Significance
The circadian clock allows an organism to anticipate daily changes imposed by the environment. The response to LPS is altered depending on time of day; however, the molecular mechanisms underlying this are unclear. We find that the clock in myeloid cells plays a role in LPS-induced sepsis by altering NF-κB activity and the induction of the microRNA miR-155. LPS causes the repression of BMAL1 via the targeting of miR-155 to two seed sequences in the 3′-untranslated region of Bmal1. Lack of miR-155 has profound effects on circadian function and circadian induction of cytokines by LPS. Thus, the molecular clock is using miR-155 as an important regulatory component to control inflammation variably across the circadian day in myeloid cells.
Keywords: inflammation, sepsis, circadian clock, miR-155, Bmal1
Abstract
The response to an innate immune challenge is conditioned by the time of day, but the molecular basis for this remains unclear. In myeloid cells, there is a temporal regulation to induction by lipopolysaccharide (LPS) of the proinflammatory microRNA miR-155 that correlates inversely with levels of BMAL1. BMAL1 in the myeloid lineage inhibits activation of NF-κB and miR-155 induction and protects mice from LPS-induced sepsis. Bmal1 has two miR-155–binding sites in its 3′-UTR, and, in response to LPS, miR-155 binds to these two target sites, leading to suppression of Bmal1 mRNA and protein in mice and humans. miR-155 deletion perturbs circadian function, gives rise to a shorter circadian day, and ablates the circadian effect on cytokine responses to LPS. Thus, the molecular clock controls miR-155 induction that can repress BMAL1 directly. This leads to an innate immune response that is variably responsive to challenges across the circadian day.
Cellular molecular clocks entrain the body to deal with periodic daily changes in the environment. They anticipate and coordinate physiological, behavioral, and biochemical responses. Clocks influence a myriad of fundamental processes such as activity, feeding behavior, body temperature, cell cycle regulation, and hormonal secretion and, as such, are central to the coordination of highly integrated response systems such as metabolism, cardiovascular homeostasis, and immune function (1).
The type and magnitude of the immune response to an inflammatory challenge alters significantly throughout the day, probably to respond optimally at times when the host is most likely to encounter pathogens or danger signals and to optimize the opportunity for resolution of inflammation and recovery (2). In mice, death from sepsis is greatest when the animals begin to transition into the active phase (3–5), and this correlates with an increase in immune cell number (6), immune cell trafficking (4, 7), and circadian gated cytokine production from immune cells (6, 8). Chronic disruption of the molecular clock in mice via jet lag leads to enhanced LPS-induced sepsis, and peritoneal macrophages harvested from these mice produce a greater amount of IL6 in response to LPS (9).
However, the molecular basis underlying the circadian control of innate immunity is still not fully understood. Evidence exists for circadian oscillations of some Toll-like receptors (TLRs) and some of their downstream effector genes (6, 10). TLR9, a receptor for CpG-rich DNA, is controlled by BMAL1 and CLOCK promoter binding (11). BMAL1 and CLOCK are basic helix–loop–helix PAS (bHLH/PAS) transcription factors that drive oscillatory gene expression and lie at the core of molecular clockworks. BMAL1 has also been shown to attenuate NF-κB activation by sequestering CLOCK. CLOCK is required for acetylation of p65, a key event for NF-κB transactivation (12) and downstream cytokine production. Rhythmic oscillation in the numbers of Ly6Chi monocytes in circulation and the magnitude of recruitment of these cells into inflamed tissue is dependent on BMAL1 (4). As cells lacking BMAL1 lose circadian expression of many clock components, the effect of BMAL1 on inflammation may be due to other clock components acting as intermediaries. For example, the expression of the clock component Rev-Erbα, a nuclear receptor that functions as a transcriptional repressor, is regulated positively by BMAL1 (13, 14). REV-ERBα has been shown to act as the rhythmic repressor of proinflammatory cytokine production, in particular via repression of IL6 (8). REV-ERBα can repress via recruitment of the NCoR–HDAC3 complex (14) but also through inhibition of enhancer-derived RNAs (15).
miRNAs are estimated to control more than 30% of the human protein-coding genome (16). Evidence that miRNAs can regulate and be regulated by aspects of the molecular clock exists in Drosophila (17) and mice (18). Cheng et al. (19) identified miR-219 and miR-132 as regulated, respectively, by the molecular clock and light impinging on the suprachiasmatic nucleus (which contains the master clock). miR-219 regulates the length of the circadian day and miR-132 modulates the phase-shifting capacity of light. REV-ERBα regulates miR-122 in the liver (20), and miR- 142–3p is controlled by the BMAL1:CLOCK heterodimer and, in turn, can target Bmal1 (21).
Here, we provide evidence that miR-155, a proinflammatory microRNA induced by TLRs (22, 23), controls Bmal1 mRNA and protein levels in myeloid cells, leading to alterations in clock function and circadian control of inflammation. In agreement with a previous study (4), we demonstrate that the time-dependent variation in the consequences of acute sepsis is reliant on the level of myeloid BMAL1. The molecular clock attenuates inflammation via its effects on NF-κB (12) and can suppress proinflammatory cytokines and miR-155. miR-155 via its effects on BMAL1 can alter circadian function, including the control of inflammatory cytokines. We identify miR-155 as a critical posttranscriptional repressor of Bmal1, providing a direct link between the molecular clock, a microRNA, and immune function in macrophages.
Results
Mice Deficient in Myeloid BMAL1 Have an Increased Risk of LPS-Induced Sepsis with Heightened Induction of the Proinflammatory microRNA miR-155 and NF-κB Activity.
