Significance
The brain comprises an immense number of cells and cellular connections. We describe the first, to our knowledge, single cell whole transcriptome analysis of human adult cortical samples. We have established an experimental and analytical framework with which the complexity of the human brain can be dissected on the single cell level. Using this approach, we were able to identify all major cell types of the brain and characterize subtypes of neuronal cells. We observed changes in neurons from early developmental to late differentiated stages in the adult. We found a subset of adult neurons which express major histocompatibility complex class I genes and thus are not immune privileged.
Keywords: human brain, single cells, RNAseq, neurons, interneurons
Abstract
The human brain is a tissue of vast complexity in terms of the cell types it comprises. Conventional approaches to classifying cell types in the human brain at single cell resolution have been limited to exploring relatively few markers and therefore have provided a limited molecular characterization of any given cell type. We used single cell RNA sequencing on 466 cells to capture the cellular complexity of the adult and fetal human brain at a whole transcriptome level. Healthy adult temporal lobe tissue was obtained during surgical procedures where otherwise normal tissue was removed to gain access to deeper hippocampal pathology in patients with medical refractory seizures. We were able to classify individual cells into all of the major neuronal, glial, and vascular cell types in the brain. We were able to divide neurons into individual communities and show that these communities preserve the categorization of interneuron subtypes that is typically observed with the use of classic interneuron markers. We then used single cell RNA sequencing on fetal human cortical neurons to identify genes that are differentially expressed between fetal and adult neurons and those genes that display an expression gradient that reflects the transition between replicating and quiescent fetal neuronal populations. Finally, we observed the expression of major histocompatibility complex type I genes in a subset of adult neurons, but not fetal neurons. The work presented here demonstrates the applicability of single cell RNA sequencing on the study of the adult human brain and constitutes a first step toward a comprehensive cellular atlas of the human brain.
The adult human brain is a tissue of immense complexity, composed of multiple cell classes, each of which can be further divided into additional subtypes based on function, topology, and molecular characteristics. Studies of the human brain are typically difficult to conduct because most samples can only be acquired postmortem, thereby limiting sample quality. As a result, efforts to understand the molecular heterogeneity of the brain have been primarily focused on nonhuman model organisms. Studies of the nonhuman brain have had a remarkable impact on our understanding of this complex organ; however, given the additional complexity of the developing and mature human cortex, findings in nonhuman models may not capture the entirety of the relevant molecular diversity in the human brain.
Efforts thus far to elucidate the heterogeneity of the human brain at the molecular level generally fall in two categories. In situ approaches (1–5) preserve the structure and topological characteristics of the tissue with single cell resolution, but are usually limited to only a handful of genes or proteins. In vitro experiments that use proteomics, next generation sequencing, or microarrays provide a more holistic picture in terms of the molecular complexity. These techniques, however, are themselves limited to the analysis of pooled populations of cells on the level of the organ or cell types that are typically defined by at best a handful of markers. Recently the development of single cell RNA sequencing (RNAseq) approaches have enabled the application of genomic approaches with single cell resolution to a variety of biological areas (6–10). While these and related approaches have been applied to fetal human brain (10), it would be of great interest to use them to understand the structure and complexity of the adult human brain.
We used single cell RNA sequencing to interrogate the heterogeneity of the adult human cortex in an approach that combines transcriptome-wide analysis with single cell resolution. Brain tissue was obtained from living subjects who underwent temporal lobectomy for medically refractory seizures and mesial temporal sclerosis (MTS). In these patients, a small piece (0.5–3 g) of anterior temporal lobe was resected during the surgical approach to deeper areas of the brain and was considered normal by all detectable means including electroencephalogram (EEG) and pathological examination. We also performed a set of experiments with cortex from prenatal brain at the age of 16–18 weeks postgestation.
We clustered single cells as members of various cell types using both an unbiased approach that integrates information from the entire transcriptome and a focused approach using prior knowledge of neuroscience, in particular, a subset of highly enriched cell-type–defining genes for different cellular populations derived from the mouse brain (11). We were able to achieve a high concordance between the two approaches, illustrating the robustness of single cell RNAseq in identifying different cellular populations without prior information on population-specific genes.
Focusing in particular on the neuronal population, we assessed heterogeneity using an analysis that applies dimensional reduction techniques to the full transcriptome of each neuron. Using this approach we were able to identify seven neuronal subcategories of cells, which broadly separate into two excitatory and five inhibitory groups. We also investigated the expression of genes conventionally used to classify interneurons in mouse, namely, glutamate decarboxylase 1 (GAD1), vasoactive intestinal peptide (VIP), calbindin 2 (CALB2), cholecystokinin (CCK), reelin (RELN), parvalbumin (PVALB), and somatostatin (SST) and found that the neuronal communities identified using our unbiased approach exhibited typical patterns of expression for this subset of classic interneuron markers.
Further analysis of these data yielded several interesting results that had escaped discovery based on bulk genomic methods. First, we were able to interrogate diversity within cortical neurons of the adult brain. Second, we performed a comparative analysis of gene expression profiles between pre- and postnatal neurons and discovered gradients of expression between quiescent and replicating fetal neurons and neuronal progenitors. Third, we directly observed unambiguous expression of major histocompatibility complex class I (MHCI) genes in a subset of adult neurons. Fourth, we have been able to document differences in cell-type prevalence and expression patterns between mice and humans.
