Significance
As powerful biological catalysts, enzymes can solve challenging problems that range from the industrial production of chemicals to the treatment of human disease. The ability to design new enzymes with tailor-made chemical functions would have a far-reaching impact. However, this important capability has been limited by our cursory understanding of enzyme catalysis. Here, we report a method that uses unbiased empirical analysis to dissect the molecular basis of enzyme function. By comprehensively mapping how changes in an enzyme’s amino acid sequence affect its activity, we obtain a detailed view of the interactions that shape the enzyme function landscape. Large, unbiased analyses of enzyme function allow the discovery of new biochemical mechanisms that will improve our ability to engineer custom biocatalysts.
Keywords: protein engineering, droplet-based microfluidics, high-throughput DNA sequencing
Abstract
Natural enzymes are incredibly proficient catalysts, but engineering them to have new or improved functions is challenging due to the complexity of how an enzyme’s sequence relates to its biochemical properties. Here, we present an ultrahigh-throughput method for mapping enzyme sequence–function relationships that combines droplet microfluidic screening with next-generation DNA sequencing. We apply our method to map the activity of millions of glycosidase sequence variants. Microfluidic-based deep mutational scanning provides a comprehensive and unbiased view of the enzyme function landscape. The mapping displays expected patterns of mutational tolerance and a strong correspondence to sequence variation within the enzyme family, but also reveals previously unreported sites that are crucial for glycosidase function. We modified the screening protocol to include a high-temperature incubation step, and the resulting thermotolerance landscape allowed the discovery of mutations that enhance enzyme thermostability. Droplet microfluidics provides a general platform for enzyme screening that, when combined with DNA-sequencing technologies, enables high-throughput mapping of enzyme sequence space.
Enzymes are powerful biological catalysts capable of remarkably accelerating the rates of chemical transformations (1). The molecular bases of these rate accelerations are often complex, using multiple steps, multiple catalytic mechanisms, and relying on numerous molecular interactions, in addition to those provided by the main catalytic groups. This complexity imposes a significant barrier to understanding how an enzyme’s sequence impacts its function and, thus, on our ability to rationally design biocatalysts with new or enhanced functions (2–4).
Comprehensive mappings of sequence–function relationships can be used to dissect the molecular basis of protein function in an unbiased manner (5). Growth selections or in vitro binding screens can be combined with next-generation DNA sequencing to generate detailed mappings between a protein’s sequence and its biochemical properties, such as binding affinity, enzymatic activity, and stability (6–9). This deep mutational scanning approach has been used to study the structure of the protein fitness landscape, discover new functional sites, improve molecular energy functions, and identify beneficial combinations of mutations for protein engineering. However, these methods rely on functional assays coupled to cell growth or protein binding, severely limiting the types of proteins that can be analyzed. For example, most enzymes of biological or industrial relevance cannot be analyzed using existing methods because they do not catalyze a reaction that can be directly coupled to cell growth. Experimental advances are needed to broaden the applicability of deep mutational scanning to the diverse palette of functions performed by enzymes.
In this paper, we present a general method for mapping protein sequence–function relationships that greatly expands the scope of biochemical functions that can be analyzed. Ultrahigh-throughput droplet-based microfluidic screening enables us to characterize the chemical activities of millions of enzyme variants. By sorting the variants based on chemical activity and performing next-generation DNA sequencing of sorted and unsorted libraries, we obtain a detailed mapping of how changes to enzyme sequence impact chemical function. We demonstrate this method using a glycosidase enzyme important in the deconstruction of biomass into fermentable sugars for biofuel production. Through comprehensive mutagenesis and functional characterization of this enzyme, we were able, with minimal bias, to discover residues crucial to function and identify mutations that enhance its activity at elevated temperatures. This approach can be applied to any enzyme whose chemical activity can be measured with a fluorogenic assay in microfluidic droplets (10–13). Our method extends the applicability of deep mutational scanning to a wide range of protein functions and reaction conditions not accessible by other high-throughput methods.
Results
High-Throughput Sequence–Function Mapping.
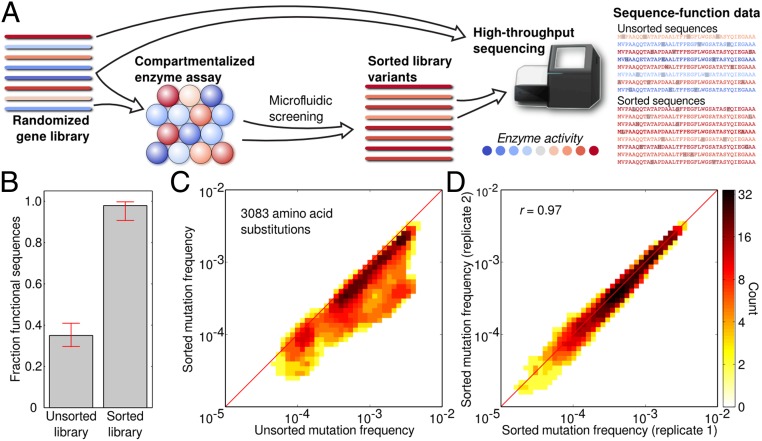
Protein sequence space is vast and an enzyme’s functional properties may depend on hundreds to thousands of molecular interactions, most of which will have never been characterized. Systematically exploring this space thus necessitates methods capable of characterizing massive numbers of sequence variants. We have developed a general method for performing millions of sequence–function measurements on an enzyme (Fig. 1A). A library of enzyme variants is expressed in Escherichia coli, and single cells are encapsulated in microfluidic droplets containing lysis reagents and a fluorogenic enzyme substrate (Fig. S1A). Upon lysis, the expressed enzyme variant is released into the droplet, allowing it to interact with the substrate. The surrounding oil acts as a barrier that keeps reagents contained within the droplets, preventing product molecules generated by one variant from mixing with those of another in a different droplet. Droplets that contain efficient variants thus rapidly accumulate fluorescent product, whereas those with inactive variants remain dim. The DNA sequences of the active variants are then recovered using a high-throughput microfluidic sorter to recover the bright droplets (14). The sorter can analyze more than 100 enzyme variants per second, reaching 1 million in just a few hours. The sorted and unsorted gene libraries are then processed using next-generation DNA sequencing and statistical analysis.
Fig. 1.
High-throughput sequence–function mapping. (A) A conceptual overview of the sequence–function mapping protocol. Individual members of a randomized gene library are assayed in aqueous microdroplets, and microfluidic screening is used to sort out the active variants. The unsorted and sorted variant pools are then analyzed using high-throughput DNA sequencing. The resulting sequence–function dataset is used to understand the functional impact of mutations. (B) Droplet-based microfluidic screening recovers functional sequences from the initial random mutagenesis library. Individual clones from the unsorted and sorted libraries were tested in a plate-based assay and were considered functional if their end-point activity was greater than 50% of Bgl3’s. Initially, only 35% of the library was functional, but after screening the fraction of functional sequences increased to 98%. Error bars represent the 90% binomial proportion confidence interval. (C) The frequency of 3,083 amino acid substitutions in the unsorted and sorted libraries. A large fraction of mutations decrease in frequency after sorting, suggesting they are deleterious to Bgl3 function. (D) Reproducibility of the sequence–function mapping protocol. Two independent experimental replicates show very good agreement in amino acid frequencies.
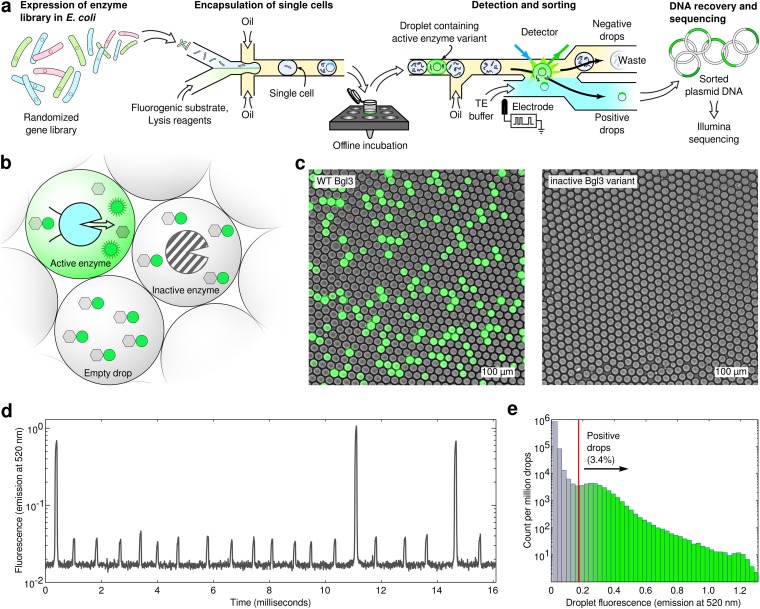
Fig. S1.