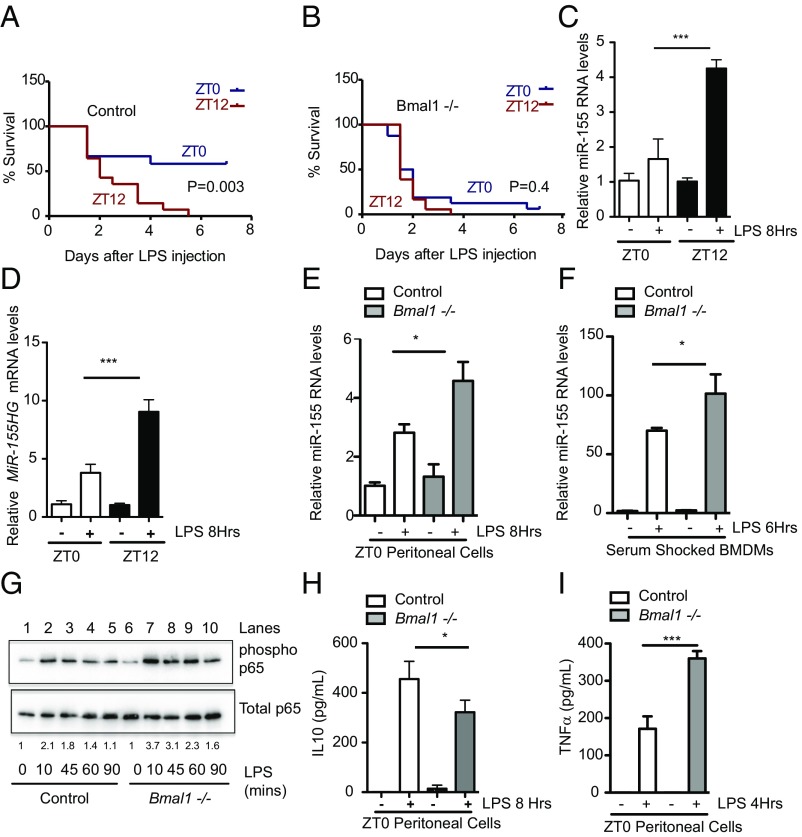
We and others (4, 8) have used the Bmal1−/−Lys-MCre mouse—in which BMAL1 is removed from the myeloid lineage (Fig. S1 A and C) and which causes suppression of the clock components Rev-Erbα and Per2 (Fig. S1C)—to study further the effect of the molecular clock on LPS challenge. As reported (8), these mice retain normal activity rhythms and period lengths and appear grossly normal. In response to LPS-induced sepsis and consistent with the distribution of lethality in previous reports (4), control animals (Bmal1+/+Lys-MCre) were protected against LPS-induced sepsis when mice were injected (25 mg/kg) at zeitgeber time 0 (ZT0) versus ZT12 (Fig. 1A, P = 0.003). In Bmal1−/−Lys-MCre mice there was no difference in survival between the ZT0 and ZT12 treated groups (Fig. 1B, P = 0.4). Zeitgeber time is a measure of time (hours) after lights go on in the mouse facility; therefore, ZT0 corresponds to zeitgeber time 0, lights on, and the beginning of the rest phase whereas ZT12 corresponds to zeitgeber time 12, lights off, and the beginning of the activity phase. These are 12 h light–dark conditions (LD). Coincident with the increase in LPS lethality in control mice, LPS-dependent elicitation of the proinflammatory microRNA miR-155 was greater in peritoneal cells isolated at ZT12 versus at ZT0 (Fig. 1C). MiR-155 maps within and is processed from the miR-155 host gene (MiR-155HG), a noncoding RNA, formerly known as Bic (24), which also showed heightened induction to LPS at ZT12 (Fig. 1D). In agreement with Gibbs et al. (8), this correlated with a range of proinflammatory cytokines. TNFα, IL6, and CXCL1 were induced to a greater extent with LPS at ZT12 versus at ZT0 (Fig. S1 D–H). The exception was the anti-inflammatory cytokine IL10, whose induction by LPS in peritoneal cells at ZT12 was lower than at ZT0 (Fig. S1 I and J). Coincident with the increase in LPS lethality, there was greater induction of miR-155 from peritoneal cells lacking BMAL1 (Fig. 1E), and this increase in LPS-induced miR-155 was also apparent in serum-shocked bone marrow-derived macrophages (BMDMs) harvested from mice lacking BMAL1 (Fig. 1F). CLOCK acetylates and thus activates p65 (a subunit of the NF-κB complex), leading to increased p65 phosphorylation in fibroblasts, and BMAL1 heterodimerization with CLOCK suppressed this activity (12). We assayed phosphorylation of p65 in peritoneal cells lacking BMAL1 versus control. Induction and maintenance of serine phosphorylation at position 536 with LPS was greater from peritoneal cells lacking BMAL1 versus control (Fig. 1G, compare lanes 2 with 7, 3 with 8, and 4 with 9). This heightened proinflammatory phenotype from peritoneal cells lacking BMAL1 was further confirmed by decreased IL10 (Fig. 1H) and increased production of TNFα (Fig. 1I), IL6, CXCL1, and MCP1/CCL2 (Fig. S2 A–E). Peritoneal cells contain a mixture of myeloid cells; therefore, we assayed macrophages directly to understand whether they were one of the cell types contributing to the enhanced proinflammatory phenotype in the absence of BMAL1. BMDMs from Bmal1−/− mice cultured in the presence of LPS for 12 h produced greater levels of IL6, CXCL1, TNFα, and MCP1/CCL2 (Fig. S2 F–I).
Fig. 1.