Results
Single Cell RNAseq Analysis Captures All of the Major Neuronal, Glial, and Vascular Cell Types in Human Cerebral Cortex.
We analyzed cortical tissue from eight adults and four embryonic samples ranging from 16 to 18 gestational weeks in age. Sample information can be found in SI Appendix, Table S1. Upon alignment of the raw RNAseq reads, we excluded cells that had fewer than 400,000 reads, reducing the initial dataset of 482 to 466 cells. We were able to get an average of 2,838,000 reads per cell, with an average mapped ratio of reads of 0.86 and a gene body coverage of 0.7 averaged across all genes with mapped reads (SI Appendix, Fig. S1).
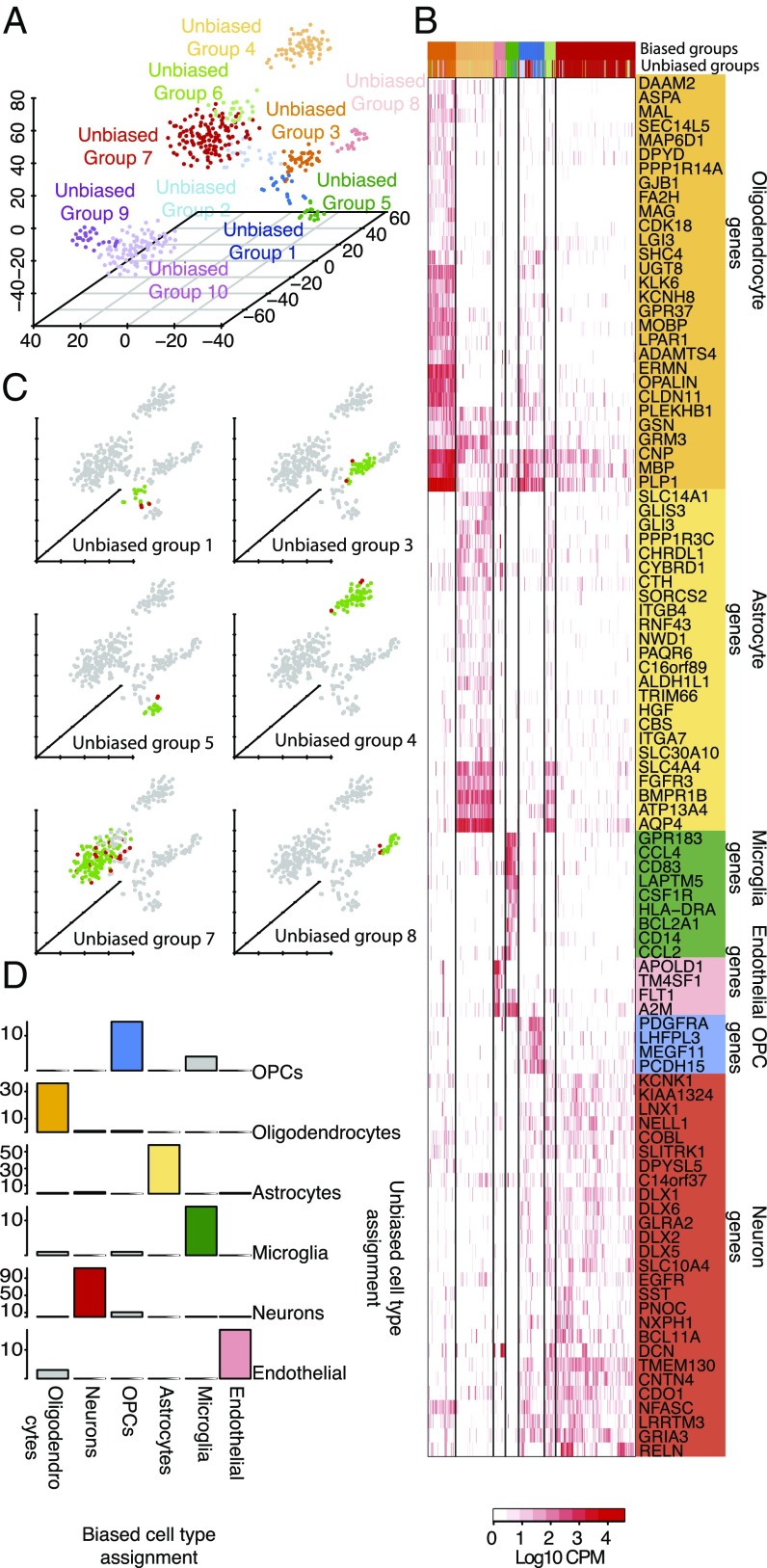
We used two complementary approaches to classify each single cell to one of the major brain cell types: astrocytes, oligodendrocytes, oligodendrocyte precursor cells (OPCs), neurons, microglia, and vascular cells. First, we used an unbiased approach to sort all 466 individual cells into distinct groups defined by the entirety of their molecular signatures as described in SI Appendix, SI Methods. This approach resulted in the identification of 10 distinct cell groups (unbiased groups) (SI Appendix, Fig. S2). A landscape of all of the cells colored by cell cluster, can be seen in Fig. 1A and SI Appendix, Table S2. Clusters 1–8 consist of adult brain cells, whereas clusters 9 and 10 consist of fetal brain cells.