Microfluidic β-glucosidase assay. (A) An overview of the microfluidic screening workflow. A library of enzyme variants is expressed in E. coli, and single cells are encapsulated in microdroplets that contain lysis reagents and a fluorogenic substrate. The droplets are incubated offline at 37 °C and reinjected onto a microfluidic sorting device. The florescence of each droplet is analyzed. If a droplet’s fluorescence meets the specified criteria, an electric pulse is used to merge its contents with the aqueous collection stream. The sorted DNA is then recovered for downstream processing. (B) The fluorogenic substrate produces a strong green fluorescence signal upon hydrolysis by a β-glucosidase. Bright droplets contain an active enzyme variant, whereas dark droplets could be empty (no E. coli) or contain an inactive enzyme variant. (C) Microscopy images of the emulsion-based enzyme assay. Both panels show an overlay of bright-field and fluorescence (FITC channel) images with the same exposure and image settings. The left panel shows droplets containing WT Bgl3, whereas the right panel shows the results using an inactive (truncated) Bgl3 variant. (D) A time trace from the photomultiplier tube (PMT) fluorescence detection system (Fig. S10). The three large peaks correspond to droplets containing WT Bgl3, whereas the remaining peaks are empty droplets. Droplets are analyzed at 1.3 kHz. (E) A histogram showing the fluorescence intensities of the Bgl3 random mutagenesis library. The red line indicates the threshold that was used for the sorting experiments.
As a demonstration of the generality and power of our sequence–function mapping method, we used it to analyze Bgl3, a β-glucosidase enzyme from Streptomyces sp. We chose Bgl3 because it catalyzes an important step in the deconstruction of biomass into fermentable sugars, it is a remarkably proficient catalyst (kcat/kuncat ∼ 1016), its structure has been solved to high resolution, and it has a simple fluorogenic assay. To enable accurate sorting of active from inactive variants, we developed an emulsion-based β-glucosidase assay that showed excellent discrimination between wild-type (WT) Bgl3 and an inactive mutant (Fig. S1 B–D). We used error-prone PCR to generate a Bgl3 mutant library with an average of 3.8 amino acid substitutions per gene. We screened this library for a total of 23 h (four separate runs), analyzing over 10 million variants, 3.4 million of which contained measurable enzymatic activity and were recovered via microfluidic sorting (Fig. S1E). To confirm enrichment of functional sequences within the sorted population, we tested a random sampling of mutants in a plate assay before and after sorting (Fig. 1B). Before sorting, ∼35% of variants were found to be functional, the remainder inactive due, presumably, to deleterious point mutations. After sorting, the fraction of functional sequences increased to 98%. The sorted sequences had an average of 2.0 amino acid substitutions per gene, approximately one-half that of the unsorted library.
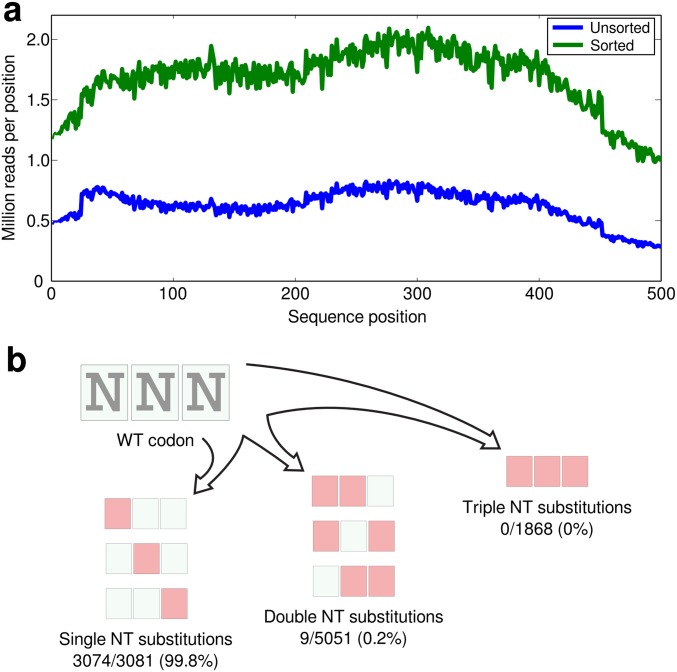
We processed the unsorted and sorted gene libraries using the Nextera XT sequencing library prep kit, sequenced using an Illumina MiSeq, version 3, 2 × 300 run, and mapped the sequence reads to the bgl3 gene using Bowtie2. The DNA sequencing showed good coverage across the entire bgl3 gene for both the unsorted and sorted libraries (Fig. S2A). The Bgl3 construct has 500 amino acid positions and therefore a total of 10,000 (500 × 20) possible amino acid substitutions including nonsense mutations. After applying sequencing quality filters, there were sufficient statistics to quantify the frequency of 3,083 (31%) of these amino acid substitutions. The remaining 6,917 substitutions were difficult to access because they require two or three nucleotide mutations within a single codon, which is a rare occurrence in libraries generated via error-prone PCR (Fig. S2B).
Fig. S2.
Sequencing and mutational coverage. (A) The sequencing coverage for the unsorted and sorted libraries. The Nextera XT kit gave roughly uniform coverage across the bgl3 gene. We observed at least 1 million reads for every position in the sorted library. (B) There are 500 positions in the Bgl3 construct and each of these positions can be mutated to 20 other amino acids (including stop codons), for a total of 10,000 possible substitutions. Of these 10,000 amino acid substitutions, 3,081 can be reached by a single nucleotide substitution, 5,051 require two nucleotide substitutions within a single codon, whereas the remaining 1,868 require all three nucleotides to be mutated. With the random mutagenesis library, we are analyzing nearly all (99.8%) of the amino acid substitutions that can be reached by a single nucleotide substitution. As expected, the coverage of amino acid substitutions requiring two or three nucleotide changes within a single codon is much lower.
The effect of an amino acid substitution can be estimated by how much its frequency changes in response to functional screening. A majority of mutations decreased in frequency in the sorted library, suggesting they are deleterious to the enzyme’s function (Fig. 1C). This observation is consistent with other studies analyzing the effects of random mutations on protein function (15–18). To further evaluate the method, we tested the reproducibility of the mapping by comparing amino acid frequencies from two independent sorting experiments (Fig. 1D). These datasets show excellent agreement (r = 0.97) across all 3,083 point mutations. Our microfluidic sequence–function mapping method was further validated on a panel of Bgl3 variants with known enzyme activities (Fig. S3).
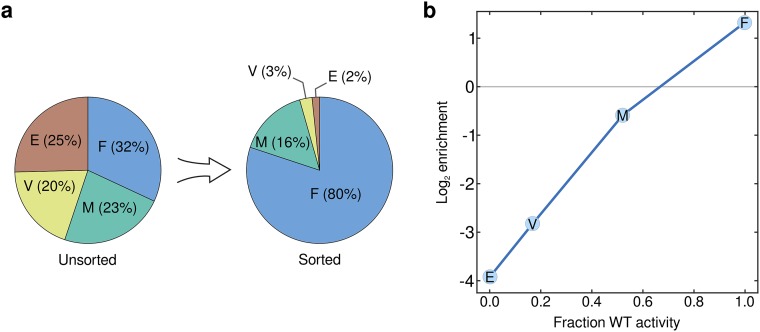
Fig. S3.
Validation of microfluidic sequence–function mapping method on a panel of Bgl3 variants. We chose four Bgl3 variants with a range of end-point enzyme activities (F288E, 0% WT activity; F288V, 17% WT activity; F288M, 52% WT activity; and F288F, 100% WT activity). These four variants were expressed separately, combined in equal proportions, and run through the microfluidic screening protocol. The library, both before and after sorting, was analyzed using Illumina sequencing. (A) The proportion of the four Bgl3 variants is roughly equal in the unsorted library, whereas the sorted library displays an increase in the WT enzyme (F) and a decrease in mutant enzymes (E, V, M). (B) The enzyme activity of the Bgl3 variants displays a strong correlation with their enrichment value in the microfluidic sequence–function mapping. The end-point activity of each enzyme variant was measured using a plate-based functional assay (SI Materials and Methods).
Site-Specific Mutational Tolerance.