The magnitude of the circadian response to sepsis and inflammation correlates with induction of the microRNA miR-155. (A) Bmal1+/+Lys-MCre (n = 12–18) or (B) Bmal1−/−Lys-MCre male mice (n = 12–18) were injected intraperitoneally with LPS (25 mg/kg) at ZT0 (blue line) or ZT12 (red line) and monitored for survival over 7 d. Wild-type peritoneal cells harvested at ZT0 and ZT12 and treated immediately ex vivo with LPS (100 ng/mL) for indicated times and analyzed for expression of (C) mature miR-155 and (D) MiR-155HG (n = 3–4). Peritoneal cells harvested at ZT0 from Bmal1+/+Lys-MCre and Bmal1−/−Lys-MCre mice and treated immediately ex vivo with LPS (100 ng/mL) for the indicated time and analyzed for expression of (E) miR-155 (n = 3–4). (F) Serum-shocked BMDMs from Bmal1+/+Lys-MCre and Bmal1−/−Lys-MCre treated with LPS (100 ng/mL) for the indicated time and analyzed for expression of miR-155 (n = 3). (G) Peritoneal cells harvested at ZT0 from Bmal1+/+Lys-MCre and Bmal1−/−Lys-MCre mice, treated immediately ex vivo with LPS (1 ng/mL) for indicated times, and analyzed by immunoblot for levels of phosphorylated p65 at Serine 536 and total p65. Blot is representative of n = 6. Values provided are relative band intensity of phospho/total 65. Peritoneal cells as in E were analyzed for protein levels of (H) IL10 and (I) TNFα. *P ≤ 0.05 and ***P ≤ 0.001.
Down-Regulation of Bmal1 upon LPS Challenge Is Coincident with miR-155 Induction in Mice.
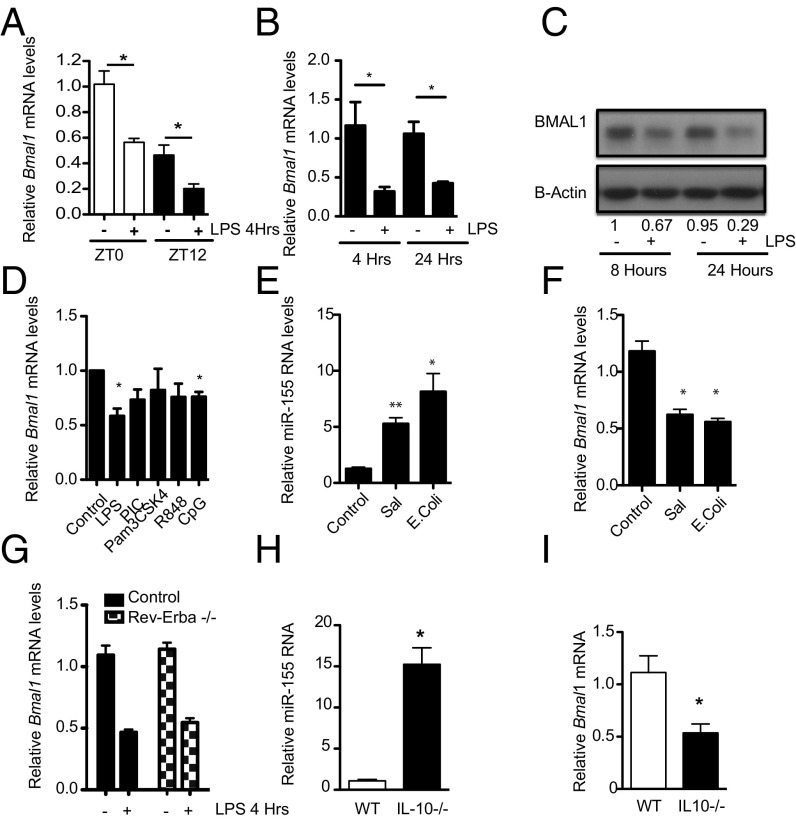
Peritoneal macrophages harvested at ZT0 and ZT12 and challenged with LPS resulted in a reduction in Bmal1 expression at both time points (Fig. 2A). Similarly, BMDMs treated with LPS had a significant reduction in Bmal1 mRNA (Fig. 2B) and protein (Fig. 2C) over time, correlating with an increase in miR-155 (Fig. S2J). Among a range of TLR agonists used at a single concentration, LPS caused significant repression of Bmal1 (Fig. 2D). Infection of the macrophage cell line, RAW264.7, with the pathogens Salmonella typhimurium and Escherichia coli also caused a significant up-regulation of miR-155 (Fig. 2E) with a corresponding down-regulation of Bmal1 (Fig. 2F). Although REV-ERBα is a potent transcriptional repressor of Bmal1 (13), it was not involved in the down-regulation of Bmal1 with LPS, as LPS-induced repression of Bmal1 was equivalent in cells lacking Rev-Erbα versus control (Fig. 2G). IL10 is known to suppress miR-155 in macrophages (25), and under steady-state conditions we observed higher levels of miR-155 in BMDMs cultured from IL10−/− mice (Fig. 2H) and significantly lower levels of Bmal1 (Fig. 2I).
Fig. 2.
Activation of macrophages represses the core clock gene Bmal1, coincident with up-regulation of MiR-155. (A) Wild-type peritoneal cells harvested at ZT0 and ZT12 and treated immediately ex vivo with LPS (100 ng/mL) for 4 h and analyzed for Bmal1 (n = 3). (B) BMDMs were exposed to LPS (100 ng/mL) for the indicated times and analyzed for Bmal1 (n = 4–5). (C) BMAL1 protein by immunoblot (representative of n = 3). Values provided are relative band intensity corrected for β-actin. (D) BMDMs were treated with LPS (100 ng/mL), Poly-I:C (PIC, 100 µg/mL), Pam3CSK4 (100 ng/mL), R848 (1 µg/mL). and CpG (1 µg/mL) for 4 h and analyzed for Bmal1 (n = 3). RAW-264 cells exposed to E. coli or Salmonella for 24 h and analyzed for expression of (E) miR-155 and (F) Bmal1 (n = 3). (G) BMDMs from wild-type and Rev-Erbα −/− mice were treated with LPS (100 ng/mL) for 4 h and analyzed for Bmal1 (n = 3). BMDMs harvested from control and Il-10−/− mice and analyzed for expression of (H) miR-155 and (I) Bmal1 (n = 8–10). *P ≤ 0.05 and **P ≤ 0.01.