Fig. 1.
(A) Single cells colored by cluster on a 3D space. Ten clusters were identified. (B) Hierarchical clustering of all adult brain cells using a subset of cell-type–enriched genes. Biased groups are groups of cells resulting from the hierarchical clustering, whereas unbiased groups are groups of cells as defined using an unbiased approach. (C) A 3D representation of all cells using the unbiased approach. For each of the unbiased groups of cells belonging to a major cell type, the agreement with the biased groups is shown, with cells classified as members of a particular cell type by both approaches colored in green and cells differentially classified shown in red. (D) Agreement between the biased and unbiased cell-type assignments for each cell type. Number of cells is shown on the y axis.
Based upon the presence of typically associated cell-type–specific markers in the top 20 enriched genes for each unbiased group (SI Appendix, Table S3) we were able to easily identify all but two of the clusters as specific groups consisting of OPCs, oligodendrocytes, astrocytes, microglia, neurons, endothelial cells, replicating neuronal progenitors, and quiescent newly born neurons. While these groups contained relatively uniform identities, the enriched genes of two remaining clusters contained genes characteristic of more than one distinct cell type; the interpretation of this finding is discussed below.
As a complementary approach, which takes into account prior biological information, we used existing RNAseq data from purified cell types of the mouse brain (11). From this data we identified the 50 genes with the highest enrichment in each bulk population of cells. We proceeded by identifying the orthologs of these genes in humans and filtered them for mean expression across all of our single cells greater than 0.1 counts per million (cpm). This resulted in a subset of 118 genes with an average enrichment for each cell type that was greater than 60-fold. The fold enrichment and the number of genes for each cell type can be seen in SI Appendix, Fig. S3. Perhaps expectedly, the mean fold enrichment for genes enriched in neuroepithelial cell types (neurons, oligodendrocytes, and astrocytes) tended to be less than that for endothelial cells and microglia—two cell types of mesoderm lineage.
We used this rubric of cell-type–defining genes to perform supervised hierarchical clustering on all single cells from the adult human brain. Fetal cells were not used in this analysis because we did not have a bulk reference from mouse or human. The results of the clustering can be seen in Fig. 1B. Clustering the single cells from this selection of cell-specific markers created seven cell groups (biased groups, n = 7), each consisting of cells previously assigned to the unbiased groups one to eight. We assigned identities to each of the biased groups using the mean expression of the genes enriched in each cell type (SI Appendix, Fig. S5). Interestingly, when we looked at the number of genes detected per cell type as defined using the biased approach, we found that neurons do seem to express a higher number of genes compared with the rest of the cell types, whereas microglia and endothelial cells seem to express the lowest number of genes (SI Appendix, Fig. S4).
Assuming that the cell-type assignments using this approach resemble the true cell types, we assessed our initial cell-type assignments made with the unbiased approach. As an example, in unbiased group four, which was initially identified as an astrocyte group containing 62 cells, 58 of 62 cells were similarly identified as members of the astrocyte group (biased group three) using the cell-type–enriched gene set (Fig. 1 C and D). This means that we were able to correctly identify astrocytes using whole transcriptome data in an unbiased way with an accuracy of 94%. Similarly, unbiased groups one (n = 18), three (n = 38), five (n = 16), seven (n = 131), and eight (n = 20), which were identified as OPCs, oligodendrocytes, microglia, neurons, and endothelial cells, respectively, contain 14 (78%), 36 (95%), 14 (88%), 113 (86%), and 17 (85%) cells belonging to biased groups consisting of identical cell types. This demonstrates that single cell RNAseq data can be used to successfully identify cell types in the brain without prior selection of genes in a straightforward manner with reasonable classification accuracy.
There were two groups to which we could initially not assign a cell identity: unbiased groups two and six. Cells belonging to unbiased group two (n = 24) seemed to be enriched for a mixture of neuronal-, oligodendrocyte-, and OPC-specific genes. Based on the cell-type assignments performed using the mouse data, it appears that the cells in this group are indeed a mixture of OPCs, oligodendrocytes, and neurons. We believe that the presence of this mixed group of cells is due to two reasons. Firstly, the genes enriched in OPCs do not show a high fold enrichment (SI Appendix, Fig. S3) and secondly, due to genes CNP, MBP, and PLP1 (Fig. 1B), three myelin-related genes, which are detected in some neurons probably due to contamination of each cell with pieces of myelin debris. This results in a somewhat noisy gene expression profile that forces OPCs, having a “weak” expression profile that looks somewhat similar to mature oligodendrocytes, and neurons, contaminated with oligodendrocyte-derived mRNA, to cluster together. An alternative explanation for the expression of genes characteristic of mature oligodendrocytes in OPCs might be that OPCs do indeed express a fraction of mature oligodendrocyte genes as previously suggested (12).
