp6A (pJ1327) and Nhp6A Δ2–12 (pJ1328) were previously described (16). See SI Methods for full details.
Molecular Modeling.
Molecular docking and graphics were implemented with 3D-DART (48) and Pymol (49).
β-Galactosidase Enzyme Assays.
A liquid β-galactosidase colorimetric enzyme assay measured lacZ expression was performed as described previously (15). The repression ratio (RR) is given as the ratio of induced/repressed expression, where induction is obtained by addition of 2 mM IPTG. Analysis of the resulting lac reporter gene expression patterns was performed as described previously (50), with fitting optimization using a simplex and inductive search hybrid algorithm (51).
Bacterial Growth and Formaldehyde Cross-Linking.
E. coli strains carrying the indicated protein expression plasmids were grown to log phase in 40 mL LB medium at 37 °C in the presence or absence of 2 mM IPTG. Cultures were pelleted at 4,000 × g for 10 min at room temperature and resuspended in 20 mL PBS (Mg2+- and Ca2+-free) before cross-linking of macromolecules by the addition of 37% (wt/vol) formaldehyde (Sigma) to a final concentration of 0.75%. Cultures were maintained at room temperature with constant gentle swirling for 20 min. Cross-linking was terminated by addition of cold, 2 M Tris⋅HCl (pH 8.0) to a final concentration of 260 mM. Cells were harvested by centrifugation, washed three times with 4 mL cold PBS, and cell pellets stored at −80 °C until ready for processing.
ChIP-exo-LMPCR Analysis.
This method was adapted for bacterial analysis based on previous eukaryotic methods (41, 42). See SI Methods for full details.
ChEC-LMPCR Analysis.
This method was adapted for bacterial analysis from previous publications (43, 44). See SI Methods for full details.
LMPCR.
λ Exonuclease and MNase cleavage sites were analyzed by adaptation of standard LMPCR methods (52, 53). See SI Methods and Fig. S7 for full details.
Supplementary Material
Acknowledgments
The authors thank Justin Peters for expert assistance. This work was supported by the Mayo Foundation and National Institutes of Health Grant GM75965 (to L.J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500412112/-/DCSupplemental.
References
- 1.Peters JP, 3rd, Maher LJ., 3rd DNA curvature and flexibility in vitro and in vivo. Q Rev Biophys. 2010;43(1):23–63. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia HG, et al. Biological consequences of tightly bent DNA: The other life of a macromolecular celebrity. Biopolymers. 2007;85(2):115–130. doi: 10.1002/bip.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond LM, Peters JP, Becker NA, Kahn JD, Maher LJ., 3rd Gene repression by minimal lac loops in vivo. Nucleic Acids Res. 2010;38(22):8072–8082. doi: 10.1093/nar/gkq755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adhya S. Multipartite genetic control elements: Communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 5.Semsey S, Virnik K, Adhya S. A gamut of loops: Meandering DNA. Trends Biochem Sci. 2005;30(6):334–341. doi: 10.1016/j.tibs.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Müller-Hill B. The lac Operon: A Short History of a Genetic Paradigm. Walter de Gruyter; Berlin: 1996. [Google Scholar]
- 7.Becker NA, Greiner AM, Peters JP, Maher LJ., 3rd Bacterial promoter repression by DNA looping without protein-protein binding competition. Nucleic Acids Res. 2014;42(9):5495–5504. doi: 10.1093/nar/gku180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellomy GR, Mossing MC, Record MT., Jr Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988;27(11):3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 9.Law SM, Bellomy GR, Schlax PJ, Record MT., Jr In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J Mol Biol. 1993;230(1):161–173. doi: 10.1006/jmbi.1993.1133. [DOI] [PubMed] [Google Scholar]
- 10.Mossing MC, Record MT., Jr Upstream operators enhance repression of the lac promoter. Science. 1986;233(4766):889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 11.Krämer H, et al. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257(1):21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 13.Oehler S, Eismann ER, Krämer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9(4):973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker NA, Kahn JD, Maher LJ., 3rd Bacterial repression loops require enhanced DNA flexibility. J Mol Biol. 2005;349(4):716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Becker NA, Kahn JD, Maher LJ., 3rd Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35(12):3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker NA, Kahn JD, Maher LJ., 3rd Eukaryotic HMGB proteins as replacements for HU in E. coli repression loop formation. Nucleic Acids Res. 2008;36(12):4009–4021. doi: 10.1093/nar/gkn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebastian NT, Bystry EM, Becker NA, Maher LJ., 3rd Enhancement of DNA flexibility in vitro and in vivo by HMGB box A proteins carrying box B residues. Biochemistry. 2009;48(10):2125–2134. doi: 10.1021/bi802269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker NA, Peters JP, Maher LJ, 3rd, Lionberger TA. Mechanism of promoter repression by Lac repressor-DNA loops. Nucleic Acids Res. 2013;41(1):156–166. doi: 10.1093/nar/gks1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han L, et al. Concentration and length dependence of DNA looping in transcriptional regulation. PLoS ONE. 2009;4(5):e5621. doi: 10.1371/journal.pone.0005621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boedicker JQ, Garcia HG, Johnson S, Phillips R. DNA sequence-dependent mechanics and protein-assisted bending in repressor-mediated loop formation. Phys Biol. 2013;10(6):066005. doi: 10.1088/1478-3975/10/6/066005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in-vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS ONE. 2006;1:e136. doi: 10.1371/journal.pone.