Bmal1 in Macrophages Is Targeted and Repressed Directly by the microRNA miR-155 Binding to Two Sites Within the 3′ UTR of Bmal1.
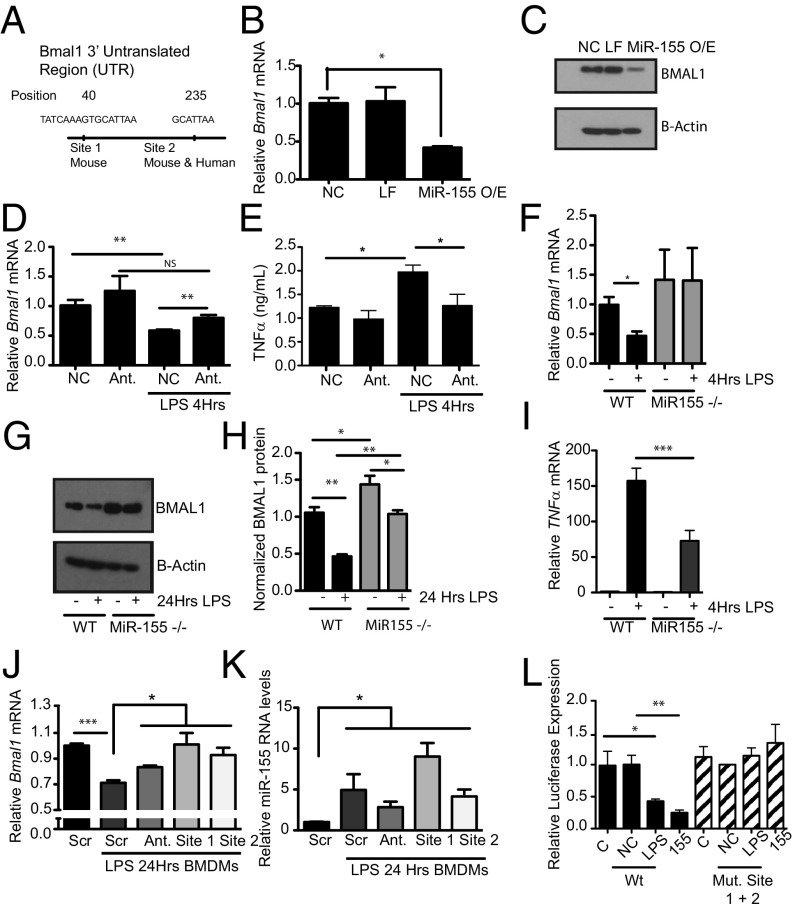
Using the software TargetScan, two predicted miR-155–binding sites (GCATTAA) were identified at positions 40 and 235 within the 3′ UTR of Bmal1 (Fig. 3A). The site at position 40 is specific to mice and the site at position 235 is conserved between mice and humans. Transfection of immortalized BMDMs (iBMDMs) with a miR-155 mimic resulted in a significant reduction in Bmal1 mRNA (Fig. 3B) and protein (Fig. 3C). Conversely, transfection of iBMDMs with an antagomir to miR-155 prevented the down-regulation of Bmal1 with LPS (Fig. 3D), and this corresponded with a reduced induction of the proinflammatory cytokine TNFα (Fig. 3E). LPS-driven reductions in Bmal1 in BMDMs did not occur in the absence of miR-155 (Fig. 3F). Clock, Cry1, and Cry2 mRNA expression were also assessed in LPS-activated macrophages, but unlike Bmal1 their expression was unaffected either by LPS or by loss of miR-155 (Fig. S3 A–C). The magnitude of repression of BMAL1 protein was attenuated in the absence of miR-155 (Fig. 3 G and H and Fig. S4A). Consistent with the effect of the miR-155 antagomir on TNFα (Fig. 3E), lack of miR-155 also suppressed the induction of Tnfα mRNA from BMDMs (Fig. 3I). Morpholinos against each of the miR-155 target sites within the Bmal1 3′ UTR [site 1 at position 40 and site 2 at position 235 (Fig. S4B)] were transfected for 24 h into BMDMs along with the antagomir to miR-155 followed by LPS treatment for 24 h. When analyzed for Bmal1 expression, the antagomir and the morpholinos blocked the ability of LPS to repress Bmal1 (Fig. 3J). The induction of mature miR-155 by LPS was not significantly affected by transfection of the morpholinos (Fig. 3K). We also cloned the Bmal1 3′ UTR into a luciferase plasmid and mutated both miR-155–binding sites. With the wild-type plasmid both LPS and miR-155 overexpression caused a reduction in luciferase (Fig. 3L, black isobars). When both sites were mutated, no reduction in luciferase was observed with either MiR-155 overexpression or LPS stimulation (Fig. 3L, striped isobars).
Fig. 3.