Cells belonging to unbiased group six, with enriched genes characteristic of neurons and astrocytes, mostly fall in biased group six using the mouse data. We noticed that in this group of cells we are indeed detecting high levels of genes such as AQP4 and GFAP (astrocyte specific) as well as THY1, SYT1, and STMN2 (neuron specific). We ruled out the possibility of contamination due to the presence of two cells in the capture chamber of the microfluidic device as a cause of the presence of these cells by examining all of the images acquired before cell lysis from the capture sites of the C1 chip (SI Appendix, Fig. S6). While it is possible that this group represents a new cell type that displays characteristics of both neurons and astrocytes, perhaps as a bipotent progenitor cell, further studies will have to be done to rule out alternative explanations such as contamination of neurons or astrocytes with cellular debri of astrocytes or neurons, respectively, or neuronal phagocytosis by astrocytes (13).
Subclustering of Adult Neurons Reveals Rich Diversity of Interneurons in Human Cerebral Cortex.
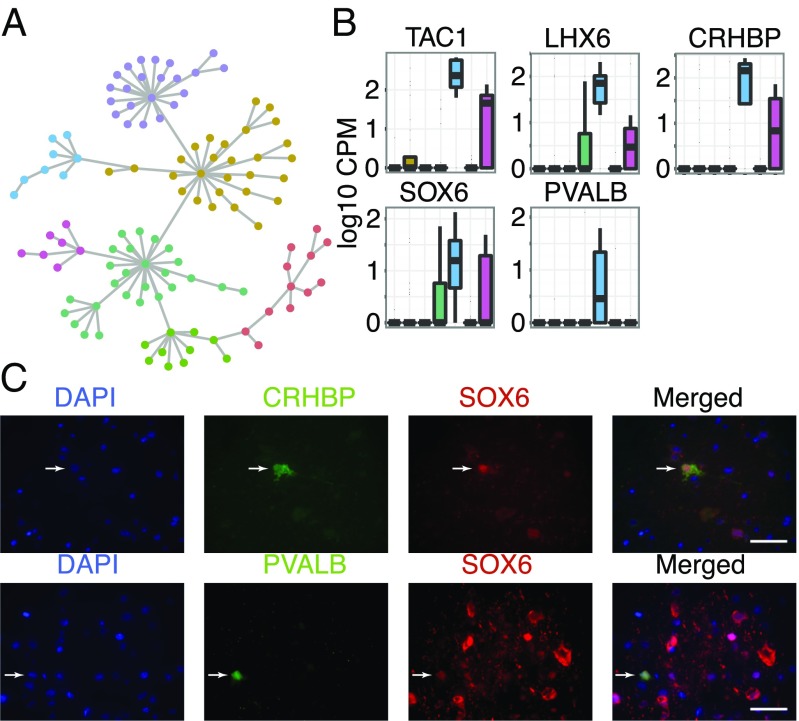
To begin to construct a molecular catalog of neuronal subtypes in the human cortex we examined neuronal subpopulations using the entire transcriptomic profile of each cell. We defined the neuronal population as the intersection of cells belonging to the neuronal groups identified using both the unbiased and biased approaches. We subdivided all neurons by constructing a matrix of pairwise distances based on their global gene expression profiles, followed by the generation of a minimum-spanning tree. We then identified densely connected networks of cells (communities) by using random walks as implemented by the Walktrap community finding algorithm (Fig. 2A and SI Appendix, SI Methods). This resulted in the identification of seven (n = 7) neuronal communities of cells. We then looked for genes that were highly correlated (Pearson correlation) and significantly overexpressed (P < 0.05) (Mann–Whitney u test) in each of the resulting communities. Each neuronal community has a unique gene expression signature, which suggests its role in the network niche. A complete list of all enriched genes per community can be found in SI Appendix, Table S4.
Fig. 2.
(A) Minimum spanning tree for all neuronal cells. Colors indicate communities of cells separated using the Walktrap community finding algorithm. (B) Enrichment of genes TAC1, LHX6, CRHBP, and SOX6 in the PVALB-expressing community of interneurons. The seven neuronal communities are shown along the x axis, consistently colored as in Fig. 2A. (C) Costaining for proteins CRHBP, SOX6, and PVALB in human cortical sections. Arrows indicate costained cells for each panel of markers. Images were taken using a 20x-objective. (Scale bar, 100μm.)
Community five is enriched for the marker PVALB and corticotropin releasing factor binding protein (CRHBP), markers for distinct populations of GABAergic neurons (14). This community is also enriched for tachykinin, precursor 1 (TAC1) as well as the transcription factor LIM homeobox 6 (LHX6) (Fig. 2B). LHX6 is closely related to the transcription factor, SRY (sex determining region Y)-box 6 (SOX6) (15), which is also enriched in the same neuronal community (Fig. 2B). We were then able to verify the coexpression of SOX6, CHRBP, and PVALB in a subpopulation of human cortical neurons via immunohistochemical stainings (Fig. 2C).