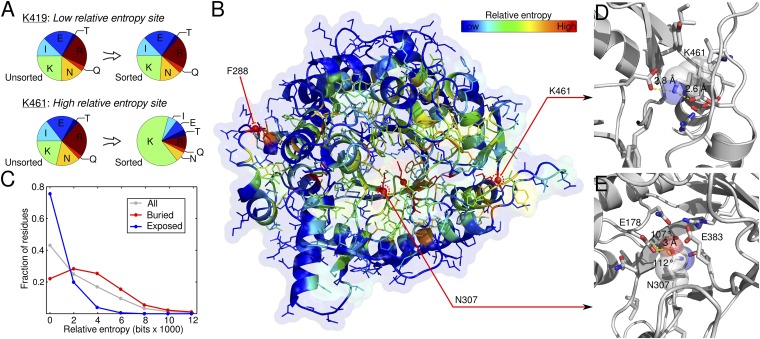
Data from millions of functional sequence variants can be used to identify residues important for enzyme function. Residues that cannot be mutated to other amino acids are likely to play a specific role required for enzyme activity. The degree to which a site can tolerate amino acid change is thus an indicator of its functional importance. The relative entropy (RE) can be used to score a residue’s mutational tolerance, because it quantifies how much the amino acid probability distribution changes between the unsorted and sorted libraries (Fig. 2A). A site whose distribution shifts significantly from random has high relative entropy, implying that a specific amino acid must reside at that position for the enzyme to remain functional.
Fig. 2.
Analysis of site-specific mutational tolerance. (A) Relative entropy (RE) describes how much the amino acid probability distribution changes in response to functional screening. The amino acid distribution of mutated codons is shown for a low RE site and a high RE site. Only synonymous substitutions are shown for the WT amino acid. The low RE site (K419) shows little change between the unsorted and sorted libraries, suggesting this position can tolerate substitutions to other amino acids. In contrast, the high RE site (K461) shows a strong shift back to the WT residue. (B) Structural patterns of mutational tolerance. The relative entropy of each site was mapped onto the Bgl3 crystal structure (Protein Data Bank ID code 1GNX). Sites with the highest relative entropies (≥99th percentile) have a red sphere at their α carbon. As expected, known functional sites, such as the catalytic residues, are highly intolerant to mutation. The analysis also reveals previously unannotated positions that are intolerant to mutation and may therefore play an important role in Bgl3 function. Three of these sites (F288, N307, and K461) are labeled in the figure. (C) The mutational tolerance of a position depends on its solvent exposure. The distribution of relative entropies for all positions is shown in gray. Buried residues [relative surface area (RSA) < 0.2] tend to have higher relative entropies and are therefore less tolerant to mutations than solvent-exposed residues (RSA ≥ 0.2). (D) Detailed view of K461 in Bgl3 structure. K461 (transparent spheres) forms salt bridges with two nearby aspartic acid residues. The short interatomic distances and their networked nature, suggests these interactions are strong and may be important for the structural stability of the enzyme. (E) Detailed view of N307 in Bgl3 structure. N307 (transparent spheres) is located directly between the enzyme’s nucleophile (E383) and the general acid/base (E178). Based on the distance and angles of the residues, N307 appears to hydrogen bond with E178, which may be important for perturbing the pKa of that group and, thus, the catalytic mechanism of the enzyme.
The mutational tolerance of a site should be related to its position in the protein’s 3D structure, because this determines the other residues with which it interacts. To investigate the relationship between enzyme structure and mutational tolerance, we mapped the relative entropy of each position onto the Bgl3 crystal structure (Fig. 2B). As expected, the catalytic nucleophile (E383) and general acid/base (E178) are both highly intolerant to mutation, falling at the 99th and 95th percentiles, respectively. We also expect core residues to be less tolerant to mutation than surface residues because the protein core tends to be well packed, forming many interresidue interactions. To support this, the α-helices that compose the TIM-barrel wall display an alternating pattern, where the interior helix face is less tolerant to mutation than the exterior face (Fig. 2B). Overall, buried residues are less tolerant to mutation than solvent-exposed residues (Fig. 2C).
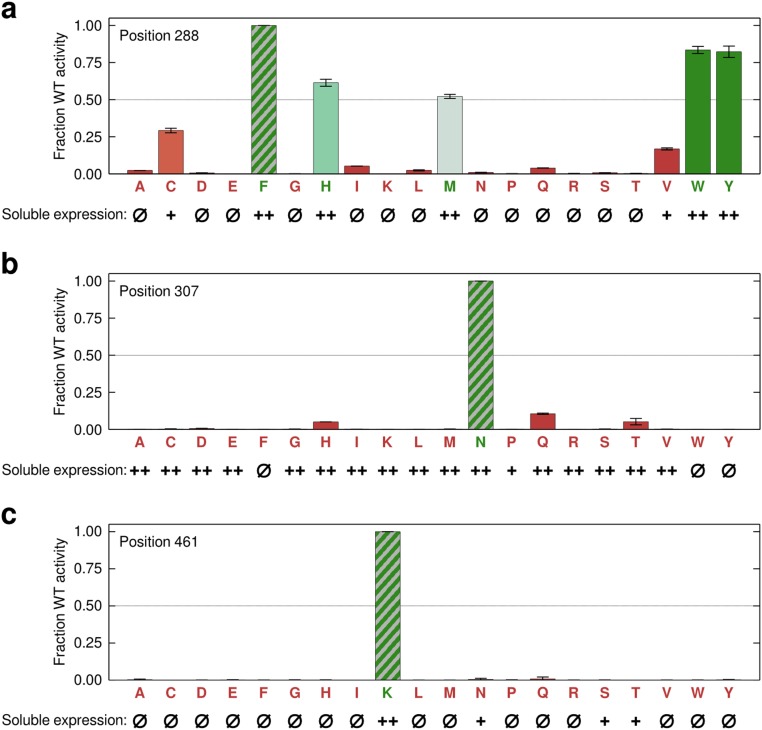
The analysis of mutational tolerance reveals sites that play an important functional role, several of which have never been described in the literature. For example, lysine 461 has the highest relative entropy of any residue (100th percentile), although, oddly, it is far from the active site (Fig. 2B). Targeted mutagenesis shows no other amino acid can be accepted at this location, validating the mutational tolerance findings (Fig. S4C). In the crystal structure, K461 is involved in networked salt bridges with two aspartic acid residues (Fig. 2D). The short distance of these interactions indicates they are strong and suggests that K461 may be important for the structural stability of the enzyme. Indeed, substitutions at this position significantly decrease the enzyme’s soluble expression (Fig. S4C).
Fig. S4.
Further validation of site-specific mutational tolerance. Sites of interest were investigated by constructing all possible amino acid substitutions and testing each mutant’s end-point activity and level of soluble expression. Activity values are shown relative to WT (striped bar). Soluble expression levels are indicated according to the following scale: “++” display WT expression levels, “+” have low expression, and “Ø” were not detected. (A) Mutagenesis of position 288 showed that only 4 of 19 (21%) of amino acid substitutions are tolerated. Inactive mutants did not express in the soluble fraction, suggesting that F288 may be important for Bgl3 stability. Because other tolerated amino acids include His, Trp, and Tyr, we hypothesize that F288 could be involved in cation–π interactions with two structurally adjacent arginine residues. (B) Position 307 cannot accept any substitutions; however, most mutants express at WT levels. This is consistent with N307 playing a role in the enzyme’s catalytic mechanism. (C) Position 461 cannot accept any mutations, and most mutants do not show any detectable soluble expression. This suggests that K461 may be important for Bgl3’s structural stability.
Asparagine 307 is another residue with high relative entropy (99th percentile) that, again, has not been described previously. N307 is located in the enzyme’s active site and appears to be hydrogen bonding with the general acid/base E178 in the crystal structure (Fig. 2E). Targeted mutagenesis at this position also shows no other amino acid is tolerated, again validating the results of the mutational tolerance map obtained with our approach (Fig. S4B). Unlike K461, substitutions at N307 demolish enzyme activity but have minimal influence on soluble expression, suggesting N307’s role in the enzyme’s catalytic mechanism. We hypothesize that N307 may act to shift the pKa of the general acid/base, which is crucial for the pKa-cycling mechanism of most retaining glycosidases (19). These results demonstrate the power of comprehensive and unbiased sequence–function mapping for investigating enzyme function and identifying important residues.
Comparison with the Natural Sequence Record.
Bgl3 is a member of glycoside hydrolase family 1 (GH1), a large enzyme family accepting a broad range of glycosylated substrates (20, 21). The sequences within the GH1 family typically differ by hundreds of mutations, providing a diverse sampling of the sequence space explored by natural evolution. By contrast, our experimental sequence–function mapping densely samples the local space of sequences within a few mutations of Bgl3. Comparing the global versus local view of sequence space may provide insight into the evolutionary constraints imposed on members of the GH1 family.