Bmal1 is repressed by the microRNA MiR-155 under basal and LPS conditions. (A) Schematic of Bmal1 3′ UTR illustrating position of the two miR-155–binding sites identified with the software TargetScan. iBMDMs transfected with a miR-155 mimic and analyzed for expression of (B) Bmal1 mRNA and (C) protein (n = 3). NC, negative control for mimic; LF, Lipofectamine; MiR-155O/E, overexpression of MiR-155 mimic. iBMDMs transfected with either a negative control for antagomir (NC) or an antagomir (Ant) to miR-155, treated with LPS (100 ng/mL), and analyzed for expression of (D) Bmal1 and (E) TNFα levels by ELISA (n = 3). WT or miR-155−/− BMDMs treated with LPS (100 ng/mL) for the indicated time and analyzed for expression of (F) Bmal1 and (G) protein levels by immunoblot. (H) Densitometry values of immunoblots from G and Fig. S4 (n = 3–4), and (I) Tnfα mRNA (n = 3). BMDMs transfected with a scrambled control morpholino (Scr), MiR-155 antagomir (Ant.) morpholino against the MiR-155 site in Bmal1 at position 40 (site 1), and morpholino against the MiR-155 site in Bmal1 at position 235 (site 2) were treated with LPS (100 ng/mL) for 24 h and analyzed for expression of (J) Bmal1 and (K) MiR-155. (L) Luciferase reporter activity from Bmal1 3′ UTR construct with LPS induction (100 ng/mL) or overexpression of MiR-155 mimic with wild-type luciferase construct or double (Mut. Site 1 + 2) mutations of miR-155–binding sites (n = 3). C, control; NC, negative control for mimic; 155, overexpression of miR-155 mimic. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
LPS-Induced miR-155 Targets BMAL1 in Human Macrophages and Adipose Tissue.
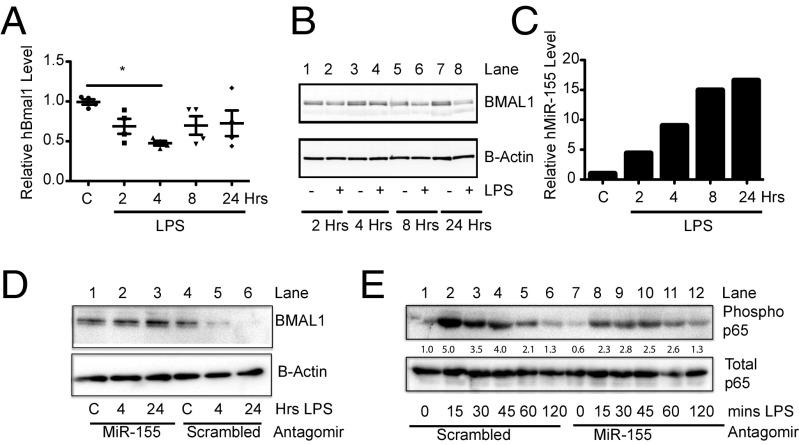
In human macrophages treated with LPS, hBMAL1 mRNA (Fig. 4A) and protein (Fig. 4B, compare lane 7 with lane 8) were reduced at 4 and 24 h, respectively, along with a progressive increase in hmiR-155 over time (Fig. 4C). This reduction in human BMAL1 with LPS was a direct effect of miR-155 as levels of BMAL1 protein in human peripheral blood mononuclear cells (hPBMCs) transfected with an antagomir to hmiR-155 were not suppressed by LPS [Fig. 4D; compare lanes 1–3 (antagomir) to 4–6 (scrambled)]. The hmiR-155 antagomir was also capable of suppressing the induction of phosphorylated p65 in PBMCs [Fig. 4E; compare lanes 2–4 (scrambled) with 8–10 (antagomir)]. Subcutaneous adipose tissue derived from 14 healthy volunteers was collected before and after an i.v. bolus of LPS (3 ng/kg) (Table 1). Gene expression analysis by microarray established that MiR-155HG was significantly increased on microarray after 4 h of exposure to LPS together with a significant reduction in BMAL1 (Table 1). An increase in PER2 and a decrease in REV-ERBα was observed in these human biopsies upon LPS activation with no change in CLOCK or RORα. Remarkably, the same effects on those genes were observed in mouse peritoneal cells harvested at ZT0 and treated with LPS for 4 h (Fig. S3 D–H). As Rev-Erbα was the only other clock gene analyzed that was repressed by LPS in humans (Table 1) and mice (Fig. S3F), we assessed whether deletion of miR-155 would effect its expression. Basal expression of Rev-Erbα was higher in peritoneal cells with miR-155 deletion, and LPS did not cause significant repression in comparison with control (Fig. S3I). Similarly, Rev-Erbα expression was not reduced with LPS in miR-155−/− BMDMs (Fig. S3J).
Fig. 4.
MiR-155 targets BMAL1 in humans and suppresses NF-κB activity. Human macrophages from four donors stimulated ex vivo with LPS (100 ng/mL) for the indicated times and analyzed for expression of (A) BMAL1 mRNA, (B) BMAL1 protein, and (C) miR-155. (D) hPBMCs were transfected with an antagomir to miR-155 or scrambled control and then treated with LPS for indicated times and immunobloted for BMAL1 and β-actin as control. Values provided are relative band intensity of BMAL1/β-Actin. (E) hPBMCs were transfected with an antagomir to miR-155 or scrambled control and then treated with LPS for indicated times and immunoblotted for phosphorylated p65 at Serine 536 and total p65 as control. Values provided are relative band intensity of phospho-p65/total p65. *P ≤ 0.05.
Table 1.
Clock gene expression from human endotoxemia study
| Gene | Pre-LPS (mean)† | SD | Post-LPS (mean)† | SD | P value‡ | Significance |
| BMAL1 | 8.4496 | 0.71 | 7.3298 | 0.84 | 1.65E-05 | *** |
| CLOCK | 7.3852 | 0.36 | 7.4554 | 0.56 | 0.6640 | |
| PER-2 | 8.3033 | 0.70 | 8.8539 | 0.42 | 0.0087 | ** |
| REV-ERBa | 9.2021 | 0.65 | 7.9089 | 0.57 | 8.11E-09 | *** |
| RORa | 7.1705 | 0.37 | 7.3213 | 0.37 | 0.1308 | |
| MiR-155HG | 5.6459 | 1.43 | 6.8753 | 1.02 | 0.0006 | *** |
**P ≤ 0.01 and ***P ≤ 0.001.
Pre-LPS and Post-LPS columns show the average expression of indicated genes across the 14 subjects and are expressed as arbitrary units.
A paired t test was performed on pre-LPS and 4-h post-LPS expression values to determine significance.