We noticed that only a small fraction of the interneurons express PVALB, which is in contrast to the mouse brain where 40% of all interneurons are expected to be PVALB positive (16). To determine whether this was due to undersampling of PVALB expressing cells or whether it could be attributed to an interspecies difference, we performed immunofluorescent stainings using NeuN, a neuronal marker, and PVALB in both mouse and human brain sections. We found that the fraction of PVALB-expressing neurons in the human brain is similar to the percentage we observe in our single cell data. In addition, we found that the fraction of PVALB-expressing neurons is significantly lower (P < 10−6) in humans (mean = 1.8%, SD = 0.73%, range = 0.3–2.91%) than in mice (mean = 5.85%, SD = 1.57%, range = 3.48–8.75%) (SI Appendix, Fig. S7). It should be noted that previous studies indicate that the number of certain interneuron types, including PVALB-positive neurons, may be decreased in affected hippocampi of epileptic patients (17, 18). Although we are examining the unaffected cortical regions of patient temporal lobe, it is possible that the reduced number of PVALB-positive neurons that we observed is a consequence of extrahippocampal pathology.
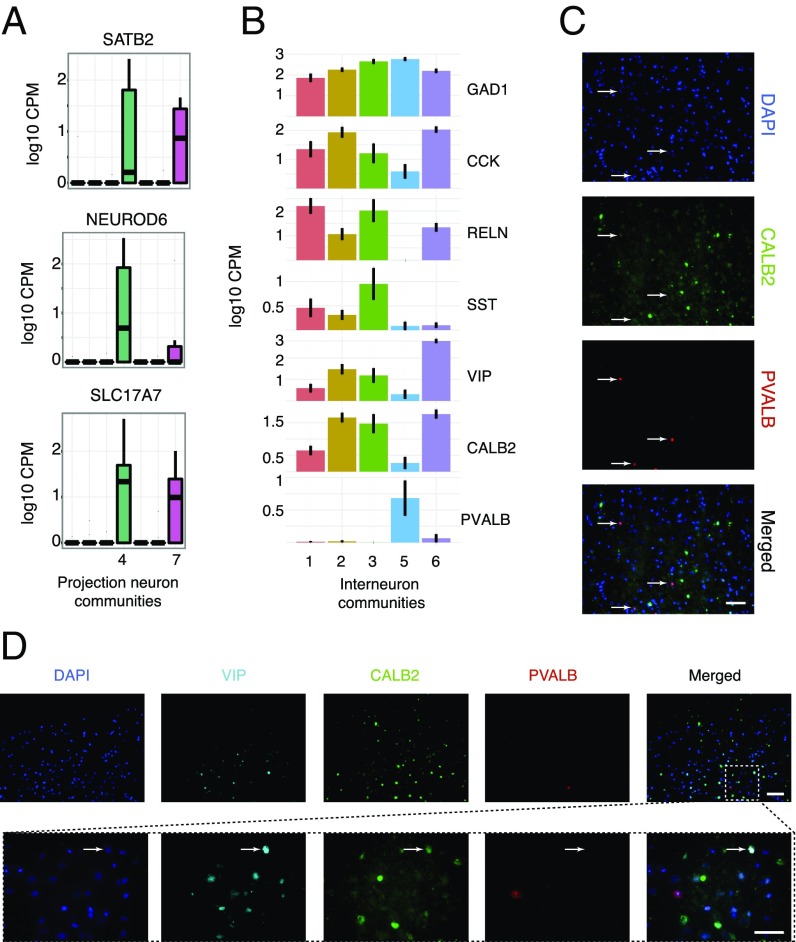
We were able to identify two excitatory communities expressing SLC17A7, NEUROD6 and SATB2 (Fig. 3A), markers for excitatory neuronal cells. The remaining communities of neurons were identified as interneurons based on the lack of expression of excitatory neuronal markers and the strong expression of inhibitory markers such as GAD1 (Fig. 2C).
Fig. 3.
(A) Expression of excitatory neuron markers in excitatory communities of neurons four and seven. (B) Expression of known inhibitory neuron markers in the interneuron communities one, two, three, five, and six. (C) Costaining for proteins PVALB and CALB2 in human cortical sections shows expression of the two proteins in distinct populations of cells. Arrows indicate PVALB-positive cells. Images were taken using a 20× objective. (Scale bar, 100 μm.) (D) Costaining for proteins VIP, PVALB, and CALB2 in human cortical sections shows VIP expression in a subset of CALB2 neurons that lack expression of PVALB. (Upper) Images were taken using a 20× objective. The white dashed box shows an area of the tissue imaged at 63×. (Scale bars, 100 μm for the 20× image and 20 μm for the 63× image.) Arrows indicate VIP/CALB2-positive cells that are negative for PVALB.
When we carefully analyzed the expression of these traditional interneuron markers (PVALB, SST, VIP, CALB2, CCK, and RELN) (Fig. 3B), we immediately noticed a hierarchy of specificity. Among the inhibitory neurons, almost all express GAD1 (the synthetic machinery required to synthesize the inhibitory neurotransmitter, GABA) as well as CCK and CALB2, with the exception of one community of neurons that exclusively expresses PVALB. The expression of PVALB and CALB2 in nonoverlapping neuronal populations was confirmed by in situ immunohistological stainings for CALB2 and PVALB (Fig. 3C), demonstrating that the two proteins are indeed detected in distinct populations of cells. In addition, we verified the coexpression of VIP in a subset of CALB2 neurons and the lack of overlap in these populations with PVALB-expressing cells (Fig. 3D).