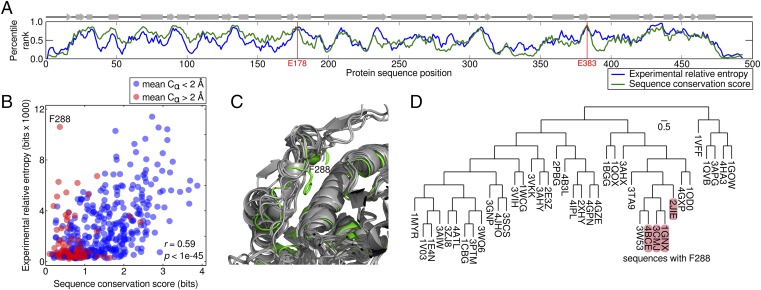
To investigate how our results compare with the natural sequence record, we used a large GH1 multiple sequence alignment to calculate a relative entropy sequence conservation score (22, 23). Bgl3’s mutational tolerance shows a strong correspondence with the observed GH1 sequence conservation. Gene-scale patterns can be visualized by taking a moving average (five-site window) of the relative entropy and sequence conservation scores across sequence positions (Fig. 3A). The experimental mutational tolerance and GH1 conservation are strikingly similar, and their patterns tend to correspond with secondary structure elements. Overall, the experimental relative entropy and the sequence conservation score display a strong, statistically significant correlation (r = 0.59, P < 1E-45; Fig. 3B), suggesting that most sites important for Bgl3 function are also important throughout the GH1 family.
Fig. 3.
Comparison with natural sequence variation. (A) Large-scale patterns of Bgl3’s mutational tolerance and the observed GH1 sequence conservation. A moving average (five-site window) of the experimental relative entropy and sequence conservation scores is plotted over sequence positions. Percentile ranks are used to plot the two scores on the same axis. The overall patterns of Bgl3 mutational tolerance and GH1 conservation are very similar and tend to correspond with secondary structure elements (displayed across the top). (B) The relationship between a site’s mutational tolerance and sequence conservation. A scatter plot of the experimental relative entropy and sequence conservation scores displays a strong correlation (r = 0.59; P < 1E-45), indicating that sites important for Bgl3 function are also important throughout the GH1 family. Outlying sites, such as F288, can be explained by structural diversity within the enzyme family. Structural diversity (mean Cα displacement) was quantified by aligning all related structures to Bgl3, calculating each structure’s Cα displacement from Bgl3 at each position, and averaging over all structures. Positions with a high experimental relative entropy, but low sequence conservation score (top, left corner) tend to come from regions with more structural diversity (red points). (C) Structural diversity may explain outlying sites. Position 288 is highly intolerant to mutation in Bgl3 (99th percentile for RE) but has little conservation in the GH1 alignment (11th percentile for sequence conservation score). An alignment of GH1 structures reveals that position 288 occurs in a structurally diverse loop. We hypothesize that F288 is important for Bgl3 function, but its interactions are not conserved throughout the GH1 family. (D) Sequence–function mapping provides a local view of sequence space. A phylogenetic tree of GH1 structures shows the few sequences that do contain F288 are closely related.
There are, however, unexpected and interesting exceptions to the correspondence between Bgl3’s mutational tolerance and GH1 sequence conservation. The most extreme is position 288, which is highly intolerant to mutation in Bgl3 (99th percentile for RE) but has little conservation in the GH1 alignment (11th percentile for sequence conservation). Targeted mutagenesis at this location again validates the sequence–function mapping results, confirming that Bgl3 can only tolerate 21% of all amino acid substitutions at position 288 (Fig. S4A). The fact that other GH1 members can accept most amino acids at position 288 suggests that Bgl3 evolution may be constrained by mutational epistasis at this site.
A closer look at GH1 structures reveals that position 288 occurs within a loop region displaying high diversity in the family (Fig. 3C). In fact, the most outlying positions (high experimental RE and low sequence conservation) occur in regions with high structural variation within the GH1 family (Fig. 3B, red points). We hypothesize that, through the course of natural evolution, Bgl3 may have evolved unique structural motifs that constrain its mutational tolerance relative to the GH1 family. We expect closely related sequences to also share these motifs and therefore to have similar residue preferences. Indeed, the phylogenetic tree of GH1 structures shows the few members that do contain F288 are closely related (Fig. 3D). Similar mutational idiosyncrasies may exist in all family members, but their conservation patterns become obscured when observing the entire family alignment.
These results highlight how sequence–function mapping provides a detailed local view of sequence space, whereas large multiple-sequence alignments provide a global perspective. A local sequence space mapping is important for applications such as protein engineering or the prediction of disease-associated mutations, because they focus on the mutational properties of the specific family member under investigation.
High-Temperature Screening Enriches for Stabilizing Mutations.
Previous work in enzyme sequence–function mapping has used in vivo assays that couple an enzyme’s function to cellular growth (7, 24–26). These in vivo selections are limited not only in the types of enzyme functions that can be analyzed, but also by the range of experimental conditions compatible with the intracellular environment. An advantage of droplet-based microfluidics is the ability to precisely control screening conditions, such as time, temperature, and concentration. Screening under altered conditions allows for enrichment of variants with enhanced unnatural properties.
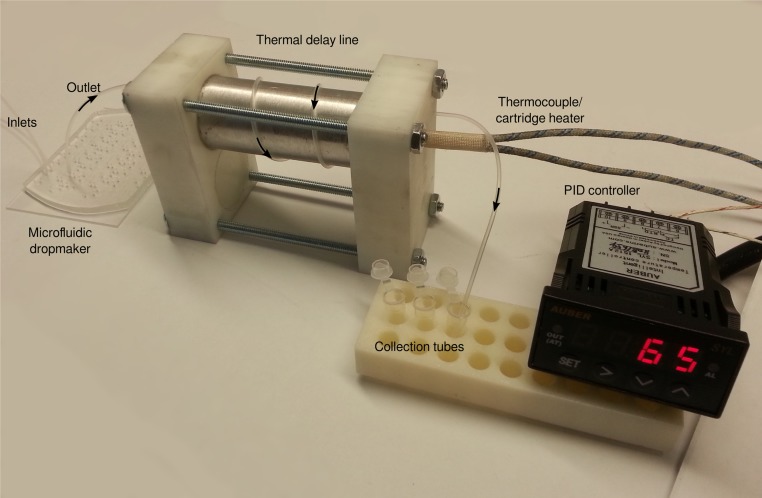
To investigate this capability, we modified the microfluidic screening protocol to include a heat challenge directly after droplet formation (Fig. S5). We hypothesized that this should enrich for mutations that increase Bgl3’s thermostability. We screened a total of 10 million enzyme variants, 2 million (20%) of which were determined to remain active and recovered via sorting. In this experiment, the heat challenge inactivated approximately one-half of the variants active in the original room temperature screen.
Fig. S5.
Thermal inactivation device used for high-temperature screening. An aluminum cylinder was interfaced with a thermocouple and a cartridge heater, and a PID controller was used to hold the cylinder at 65 °C. The microemulsions were made using a microfluidic droplet maker and immediately flowed through a thermal delay line coiled around the heated cylinder. The length of polyethylene tubing was adjusted to incubate the droplets for ∼10 min. After the heat challenge, the emulsion was collected and processed using the standard workflow.