Altered Circadian Function and Clock-Controlled Cytokine Production in Mice Lacking miR-155.
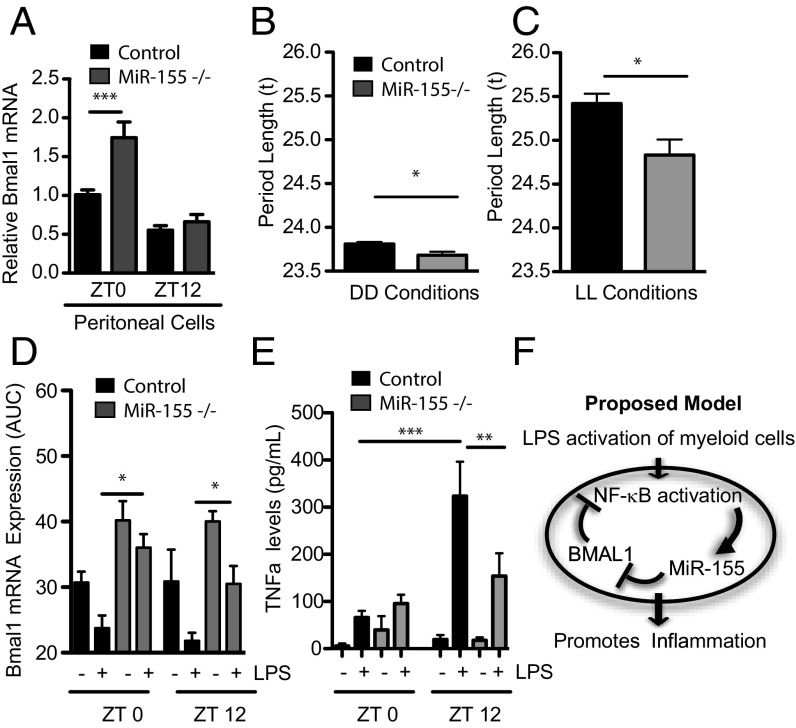
Peritoneal cells were extracted at ZT0 and ZT12 to assay circadian expression under steady-state conditions. miR-155 acts as a repressor of Bmal1 even under steady-state conditions as its deletion caused higher basal levels of Bmal1 at ZT0 (Fig. 5A). Given this observation, we assayed a free-running period in mice lacking miR-155. Over the course of a 4-wk-free run in constant darkness (DD), the free-running period was shortened in miR-155−/− mice compared with WT (t of 23.81 ± 0.02 h in WT compared with 23.66 ± 0.04 h in miR-155−/− animals; P = 0.013; Fig. 5B and Fig. S5). To examine whether this difference in τ would persist in constant light, animals were then transferred into constant light (LL) for 4 wk. Again, the miR-155−/− animals displayed a significantly shorter free-running period than the controls (t of 25.42 ± 0.11 h in WT compared with 24.83 ± 0.17 h in miR-155−/− animals; P = 0.013; Fig. 5C and Fig. S5).
Fig. 5.
Altered clock function and clock gated cytokine responses in mice lacking MiR-155. (A) Peritoneal cells harvested from WT and miR-155−/− mice at ZT0 or ZT12 were immediately lysed and analyzed for Bmal1 (n = 3–4). Period length of control and miR-155–deficient mice in (B) constant dark conditions (DD) and (C) constant light conditions (LL) (n = 9–14). (D) Peritoneal macrophages harvested from WT and miR-155 mice at ZT0 and at ZT12 and treated immediately ex vivo with LPS (100 ng/mL) for 4, 8, and 24 h and analyzed for expression of Bmal1 by area under a curve (AUC). (E) Peritoneal macrophages were harvested from WT and miR-155−/− mice at ZT0 and at ZT12 and treated immediately ex vivo with LPS (100 ng/mL) for 24 h, and supernatants were analyzed by ELISA for TNFα (n = 4). (F) Schematic model to depict the circadian effect on LPS activation via the circadian control of miR-155 on Bmal1 in myeloid cells. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Consistent with previous results, peritoneal cells from miR-155−/− mice harvested at ZT0 and ZT12 and treated with LPS for up to 24 h were unable to repress Bmal1 in comparison with wild-type cells (Fig. 5D). Strikingly, the circadian effect on TNFα production (i.e., higher induction from ZT12-treated cells versus ZT0-treated cells) was lost from cells lacking miR-155, with similar production of TNFα by LPS at both ZTs (Fig. 5E).
Discussion
Previous reports of a rhythm in the susceptibility of mice to LPS, Streptococcus pneumoniae, Listeria monocytogenes, and S. typhimurium challenge (3–5, 26) suggest that the molecular clock may play a fundamental role in controlling the mammalian immune response. Susceptibility to lethality is greatest when the challenge occurs close to the transition into the activity phase. Gibbs et al. demonstrated that this period of susceptibility correlates with a heightened production of proinflammatory cytokines with activation of REV-ERBα, a BMAL1 target, attenuating inflammation, in particular via repression of IL6 (8). A more recent study reported that Bmal1 in the myeloid lineage controls the number of circulating Ly6Chi monocytes (4). When Bmal1 was absent from these cells, their recruitment to the site of infection was amplified as BMAL1 directly represses Ccl2 transcription. However, identifying the mechanisms that intersect the immune and clock systems have been challenging.
Here we show that the protection from LPS-induced lethality at ZT0 in comparison with ZT12 correlates with reduced induction of the proinflammatory microRNA miR-155, but increased induction of the anti-inflammatory cytokine IL10. The absence of BMAL1 in the myeloid lineage sensitizes the mice to LPS along with a corresponding increase in miR-155, but a decreased production of IL10. The heightened proinflammatory milieu of myeloid cells may in part be responsible for the enhanced lethality observed in Bmal1-deficient mice. However, as has been reported, uncontrolled trafficking of monocytes (4) and possibly other myeloid populations such as neutrophils, also known to oscillate (7), may impact on the phenotype in Bmal1-deficient mice.