The remaining communities display an interesting combination of layer-enriched populations. First, we observed a community of neurons enriched for paired box 6 (PAX6), a gene that has been suggested as a layer I interneuron marker (7). Our data further support this hypothesis with the strong coexpression of RELN in the same community (SI Appendix, Fig. S8). We next observed a neuronal community enriched for VIP and tachykinin 3 (TAC3), two genes that are expressed in neurons enriched in layers two to four. Additionally, we found a subpopulation of neurons exhibiting coexpression of genes complexin 3 (CPLX3); secreted protein, acidic, cysteine-rich (osteonectin) (SPARC); and synaptic vesicle glycoprotein 2C (SV2C) (SI Appendix, Fig. S8), which has been shown to be a human-specific marker for layer three neurons (19). To further investigate CPLX3 as a potential marker of an interneuron subpopulation, we validated specific expression of CPLX3 in a subset of NeuN-positive cells in human cortical slices (SI Appendix, Fig. S8).
Differential Analysis of Adult and Fetal Neurons.
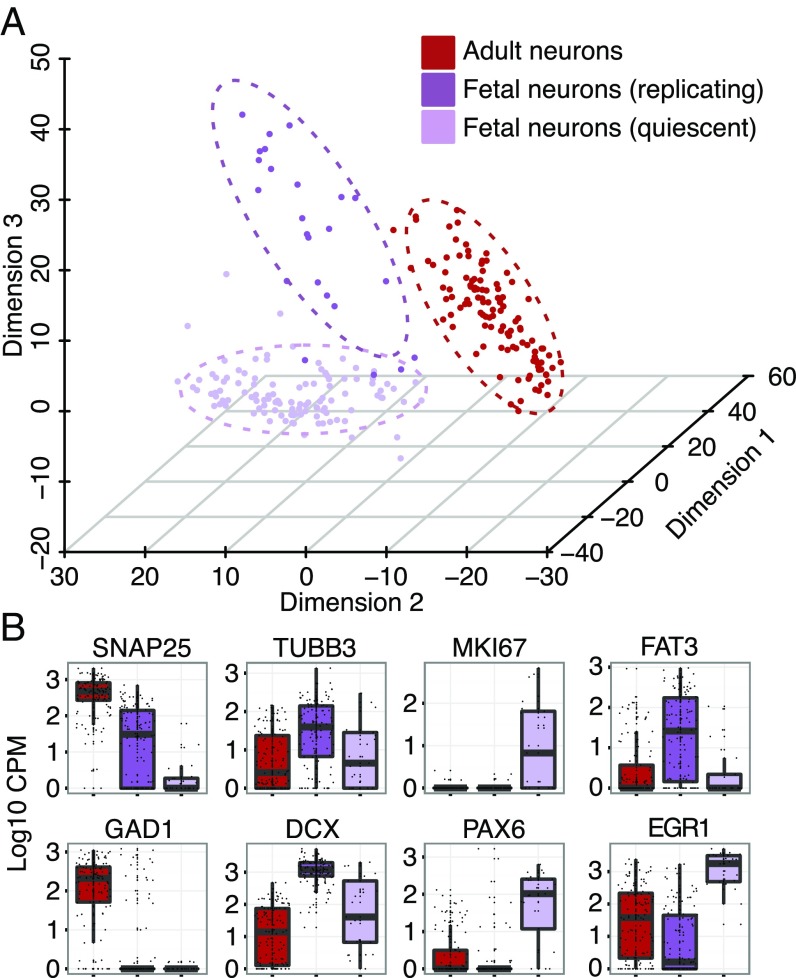
From the classification of all cells (Fig. 1A), it appears that neuronal cells of the fetal brain are considerably distant from any cell type in the adult brain. We investigated the differences between pre- and postnatal neuronal cells by performing a principal component analysis (PCA) on these two populations (Fig. 4A). This identified the same three groups of cells as we saw in our initial analysis. These include adult neurons, expressing high levels of SNAP25 and GAD1, as well as actively dividing fetal neuronal progenitors (MKI67+ and PAX6+) and quiescent subplate fetal neurons (MKI67−, DCX+, and TUBB3+) (Fig. 4B). Genes with the highest contribution to the three first principal components of adult and fetal neurons can be found in SI Appendix, Table S5.
Fig. 4.
(A) PCA on adult and fetal neuronal cells for the first three principal components. (B) Boxplots show expression levels of known markers and newly identified markers for adult neurons, fetal neurons, and neuronal progenitors.