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czapla L, Peters JP, Rueter EM, Olson WK, Maher LJ., 3rd Understanding apparent DNA flexibility enhancement by HU and HMGB architectural proteins. J Mol Biol. 2011;409(2):278–289. doi: 10.1016/j.jmb.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouvière-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 24.Swinger KK, Rice PA. IHF and HU: Flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14(1):28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Masse JE, et al. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol. 2002;323(2):263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 26.Travers AA, Ner SS, Churchill ME. DNA chaperones: A solution to a persistence problem? Cell. 1994;77(2):167–169. doi: 10.1016/0092-8674(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 27.Travers AA. Priming the nucleosome: A role for HMGB proteins? EMBO Rep. 2003;4(2):131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi ME. Prokaryotic HU and eukaryotic HMG1: A kinked relationship. Mol Microbiol. 1994;14(1):1–5. doi: 10.1111/j.1365-2958.1994.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 29.Ross ED, Hardwidge PR, Maher LJ., 3rd HMG proteins and DNA flexibility in transcription activation. Mol Cell Biol. 2001;21(19):6598–6605. doi: 10.1128/MCB.21.19.6598-6605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCauley MJ, Zimmerman J, Maher LJ, 3rd, Williams MC. HMGB binding to DNA: Single and double box motifs. J Mol Biol. 2007;374(4):993–1004. doi: 10.1016/j.jmb.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, McCauley MJ, Maher LJ, 3rd, Williams MC, Israeloff NE. Mechanism of DNA flexibility enhancement by HMGB proteins. Nucleic Acids Res. 2009;37(4):1107–1114. doi: 10.1093/nar/gkn1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCauley MJ, Rueter EM, Rouzina I, Maher LJ, 3rd, Williams MC. Single-molecule kinetics reveal microscopic mechanism by which high-mobility group B proteins alter DNA flexibility. Nucleic Acids Res. 2013;41(1):167–181. doi: 10.1093/nar/gks1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czapla L, Swigon D, Olson WK. Effects of the nucleoid protein HU on the structure, flexibility, and ring-closure properties of DNA deduced from Monte Carlo simulations. J Mol Biol. 2008;382(2):353–370. doi: 10.1016/j.jmb.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16(12):3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei J, Czapla L, Grosner MA, Swigon D, Olson WK. DNA topology confers sequence specificity to nonspecific architectural proteins. Proc Natl Acad Sci USA. 2014;111(47):16742–16747. doi: 10.1073/pnas.1405016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borowiec JA, Zhang L, Sasse-Dwight S, Gralla JD. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- 37.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70(6):2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krämer H, Amouyal M, Nordheim A, Müller-Hill B. DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988;7(2):547–556. doi: 10.1002/j.1460-2075.1988.tb02844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lia G, et al. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc Natl Acad Sci USA. 2003;100(20):11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boles TC, White JH, Cozzarelli NR. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990;213(4):931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 41.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147(6):1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee HS, Pugh BF. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol. 2012;Chapter 21:24. doi: 10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid M, Durussel T, Laemmli UK. ChIC and ChEC; Genomic mapping of chromatin proteins. Mol Cell. 2004;16(1):147–157. doi: 10.1016/j.molcel.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Goetze H, et al. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol Cell Biol. 2010;30(8):2028–2045. doi: 10.1128/MCB.01512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262(26):12570–12574. [PubMed] [Google Scholar]
- 46.Marathe A, Bansal M. An ensemble of B-DNA dinucleotide geometries lead to characteristic nucleosomal DNA structure and provide plasticity required for gene expression. BMC Struct Biol. 2011;11:1. doi: 10.1186/1472-6807-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26(16):3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Dijk M, Bonvin AM. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009;37(Web Server issue):W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrodinger LLC 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
- 50.Peters JP, et al. Quantitative methods for measuring DNA flexibility in vitro and in vivo. Methods Enzymol. 2011;488:287–335. doi: 10.1016/B978-0-12-381268-1.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Offord C, Bajzer Z. A hybrid global optimization algorithm involving simplex and inductive search. In: Alexandrov VN, Dongarra JJ, Juliano BA, Renner RS, Tan CJK, editors. Computational Science-ICCS 2001. Springer; Berlin: 2001. pp. 680–688. [Google Scholar]
- 52.Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- 53.Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 54.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22(14):3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.