A number of mechanisms may explain the circadian control on miR-155 induction by LPS. First, the promoter of the MiR-155HG contains an NF-κB site (25, 27), and NF-κB inhibitors attenuate the induction of MiR-155HG (28). Splenger et al. revealed that in fibroblasts CLOCK acetylates and thus activates p65 via increased levels of p65 phosphorylation and that BMAL1 heterodimerization with CLOCK suppressed this activity (12). We also confirm the effect of BMAL1 on NF-κB activity in peritoneal cells, so BMAL1 could regulate miR-155 and some proinflammatory cytokines including TNFα (29) via its effects on p65. The transcription factor ETS2 is responsible for miR-155 induction in LPS-activated macrophages, and IL10, via inhibition of ETS2, can suppress miR-155 induction (25). In addition to our observations that myeloid cells lacking BMAL1 produce less IL10, we observe also that IL10−/− BMDMs have high levels of miR-155 and low levels of Bmal1 under steady-state conditions. Therefore, BMAL1, through its regulation of IL10, could inhibit miR-155. This could be controlled either directly by BMAL1 or via another clock component such as Rev-Erbα. Altogether, our results confirm that BMAL1 within myeloid cells can attenuate production of miR-155 and proinflammatory cytokines in response to LPS, an effect that might impact on leukocyte activation and trafficking during sepsis occurrence.
However, it is important to note that, although BMAL1 can regulate the p65 subunit of NF-κB, not all LPS-induced NF-κB–regulated genes have a circadian gating (8). This could be because there are multiple clock-regulated components that code responses in a cell-specific manner (including REV-ERBα, which will repress certain genes) and because p65 is controlled by several other proteins other than CLOCK. It is also possible that there is circadian regulation of chromatin architecture, which limits accessibility for NF-κB. These aspects require further analysis.
Given the importance of BMAL1 in modulating the inflammatory response, we next considered the effect of LPS on Bmal1. We hypothesized that for LPS to elicit its effect on the inflammatory process it would attenuate BMAL1. LPS and bacterial infections significantly decreased the levels of Bmal1 mRNA and protein, an event coincident with increased levels of miR-155. miR-155 was a strong candidate as a regulator of Bmal1 ab initio, given that it is up-regulated rapidly upon TLR activation (30) and that its induction is dependent on the level of BMAL1 in myeloid cells. Indeed, suppression of Bmal1 mRNA and protein upon LPS activation is lost or attenuated in macrophages lacking miR-155, which also suppresses induction of Tnfα transcript. This was confirmed further with an antagomir to miR-155, which protects against suppression of Bmal1 by LPS while also attenuating TNFα induction. Some repression of BMAL1 protein to LPS was still detectable in the absence of miR-155. This suggests that, in addition to the posttranscriptional control that miR-155 exerts on Bmal1, there may exist posttranslational modifiers acting at the protein level. Morpholinos generated against the two miR-155–binding sites in the Bmal1 3′ UTR each inhibit the ability of LPS to repress Bmal1 in BMDMs, confirming further the direct effect of miR-155 on Bmal1. Therefore, controlled removal of macrophage Bmal1 by miR-155 is required to mount an acute inflammatory response.
We found that LPS repressed human BMAL1 in macrophages with a corresponding increase in hMiR-155 when treated in vitro. The human BMAL1 3′ UTR has only one miR-155–binding site in comparison with the mouse sequence that has two. However, we could completely block the repression of human BMAL1 by LPS using an antagomir to human miR-155. We could also suppress the level of p65 phosphorylation using the antagomir to human miR-155. This confirms further that modulation of the Bmal1/miR-155 axis can affect the downstream activity of NF-κB and the inflammatory process. An increase in the hMiR-155HG and a decrease in hBMAL1 was also evident in the adipocyte biopsies obtained from patients treated with low-dose LPS. Indeed, there is remarkable translational concordance between the effects on Clock, Period2, Rev-Erbα, and RoRα observed in human adipocytes with those seen on mouse peritoneal cells treated with LPS. Rev-Erbα was the only other clock gene analyzed that was repressed by LPS. The repression of Rev-Erbα by LPS was in part protected with loss of miR-155, whereby BMAL1 would remain high. Therefore, some of the proinflammatory effects featured with loss of BMAL1, such as increased IL6, may be due to loss of REV-ERBα.
Under steady-state conditions, miR-155 also negatively regulates Bmal1 expression at ZT0. This led us to investigate whether miR-155 would have effects on the central clock. It is intriguing that miR-155−/− mice display a shorter period length. Although this observation requires further investigation, it is consistent with the role of miR-155 as a transcriptional repressor of Bmal1, as mice lacking Rev-Erbα, a potent transcriptional repressor of Bmal1, have a similar, albeit more pronounced, effect on activity (13). In addition, lack of miR-155 in myeloid cells ablates the circadian effect on evoked TNFα normally observed between ZT0 and ZT12.
Collectively, these studies identify miR-155 as an important regulatory component of circadian function and provide a description of a previously unidentified mechanism by which the circadian clock controls the innate immune response. miR-155 is believed to potentiate inflammation in macrophages, in part through its effects on the TLR4 repressors Ship1 and Socs1 (31, 32) and its ability to stabilize Tnfα (28). miR-155 is controlled by the molecular clock, leading to its circadian induction. We show also that high levels of miR-155 and the consequent targeting of BMAL1 might lead to a proinflammatory state through activation of the NF-κB complex. To our knowledge, this is the first report of a miRNA, integral to the immune system, affecting the temporal and inflammatory variability of the molecular clock. Furthermore, BMAL1 or its targets such as REV-ERBα negatively regulate innate immunity such that LPS must repress BMAL1 itself via miR-155 should BMAL1 be present at a particular time of day. Innate immunity therefore uses the control of BMAL1 by miR-155 to control the circadian inflammatory response in myeloid cells. Our findings provide insight into the temporal control of inflammation, which could have consequences for our understanding of the pathogenesis of inflammation and infectious diseases where circadian regulation is known to be important.