In an attempt to identify gradients of gene expression between replicating and quiescent populations, we constructed minimum spanning trees using the pairwise distances between all fetal cells. We then identified the longest path across the trees (SI Appendix, Fig. S9) and looked for genes with a high correlation across the path in both directions. We were able to identify two genes with a high correlation coefficient. The first, FAT atypical cadherin 3 (FAT3), (Fig. 4B and SI Appendix, Fig. S9) decreases from the quiescent to the replicating population, whereas early growth response 1 (EGR1) follows the opposite trend. FAT3 has been previously shown to coordinate dendrite number and neuronal orientation during development by mediating cell–cell interactions (20). EGR1 (Fig. 4B) is a transcription factor that is associated with most forms of neuronal plasticity (21) and is up-regulated in most cells of the replicating population. In addition, EGR1 has been previously identified as a candidate target of Notch signaling specifically in human radial glia (10).
HLA Expression in Human Adult and Fetal Neurons.
The central nervous system (CNS) was long believed to be immunologically inert, consisting of cells that were both postmitotic and vulnerable to activated immune cells (22). Nonetheless, over the last few years there has been accumulating evidence in mouse that the CNS is immune competent as well as interactive with the immune system (22). MHCI proteins are expressed in the adult mouse brain (23, 24) and it has been recently reported that MHCI proteins are also expressed in mouse neurons and neuronal progenitors of the prenatal brain, with a possible role in modeling brain cell synapses (25, 26). Nonetheless, neuronal expression of MHCI molecules in the human brain has only been reported in a handful of studies (27) using reagents with limited specificity.
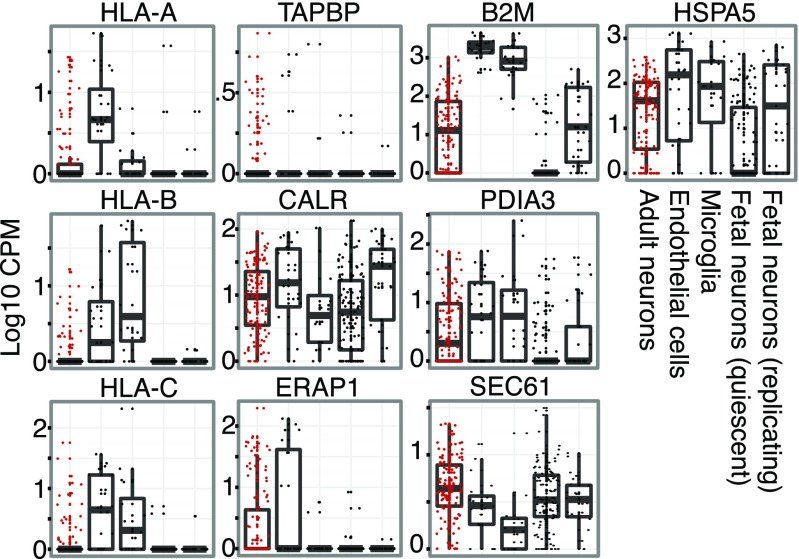
We analyzed the expression of 14 genes involved in the MHC type I pathway, major histocompatibility complex, class I, A (HLA-A), B (HLA-B), and C (HLA-C); beta-2-microglobulin (B2M); as well as tapasin (TAPBP); calreticulin (CALR); endoplasmic reticulum aminopeptidase 1 (ERAP1); heat shock 70 kDa protein 5 (HSPA5); protein disulfide isomerase family A, member 3 (PDIA3); transporter 2, ATP-binding cassette, sub-family B (TAP2); and the members of the SEC61 complex (SEC61A1, SEC61A2, SEC61B, and SEC61G), in neuronal cells of the adult brain as well as pre- and postmitotic neuronal progenitors from prenatal brain. We also included endothelial cells and microglia as positive controls for MHCI expression (Fig. 5). Our results show that MHCI genes are clearly expressed in a subset of neurons in the adult brain. Neurons do not express MHCI genes highly at a population level, but there is a significant subset of neurons that express the genes to the same level as endothelial cells and microglia. We also found that neurons expressing MHC-related genes do so in a coordinated fashion with most neurons expressing between six and eight of the investigated genes at the same time (SI Appendix, Figs. S10 and S11). Our results suggest that a subset of adult brain neurons are not immunologically inert, and demonstrate the power of single cell measurements given that the expression of these genes would likely be undetected at a population level.
Fig. 5.
Expression of selected genes of the MHCI pathway for all fetal and adult neurons, endothelial cells, and microglia. For the SEC61 complex, the average of SEC61A1, A2, B, and G is shown.
In contrast with previous findings (25) reporting widespread MHC protein expression in midgestation mouse embryos, we found that both of the human fetal cell populations lack expression of HLA-A, -B, and -C. We did observe expression of B2M in the quiescent subset of cells, which could potentially be explained by functions of B2M unrelated to the MHCI pathway. Furthermore, fetal cells lack expression of MHCI-associated genes such as TAPBP and ERAP1, whereas they express genes coding for proteins with more diverse functions such as CALR, HSPA5, PDIA3, and members of the SEC61 complex. It should be noted that the gestational age of our embryonic samples was between 16 and 18 wk and it is possible that fetal expression of MHCI in human neurons, as opposed to mice, starts at a later developmental point.
Discussion
The work presented here describes the heterogeneity of the adult human cortex as measured by single cell RNA sequencing. This technique interrogates the whole transcriptome of the smallest biologically meaningful unit, the single cell. Although we sampled cells belonging to each of the major cell types in the brain, we focused our analysis on what we identified as the neuronal population of cells to address the diversity of this heterogeneous population at single cell resolution.