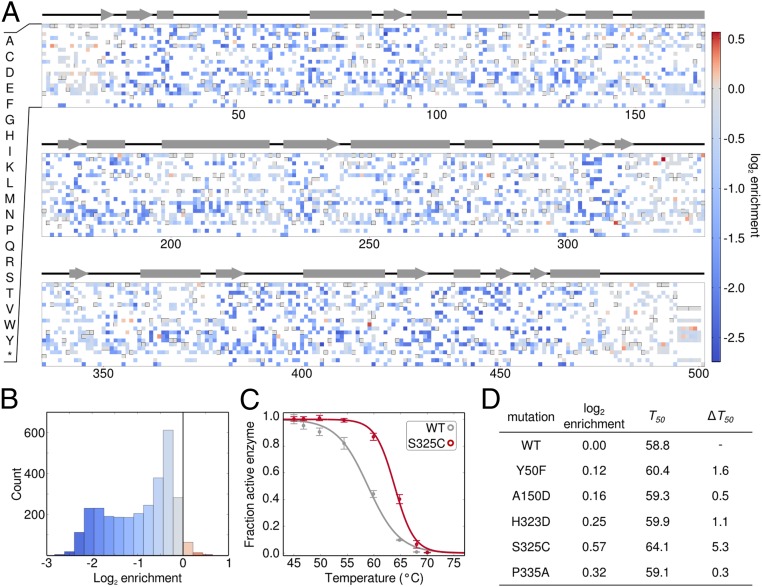
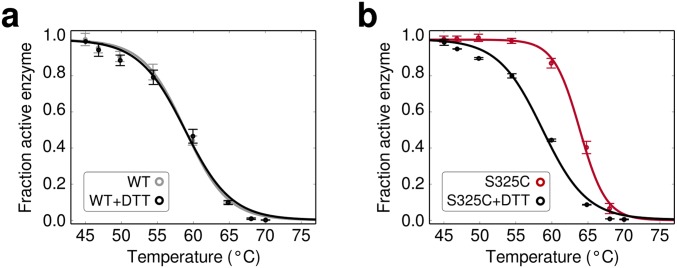
To observe the effects of the heat challenge on the functional space of enzyme sequences, we plotted the enrichment value for every observed amino acid substitution along the length of the enzyme (Fig. 4A). Overall, most mutations (97%) decreased in frequency (blue), but a small number showed positive enrichment values (red, Fig. 4B). The mutation with the greatest enrichment was S325C, located in an unresolved loop of the Bgl3 structure. This mutant was constructed and characterized and, indeed, found to yield a 5.3 °C increase in thermostability (Fig. 4C). We believe S325C is involved in a disulfide bond because performing the thermostability measurements in the presence of the reducing agent DTT abolishes the stability enhancement (Fig. S7). Identifying single mutations with such dramatic stability improvements is very difficult using other protein engineering methods. Other substitutions with positive enrichment values also increase the enzyme’s thermostability (Fig. 4D and Fig. S8). This simple protocol allows the identification of thermostabilizing mutations and can be adapted to enrich for a variety of additional properties by screening under different conditions.
Fig. 4.
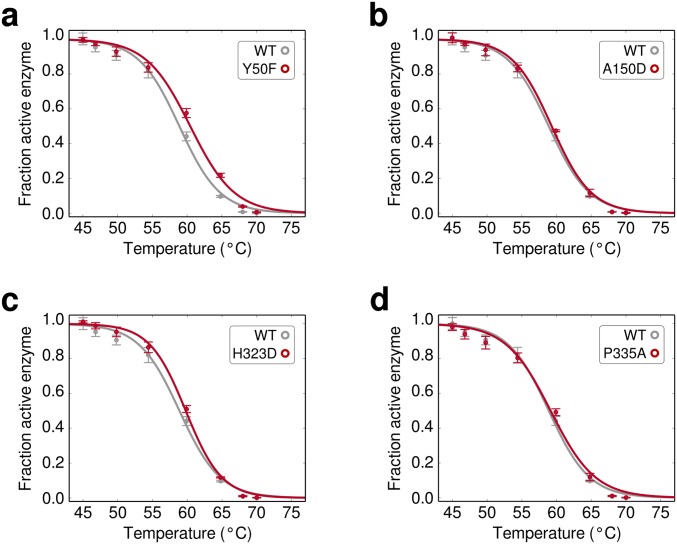
Identification of stabilizing point mutations. (A) High-temperature screening enriches for stabilizing mutations. The enrichment value of 2,956 amino acid substitutions plotted over sequence positions. Amino acids that were not observed are colored as white and the WT residue is colored gray with a box around it. (B) The overall distribution of enrichment values. Only 3% of substitutions have a positive enrichment value. (C) Thermal inactivation curves for WT Bgl3 and the mutant with the highest enrichment value. S325C increases the T50 of the enzyme by 5.3 °C. (D) Enriched mutations confer enhanced thermostability. A panel of five mutations was chosen based on their enrichment value and its reproducibility over experimental replicates. The enriched mutations were experimentally characterized, and all showed moderate-to-large increases in thermostability. The magnitudes of the stability increases depend on the assay conditions and tend to be lower when tested under conditions different from the screen (Fig. S6).
Fig. S7.
Effect of reducing agents on enzyme thermostability. (A) The 5 mM DTT has virtually no effect on WT Bgl3’s thermostability. (B) In contrast, S325C is significantly destabilized in the presence of 5 mM DTT. In the presence of DTT, S325C’s T50 value becomes nearly identical to WT (58.6 °C).
Fig. S8.
(A–D) Thermal inactivation curves for enriched Bgl3 mutants. All mutants were assayed in conditions that matched the original microfluidic screening protocol.
Fig. S6.
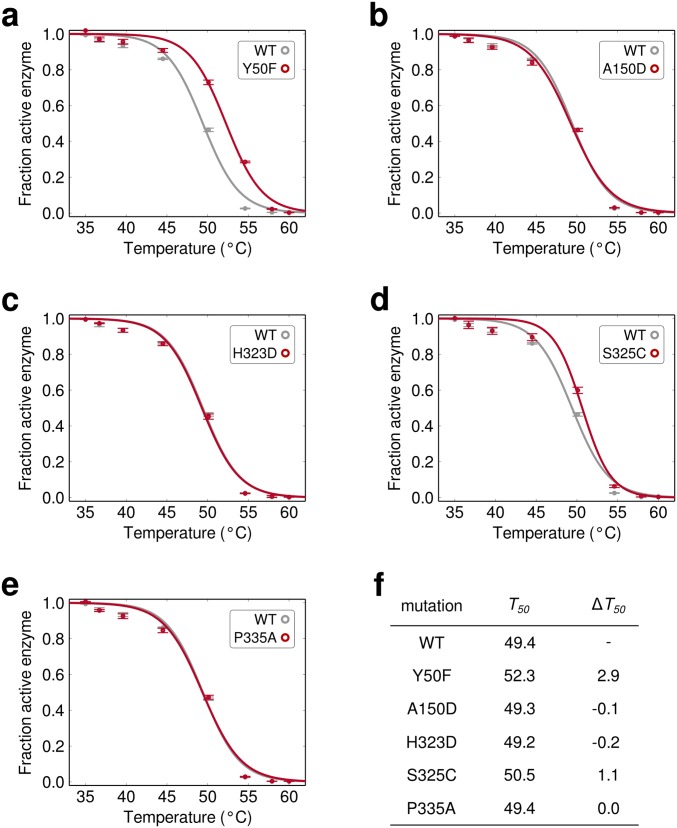
Thermostability of Bgl3 mutants in 100 mM potassium phosphate, pH 7. Cells were lysed using sonication rather than detergents and lysozyme. (A–E) Thermal inactivation curves for enriched Bgl3 mutants. (F) Summary of thermostability measurements in 100 mM potassium phosphate, pH 7. All measurements were performed in at least triplicate, and the median T50 values are reported. The absolute T50 values decrease when assayed in pure buffer, which we attribute to a stabilizing effect caused by the lysis detergents used in the microfluidic screen. In addition, the magnitudes of the stability increases (ΔT50) tend to be lower when tested under conditions different from the original screening conditions.
Discussion
Deep mutational scanning is a powerful tool for exploring the molecular basis of protein function (7, 15, 25, 26). However, restrictions on functional assays have limited its general applicability, particularly for enzymes. We have presented a method for characterizing millions of enzyme variants by compartmentalizing reactions in aqueous microdroplets. The assays use an optical readout and can therefore be readily adapted to the numerous classes of enzymes with fluorescence-based activity assays.
Our experimental protocol enabled the analysis of over 1 million Bgl3 variants, and we used the resulting sequence–function map to evaluate the enzyme’s tolerance to mutation. This unbiased analysis discovered sites within the enzyme that cannot tolerate mutations and are therefore likely to play an important role in Bgl3 function. Alternately, sites with a high tolerance to mutation are important for protein evolution and engineering because they can accept diversification while still maintaining catalytic function; this provides the protein engineer with flexibility in enhancing certain properties while maintaining others. The sequence–function mapping approach provides a local view of protein sequence space that can identify important interactions overlooked by large alignments of homologous sequences.
Droplet-based microfluidic screening provides a flexible platform for assaying enzyme activity over a broad range of reaction conditions (10–13). We adapted our screening protocol to include a heat challenge and enriched for mutations that increase the enzyme’s thermostability. An alternative approach for identifying stabilizing mutations from high-throughput sequence–function data was recently developed that involved scoring a residue’s ability to rescue the deleterious effects of other mutations (27). However, the droplet-based screening approach is extremely versatile and could theoretically be used to identify variants with enhanced properties including increased kcat (reduced reaction time), decreased Km (reduced substrate concentration), increased tolerance to biomass pretreatments (increased ionic liquid concentration), and reduced product inhibition (increased glucose concentration). Systematically mapping multiple enzyme properties will allow us to evaluate the trade-offs between properties and enable multiobjective protein engineering.