Materials and Methods
LPS Survival in Mice.
Mice were maintained in a controlled environment for 1 wk before the LPS study. Mice were injected either at ZT0 or ZT12 with LPS derived from E. coli serotype 055:B5 (Sigma Aldrich) in sterile PBS at 25 mg/kg by the i.p. route and monitored for 6–8 d. All animal studies were performed in accordance with the guidelines approved by the Institute for Animal Care and Use Committee at the University of Pennsylvania and the Animal Research Ethics Committee at Trinity College Dublin.
Peritoneal Exudate Cells.
Mice were euthanized at indicated times and cells were collected by peritoneal lavage. Cells were seeded at 1 × 106 cells per well in serum-free media, and after 45 min nonadherent cells were washed out and attached cells were harvested immediately for RNA and protein analysis, or LPS was added to the attached cells for specified times and then cells were harvested for RNA, and protein and supernatants were harvested for ELISAs.
Transfections.
Wild-type iBMDMs, a gift from Douglas Golenbock, University of Massachusetts Medical School, Worchester, MA, were seeded at 1 × 105 cells/well. On the next day, cells were transfected with 1 μM of miR-155 premiR or the scrambled oligonucleotide premiR negative control (Ambion) using Lipofectamine 2000 (Invitrogen) for 8 h. Media were replaced with fresh antibiotic-free media, and cells were harvested 24 h later. For the antagomir studies in iBMDMs and hPBMCs, 50 nM of the miR-155 antagomir or negative control (Ambion) was used. For BMDMs, morpholinos against the two miR-155–binding sites (1 µM) were transfected with the reagent Endo-Porter (33). Twenty-four hours later, cells were treated to LPS (100 ng/mL) and harvested. For luciferase reporter studies, the complete 3′ UTR of Bmal1 containing the two predicted miR-155–binding sites or with the sites mutated was inserted into the dual luciferase PsiCheck2 reporter vector (Promega). iBMDM cells were plated in a six-well format and cotransfected using Lipofectamine 2000, and luciferase activity was measured after 48 h.
Human Subjects Study Protocol.
The institutional review board of the University of Pennsylvania approved the protocol and all subjects gave written informed consent.
Data Analysis.
Results are presented as mean ± SEM. Statistical analysis was performed using Prism 5. Differences were compared by using analysis of variance followed by Student–Newman–Keuls post hoc analysis and/or paired or unpaired Student’s t test, as appropriate. Significance values are indicated as *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. For survival data, a log-rank (Mantel–Cox) test was used.
Supplementary Material
Acknowledgments
We thank Rebecca Leyland for assistance with Bic/MiR-155−/− cells and Jennifier Jager and Mitch Lazar for providing Rev-Erbα−/− cells. This work was supported by grants from the European Research Council (268155_MicroInnate to L.A.J.O.), the Science Foundation Ireland (13/SIRG/2130 to A.M.C.), NIH (HL097800 to G.A.F.), and the European Community Seventh Framework Programme TIMER Project. C.T.F. was supported by the “Science without Borders” programme from Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil. G.A.F. is the McNeil Professor of Translational Medicine and Therapeutics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501327112/-/DCSupplemental.
References
- 1.Yang G, et al. Knitting up the raveled sleave of care. Sci Transl Med. 2013;5(212):212rv213. doi: 10.1126/scitranslmed.3007225. [DOI] [PubMed] [Google Scholar]
- 2.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen KD, et al. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: Influence of light and adrenocortical secretions. Science. 1973;182(4109):285–287. doi: 10.1126/science.182.4109.285. [DOI] [PubMed] [Google Scholar]
- 6.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheiermann C, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37(2):290–301. doi: 10.1016/j.immuni.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs JE, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castanon-Cervantes O, et al. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. 2010;185(10):5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froy O, Chapnik N. Circadian oscillation of innate immunity components in mouse small intestine. Mol Immunol. 2007;44(8):1954–1960. doi: 10.1016/j.molimm.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36(2):251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spengler ML, et al. Core circadian protein CLOCK is a positive regulator of NF-κB-mediated transcription. Proc Natl Acad Sci USA. 2012;109(37):E2457–E2465. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 14.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 15.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 17.Kadener S, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23(18):2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta N, Cheng HY. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J Mol Biol. 2013;425(19):3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Cheng HY, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan X, et al. Clock-controlled mir-142-3p can target its activator, Bmal1. BMC Mol Biol. 2012;13:27. doi: 10.1186/1471-2199-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy CE, et al. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem. 2010;285(27):20492–20498. doi: 10.1074/jbc.M110.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazari-Jahantigh M, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122(11):4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274(1-2):157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 25.Quinn SR, et al. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J Biol Chem. 2014;289(7):4316–4325. doi: 10.1074/jbc.M113.522730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellet MM, et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci USA. 2013;110(24):9897–9902. doi: 10.1073/pnas.1120636110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson RC, Vardinogiannis I, Gilmore TD. Identification of an NF-κB p50/p65-responsive site in the human MIR155HG promoter. BMC Mol Biol. 2013;14:24. doi: 10.1186/1471-2199-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bala S, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNFalpha) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286(2):1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakhov AN, Collart MA, Vassalli P, Nedospasov SA, Jongeneel CV. Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J Exp Med. 1990;171(1):35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117(16):4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summerton JE. Endo-Porter: A novel reagent for safe, effective delivery of substances into cells. Ann N Y Acad Sci. 2005;1058:62–75. doi: 10.1196/annals.1359.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.