Experimentally mapping protein sequence space requires high-throughput library synthesis, screening, and sequencing, any of which could be a bottleneck. From this work, we found library construction and sequencing to be more limiting than microfluidic screening. Our random mutagenesis library contained 6 million unique variants (colony-forming units), and the transformation efficiency limited the size of this library. The microfluidic sorter analyzed over 10 million enzyme variants in 23 h, and the throughput of more recent sorter designs is more than an order of magnitude faster (28)—enabling the screening of libraries beyond 108 variants. Although Illumina DNA sequencers can provide a large number of sequencing reads, read length is currently limited to ∼600 bp, about one-third of the bgl3 gene. A number of new methods to generate longer read lengths have recently been developed (29, 30) and would allow a pairwise analysis by correlating the effects of mutations at distant sequence positions.
Our method relies on a microfluidic droplet sorter that requires specialized instrumentation not typically found in a biochemistry laboratory. However, an alternative to screening enzyme variants in water-in-oil droplets is to screen using water-in-oil-in-water double emulsions (31). Double-emulsion droplets also provide microcompartments with which to test individual enzyme variants but can be generated using commercially available microfluidic systems (Dolomite Microfluidics) and sorted using standard cell sorters (32). This should provide an easily adoptable and widely available solution for implementing our sequence–function mapping method.
Our method could potentially be applied to a large number of different enzyme classes. In addition to glycosidases, emulsion-based methods have been used to screen DNA/RNA polymerases, oxidoreductases, sulfatases, peroxidases, esterases, proteases, and even ribozymes (10, 11, 33–37). The greatest challenge with emulsion-based screening is finding a fluorescent assay for one’s particular enzyme of interest. It is important to note that some small-molecule dyes readily exchange between emulsion droplets and limit the ability to resolve functional differences (38).
The ability to rationally engineer enzymes will have a far-reaching impact on areas that range from medicine and agriculture to environmental protection and industrial chemistry. However, enzyme function involves an extraordinarily complex balance of numerous physical interactions, which has limited the design of tailor-made enzymes. Large sequence–function datasets will provide an increasingly detailed view of the determinants of enzyme function. When combined with methods from statistics and machine learning, protein design rules can be extracted and applied in an automated manner (39). Given the rapid pace of advances in high-throughput experimentation, data-driven protein engineering may be able to outpace more traditional physics-based methods.
Materials and Methods
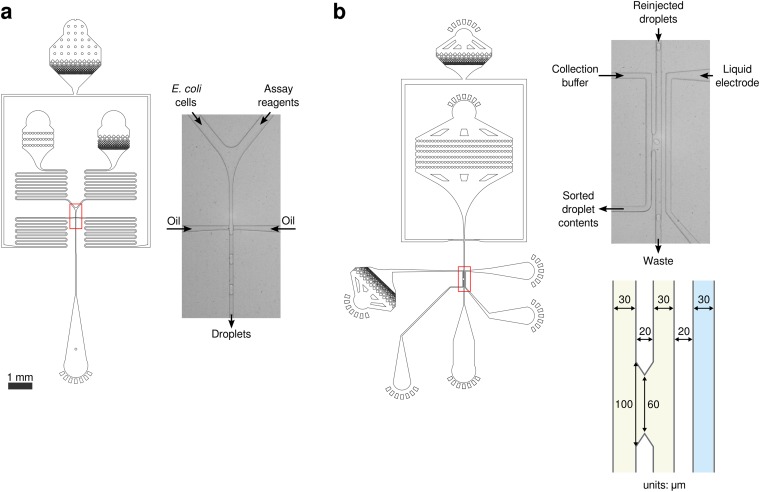
All microfluidic devices were fabricated in-house using standard soft lithography techniques (Fig. S9). Photomasks were used to pattern layers of photoresist (SU-8 3025) on a silicon wafer, and uncured polydimethylsiloxane (PDMS) (11:1 polymer–to–cross-linker ratio) was poured over the mold. The PDMS was cured at 80 °C for 1 h, extracted from the mold with a scalpel, and access holes were punched using a 0.75-mm biopsy core. The devices were then bonded to glass slides after a plasma surface treatment. The device channels were made hydrophobic by flushing with Aquapel (Pittsburgh Glass Works) and then baking for an additional 10 min at 80 °C. Microfluidic fluorescence measurements were performed using a custom-built fluorimeter (Fig. S10).
Fig. S9.
Illustrations and microscopy images of the microfluidic devices used in this work. Both devices had channels 20 μm tall. (A) Droplet maker device with a microscopy image of the device in operation. The droplet-making junction is an intersection of two 15-μm-wide channels. The cell suspension enters the left inlet, and the assay reagents, the right inlet. Immediately after these two aqueous streams combine, the oil pinches off monodisperse droplets. The serpentine channels act as flow resistors that dampen pressure fluctuations. (B) Sorting device with microscopy image of device in operation and detailed device dimensions. Close-packed droplets are reinjected onto the chip and spaced with additional oil. If a droplet meets the desired fluorescence criteria, then a series of electric pulses is applied to the collection buffer. The applied electric field destabilizes the interface between the droplet and the adjacent aqueous stream, and surface tension pulls the sorted droplet into the aqueous collection stream. The liquid electrode serves as a ground and an electrostatic shield. Detailed dimensions of the sorting junction are shown below the image.
Fig. S10.
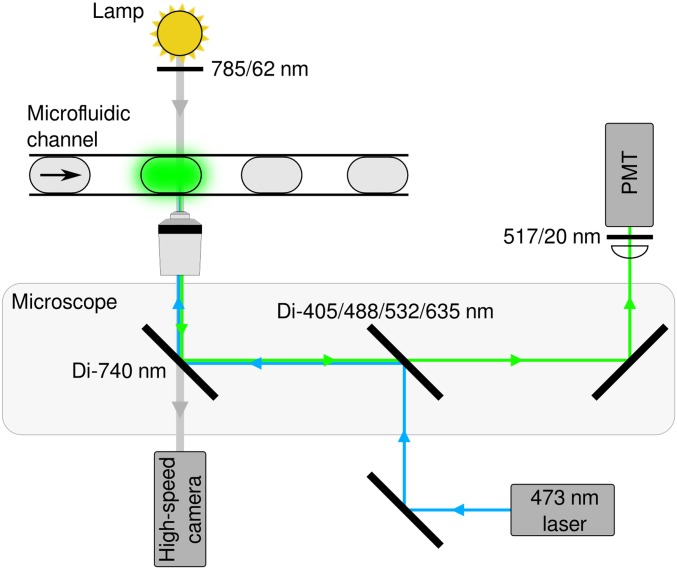
Fluorescence detection system. The fluorescence of each droplet is analyzed using an epifluorescence microscope. A 473-nm laser is used to excite each droplet, and the fluorescence emission is measured using a photomultiplier tube (PMT) with a 517-nm bandpass filter. Simultaneously, an incandescent lamp is used for high-speed, bright-field imaging of the microfluidic channel.
SI Materials and Methods
Construction of Bgl3 Random Mutagenesis Library.
The bgl3 gene was cloned into the pET-22b (Novagen) expression vector and used as a template for error-prone PCR. Error-prone PCR was performed following a protocol where MnCl2 is used to tune the mutation rate of Taq polymerase (40). We determined that a final concentration of 100 μM MnCl2 yielded ∼4 amino acid substitutions per gene. After 15 PCR cycles, the reaction was treated with DpnI overnight and purified with a DNA spin column (Zymo Research).
The mutagenized bgl3 insert was cloned back into pET-22b using circular polymerase extension cloning (CPEC) (41). The CPEC reaction was purified and concentrated using a DNA spin column (Zymo Research) and used to transform electrocompetent BL21(DE3) Escherichia coli cells (Lucigen). The transformed cells were recovered in expression recovery media (Lucigen) at 37 °C for 1 h. Several dilutions of the transformation were plated to determine the total library size and the remainder used to inoculate a 50-mL LB-carbenicillin culture. Once the culture reached a measurable OD600, freezer stocks were made by combining with 50% (vol/vol) glycerol and the library was stored at −80 °C until use. The final library contained 6 million unique transformants. Ten individual clones were sequenced to determine the library’s mutation rate of 3.8 amino acid substitutions per gene. The library displayed the expected mutational biases for error-prone PCR.
Microfluidic Screening of Bgl3 Library.
A glycerol stock of the Bgl3 library was used to inoculate a 5-mL MagicMedia (Invitrogen) expression culture. This library was expressed overnight, pelleted, and resuspended in assay buffer (100 mM potassium phosphate, pH 7). A 2× cell solution was made by diluting the cell suspension to an OD600 of 0.05 in assay buffer. Assay reagents at 2× concentration were combined to a final concentration of 0.6× BugBuster (Novagen), 60 kU/mL rLysozyme (Novagen), 200 μM fluorescein di-(β-d-glucopyranoside) (Sigma) in 100 mM potassium phosphate, pH 7. A relatively low substrate concentration (∼100–1,000× enzyme concentration) was chosen to allow most reactions to go to completion and to identify all active variants even if they have diminished total activity.
Microdroplets containing expressed enzyme variants were generated using a coflow droplet maker device (Fig. S9A). Equal volumes of 2× cells and 2× assay reagents were combined by the device and emulsions generated using fluorinated oil (HFE 7500) containing 2% (wt/wt) PEG–perfluoropolyether amphiphilic block copolymer surfactant (RAN Technologies) in a flow focus droplet maker. Both aqueous inlets were injected at 150 μL/h and the fluorinated oil at 700 μL/h. At these flow rates, each droplet has a volume of ∼8 pL, and, on average, 1 in 10 contains a single E. coli cell. Under these lysis conditions, E. coli cells fully rupture and solubilize within a few seconds. The droplets were collected into a syringe and incubated at 37 °C for 1 h.
After incubation, the droplets were sorted using selective electrocoalescence with an aqueous collection stream (Fig. S9B). A 473-nm laser was focused onto the channel just upstream of the sorting junction, each droplet was individually excited, and its fluorescence emission measured using a spectrally filtered PMT (Hamamatsu Photonics) at 520 nm (Fig. S10). A field-programmable gate array card controlled by custom LabVIEW code analyzed the droplet signal at 200 kHz, and if it detected sufficient fluorescence (Fig. S1 D and E), a train of seven 100-V, 40-kHz pulses was applied by a high-voltage amplifier (Trek). This pulse destabilized the interface between the droplet and the adjacent aqueous stream, causing the droplet to merge with the stream via a thin-film instability, after which the droplet contents were injected into the collection stream via its surface tension (14). The contents of the sorted droplets were collected in a microcentrifuge tube for further processing. Droplets were analyzed at 1,300 per s, and, because 1 in 10 droplets contained a cell, cells were analyzed at ∼130 per s.
The Bgl3 library was sorted on 4 separate days for about 6 h each day. During each of these runs, we analyzed ∼27 million droplets containing ∼2.7 million cells. The droplet fluorescence intensity distribution (Fig. S1E) shows two peaks that correspond to inactive and active populations, and the sorting threshold was chosen at the minimum between these two peaks. Approximately 900,000 individual droplets containing active cells were sorted during each run. In total, we analyzed over 10 million cells and recovered ∼3.4 million active variants, which fed into the sequence–function mapping pipeline.
For the screen containing a heat challenge, a proportional–integral–derivative (PID)-controlled heating element was added in-line directly after droplet formation (Fig. S5). This allowed us to heat the droplets at 65 °C for ∼10 min. Using this protocol, we analyzed 100 million droplets containing ∼10 million cells and recovered ∼2 million active variants.
Recovery of Sorted DNA.
The contents of the sorted droplets were collected from the microfluidic chip, and DNA was recovered using a DNA spin column (Zymo Research). The eluted DNA was transformed into high efficiency 10G SUPREME Electrocompetent E. coli cells (Lucigen), and transformed cells were cultured in expression recovery media (Lucigen) at 37 °C for 1 h. Several dilutions of the transformation were plated to determine the total number of transformants and the remainder used to inoculate a 50-mL LB-carbenicillin culture. Once the culture reached a measurable OD600, freezer stocks were made by combining the culture with 50% (vol/vol) glycerol and stored at −80 °C. For these transformations, we typically obtained 1–10 times more transformants (colony-forming units) than sorted droplets that entered the protocol, suggesting good sampling of the genetic diversity within sorted population.
Illumina Library Preparation and Sequencing.
The gene libraries before and after sorting were used to prepare an Illumina sequencing library. All samples were processed in parallel and sequenced on the same run to minimize potential biases. Individual sorting runs were prepared as separate sequencing libraries to allow for internal validation of reproducibility. Because current Illumina MiSeq kits can only sequence 600 bp (approximately one-third of bgl3 gene), the genes were randomly fragmented and sequenced.
A library’s glycerol stock was used to inoculate an overnight LB culture and the plasmid DNA was miniprepped. A 2-kb fragment containing the bgl3 gene was cut out of the pET-22b vector using the SgrAI and DraIII sites and gel extracted. These gel-extracted inserts were used as inputs to the Nextera XT DNA Sample Prep Kit (Illumina). Each sample was barcoded using a different index primer. A low SPRI bead ratio (0.4×) was used to select for longer sequence fragments in an attempt to obtain pairwise mutation information from sites distant in the gene. The resulting libraries were quantified using a high-sensitivity Bioanalyzer chip (Agilent), a Qubit Assay Kit (Invitrogen), and finally quantitative PCR (Kapa Biosystems). The average sequence fragment was ∼1,400 bp. All libraries were pooled in equimolar proportions and sequenced using a MiSeq, version 3, 2 × 300 run with a 5% PhiX control spike-in.
Analysis of Illumina Sequencing Data.
Paired-end DNA-sequencing reads were mapped to the bgl3 gene using Bowtie2’s very-sensitive–local alignment setting (42). Typically, 80–90% of the paired-end reads aligned concordantly exactly one time. The resulting SAM files were parsed to count the amino acids observed at each Bgl3 position. Reads with a Phred quality score (Q score) of less than 30 were excluded from the analysis.
The frequency of each amino acid at each position was calculated by dividing the number of times the amino acid was observed by the total number of observations at that position. Amino acids with less than 10 total observations at a given position were considered insignificant and excluded from the analysis. After this filter, there were good statistics on the 500 wild-type (WT) amino acids plus 3,083 amino acid substitutions. The frequency of WT amino acids was significantly larger than the substitutions because mutations only occur ∼1% of the time.
The relative entropy of a specific site is given by the following:
where the sum is over all 20 amino acids, and fsort,a and funsort,a are the frequencies of amino acid a in the sorted and unsorted libraries, respectively. If either fsort,a or funsort,a are equal to zero, then amino acid a is excluded from the summation to prevent infinite values.
The enrichment of a substitution to amino acid a is given by the following:
where fsort,a and funsort,a are the frequencies of amino acid a in the sorted and unsorted libraries, respectively.
Analysis of Natural Glycoside Hydrolase Family 1 Sequences.
The sequences of other glycoside hydrolase family 1 members were downloaded from the National Center for Biotechnology Information Protein database using GenBank accession numbers from the Carbohydrate Active Enzymes (CAZy) GH1 database (43). Sequences containing less than 30% sequence identity with Bgl3 were removed, and the remaining 1,300 sequences aligned using the MUSCLE multiple sequence alignment program (44). The frequency of each amino acid at each Bgl3 site was calculated by dividing the number of times the amino acid was observed by the total number of observations at that position. Gaps in the alignment were excluded from the analysis.
The sequence conservation score describes how much the amino acid distribution at a given site in the multiple sequence alignment (MSA) differs from a general, background amino acid distribution. This is quantified using the MSA relative entropy (REMSA) (22, 23):
where the sum is over all 20 amino acids, fmsa,a is the frequency of amino acid a at a particular position in the MSA, and fbg,a is the background amino acid frequency of amino acid a taken from all positions in the MSA. If fmsa,a is equal to zero, then amino acid a is excluded from the summation to prevent infinite values. The REMSA is different from the relative entropy used to analyze the experimental mutational data because it describes how the MSA’s amino acid distribution differs from a fixed background amino acid distribution.
We generated the glycoside hydrolase family 1 phylogenetic tree (Fig. 3D) by taking the sequences of all GH1 entries in the Protein Data Bank. Redundant sequences containing greater than 90% sequence identity were removed. The remaining 39 sequences were then processed using the Phylogeny.fr web server (45).
Cloning of Individual Mutations.
Individual mutations for follow-up analyses were cloned using the QuikChange Lightning kit (Agilent) and transformed into Bl21 (DE3) (Lucigen). A single colony was grown overnight, miniprepped, and gene sequence was verified using Sanger sequencing with the T7 promoter and T7 terminator primers.
Plate-Based Functional Assay.
The fraction of functional sequences was determined for the initial library, the sorted library, and the site-specific libraries using a plate-based functional assay. Single colonies were picked into a 96–deep-well plate containing 500 μL of MagicMedia (Invitrogen), and these cultures were expressed overnight, shaking at 37 °C. The next day, the cells from the expression culture were pelleted and resuspended in 200 μL of assay buffer (100 mM potassium phosphate, pH 7). The 2× assay reagents were combined to a final concentration of 0.6× BugBuster (Novagen), 60 kU/mL rLysozyme (Novagen), 2 mM 4-methylumbelliferyl-β-d-glucopyranoside (Sigma) in 100 mM potassium phosphate, pH 7. A volume of 75 μL of the cell suspension was combined with 75 μL of the 2× assay reagents and allowed to react for 15 min at room temperature. Then 100 μL of 1 M Tris, pH 9.5, was added to each reaction, and the fluorescence was measured with an excitation of 380 nm and an emission of 450 nm. A sequence was considered functional if its end-point activity was at least 50% of Bgl3’s.
Thermostability Measurements.
A Bgl3 variant was expressed overnight, shaking at 37 °C in a 5-mL MagicMedia (Invitrogen) culture. The cells from the expression culture were pelleted and frozen. The cell pellets were resuspended in lysis buffer [0.3× BugBuster (Novagen), 30 kU/mL rLysozyme (Novagen), and 50 U/mL DNase I (New England Biolabs) in 100 mM potassium phosphate, pH 7]. Serial dilutions of the lysate were performed to determine the linear range of the enzyme assay, and all samples were diluted in lysis buffer to be within the linear range and have similar end-point activities.
The diluted cell extracts were arrayed into 96-well PCR plates. Using a gradient thermocycler, the samples were heated over multiple temperatures (typically 45–70 °C) for 10 min. After the heat step, the remaining functional enzyme was quantified by adding the substrate 4-methylumbelliferyl-β-d-glucopyranoside (Sigma) to a final concentration of 1 mM. After reacting for 15 min, the fluorescence was measured with an excitation and emission of 380 and 450 nm, respectively. The T50 (temperature where 50% of the protein is inactivated in 10 min) was determined by fitting a shifted sigmoid function to the thermal inactivation curves. All measurements were performed in at least triplicate with the median T50 values reported.
Acknowledgments
We thank R. A. Heins for providing the bgl3 gene and useful feedback. We acknowledge J. Fraser, P. Babbit, and T. Kortemme for helpful discussions and feedback on the manuscript. P.A.R. is supported by the National Institute of General Medical Sciences of the NIH under Award F32GM107107, the University of California President’s Postdoctoral Fellowship Program, and the Burroughs Wellcome Fund Postdoctoral Enrichment Program. T.M.T. is supported by the National Science Foundation Graduate Research Fellowship under Grant 1144247. This work was funded by a National Science Foundation CAREER Award (DBI-1253293), the NIH New Innovator Award (AR068129-01) and an R21 (HG007233-01), the Defense Advanced Research Projects Agency Living Foundries Program (HR0011-12-C-0065), a Research Award from the California Institute for Quantitative Biosciences, and the Bridging the Gap Award from the Rogers Family Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422285112/-/DCSupplemental.
References
- 1.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34(12):938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 2.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19(10):1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassila JK, Baker D, Herschlag D. Origins of catalysis by computationally designed retroaldolase enzymes. Proc Natl Acad Sci USA. 2010;107(11):4937–4942. doi: 10.1073/pnas.0913638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frushicheva MP, Cao J, Chu ZT, Warshel A. Exploring challenges in rational enzyme design by simulating the catalysis in artificial kemp eliminase. Proc Natl Acad Sci USA. 2010;107(39):16869–16874. doi: 10.1073/pnas.1010381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler DM, Fields S. Deep mutational scanning: A new style of protein science. Nat Methods. 2014;11(8):801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7(9):741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA. 2011;108(19):7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30(6):543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491(7422):138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kintses B, et al. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem Biol. 2012;19(8):1001–1009. doi: 10.1016/j.chembiol.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Granieri L, Baret JC, Griffiths AD, Merten CA. High-throughput screening of enzymes by retroviral display using droplet-based microfluidics. Chem Biol. 2010;17(3):229–235. doi: 10.1016/j.chembiol.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Fallah-Araghi A, Baret J-C, Ryckelynck M, Griffiths AD. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip. 2012;12(5):882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- 14.Fidalgo LM, et al. From microdroplets to microfluidics: Selective emulsion separation in microfluidic devices. Angew Chem Int Ed Engl. 2008;47(11):2042–2045. doi: 10.1002/anie.200704903. [DOI] [PubMed] [Google Scholar]
- 15.Jacquier H, et al. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc Natl Acad Sci USA. 2013;110(32):13067–13072. doi: 10.1073/pnas.1215206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci USA. 2004;101(25):9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom JD, et al. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci USA. 2005;102(3):606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444(7121):929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 19.Zechel DL, Withers SG. Glycosidase mechanisms: Anatomy of a finely tuned catalyst. Acc Chem Res. 2000;33(1):11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- 20.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 21.Marana SR. Molecular basis of substrate specificity in family 1 glycoside hydrolases. IUBMB Life. 2006;58(2):63–73. doi: 10.1080/15216540600617156. [DOI] [PubMed] [Google Scholar]
- 22.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: Evolutionary units of three-dimensional structure. Cell. 2009;138(4):774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan BJ, et al. Stabilizing proteins from sequence statistics: The interplay of conservation and correlation in triosephosphate isomerase stability. J Mol Biol. 2012;420(4-5):384–399. doi: 10.1016/j.jmb.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adkar BV, et al. Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure. 2012;20(2):371–381. doi: 10.1016/j.str.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Wu NC, et al. Systematic identification of H274Y compensatory mutations in influenza A virus neuraminidase by high-throughput screening. J Virol. 2013;87(2):1193–1199. doi: 10.1128/JVI.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagenaar TR, et al. Resistance to vemurafenib resulting from a novel mutation in the BRAFV600E kinase domain. Pigment Cell Melanoma Res. 2014;27(1):124–133. doi: 10.1111/pcmr.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araya CL, et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci USA. 2012;109(42):16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sciambi A, Abate AR. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip. 2015;15(1):47–51. doi: 10.1039/c4lc01194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods. 2010;7(2):119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundin S, et al. Hierarchical molecular tagging to resolve long continuous sequences by massively parallel sequencing. Sci Rep. 2013;3:1186. doi: 10.1038/srep01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aharoni A, Griffiths AD, Tawfik DS. High-throughput screens and selections of enzyme-encoding genes. Curr Opin Chem Biol. 2005;9(2):210–216. doi: 10.1016/j.cbpa.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Lim SW, Abate AR. Ultrahigh-throughput sorting of microfluidic drops with flow cytometry. Lab Chip. 2013;13(23):4563–4572. doi: 10.1039/c3lc50736j. [DOI] [PubMed] [Google Scholar]
- 33.Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA. 2001;98(8):4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beneyton T, Coldren F, Baret J-C, Griffiths AD, Taly V. CotA laccase: High-throughput manipulation and analysis of recombinant enzyme libraries expressed in E. coli using droplet-based microfluidics. Analyst (Lond) 2014;139(13):3314–3323. doi: 10.1039/c4an00228h. [DOI] [PubMed] [Google Scholar]
- 35.Ma F, Xie Y, Huang C, Feng Y, Yang G. An improved single cell ultrahigh throughput screening method based on in vitro compartmentalization. PLoS One. 2014;9(2):e89785. doi: 10.1371/journal.pone.0089785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu R, Martinez R, Prodanovic R, Klein M, Schwaneberg U. A flow cytometry-based screening system for directed evolution of proteases. J Biomol Screen. 2011;16(3):285–294. doi: 10.1177/1087057110396361. [DOI] [PubMed] [Google Scholar]
- 37.Ryckelynck M, et al. Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions. RNA. 2015;21(3):458–469. doi: 10.1261/rna.048033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skhiri Y, et al. Dynamics of molecular transport by surfactants in emulsions. Soft Matter. 2012;8(41):10618–10627. [Google Scholar]
- 39.Romero PA, Krause A, Arnold FH. Navigating the protein fitness landscape with Gaussian processes. Proc Natl Acad Sci USA. 2013;110(3):E193–E201. doi: 10.1073/pnas.1215251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloom JD, et al. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 2007;5:29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6(2):242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]