In addition to demonstrating that the various cell types of the brain could be accurately resolved using unbiased analysis of whole transcriptome information, our dataset allowed us to answer several different questions. First, what types of neurons could be identified using a complete transcriptomic profile for every cell? Second, how do these relate to subtypes of neuronal cells defined by known genes? Third, what are the expression patterns of these known genes and how could they be used to identify distinct nonoverlapping populations? Fourth, what are the differences between mature postnatal neurons and neuronal progenitors of the prenatal brain? Fifth, are MHC type I genes expressed in a subpopulation of adult neurons?
We identified various cell types using a two-pronged approach: we first applied an unbiased approach using information from the whole transcriptomes of 466 cells followed by a biased comparison of these cells with a small set of highly cell-type–enriched genes derived from purified mouse brain samples. We found the two methods to be in good agreement and conclude that the unbiased approach can be used when information on expected cell-type–specific gene expression information is not available.
We then performed a subclassification of 113 adult human neurons to additional subtypes. This approach complements prior work focused on physical and morphological characteristics of neurons. Here, however, we were able to supplement traditional neuronal classifications that are based on a subset of functionally relevant genes with an entire transcriptomic dataset for each of these cells. These data allowed us to examine all of the genes that are expressed in classically defined neuronal subpopulations and to compare these to work performed in nonhuman model organisms. We observed that CCK, which in the mouse brain is used to define a small subset of neurons with certain electrophysiological characteristics, is ubiquitously expressed at the mRNA level in most human neuronal cells. We also noticed that PVALB expression is relatively limited in humans. In mice, PVALB has been previously reported to demarcate a discrete subpopulation of interneurons that accounts for 40% of all of the neocortical interneurons, whereas SST and Htr3a each account for 30%, respectively, in separate subpopulations (28). In our data, we indeed see that SST is expressed in 28% of the neurons in agreement with the existing literature. In addition, we observed a previously unreported discrepancy in the proportion of PVALB-expressing neurons between humans and mice. This finding, however, needs further validation in epileptic-free patient samples to refute the possibility that the number of PVALB-positive interneurons is reduced in patients with temporal lobe epilepsy.
Subclustering all neurons with whole transciptome data generated clear groups of excitatory and inhibitory neurons. Within the population of inhibitory interneurons, we observed widespread expression of CCK and CALB2. We found that PVALB-expressing neurons are distinct from all other interneuron subtypes and that VIP demarcates a subset of CALB2 cells. In addition, we found the genes CRHBP, LHX6, SOX6, and TAC1 to be expressed in a subset of PVALB neurons and the gene TAC3 in a subset of VIP-positive cells. We identified a subset of neurons most likely belonging to cortical layer I that expresses RELN and PAX6, which is typically considered a marker for neuronal progenitors but has been recently shown to be a layer I marker in mice. Finally, we show that CPLX3 is a marker for a distinct subpopulation of NeuN-positive cells and is usually coexpressed with SPARC. Our findings demonstrate that transcriptome-based classifications are consistent with previous classifications based on morphological and electrophysiological data and will likely continue to be refined as more data become available.
We also investigated the expression of MHCI genes that have been the subject of some controversy. These genes have been implicated to play a role in the complexity of neuronal connectivity, and we found that they are indeed expressed in a subpopulation of adult neurons. Furthermore, we notice that fetal neurons lack expression of MHCI-related genes at the gestational stage we investigated, in contrast to what has been previously shown in mouse embryos. This discrepancy suggests a later onset of MHCI expression during human brain development.
The work presented here demonstrates the applicability of single cell RNAseq to the analysis of cellular heterogeneity of the brain, one of the most complex tissues of the human body. These results lay the groundwork for the construction of a cellular map of the human brain that can be completed through the analysis of a larger number of cells from different anatomical regions of the brain. Such a map will help us identify specific markers for neuronal, glial, and vascular subtypes that we can link with information on location, connectivity, and electrophysiological properties in an attempt to fully elucidate the cellular complexity of the human brain.
Materials and Methods
All methods and relevant materials are discussed in detail in SI Appendix, SI Methods. The single cell analysis pipeline consisted of the following steps. Brain tissue was dissociated and single cell suspensions were loaded on a medium-sized C1 Single-Cell Auto Prep Array for mRNA Seq available (Fluidigm). cDNA was converted into sequencing libraries using a Nextera XT DNA Sample Preparation Kit (Illumina). Raw sequencing reads were aligned using STAR and per gene counts were calculated using HTSEQ. Gene counts were further analyzed using R. Patients were consented for the acquisition of specimens through a process approved by the Stanford Hospital Institutional Review Board.
Supplementary Material
Acknowledgments
The authors thank Norma Neff and Gary Mantalas for assistance with sequencing and Carla Shatz and Winston Koh for helpful discussions. This study was supported by National Institutes of Health Grants R01NS081703 and R01MH099555 (to B.A.B.), U01-HL099999 and U01-HL099995 (to S.R.Q.), and California Institute for Regenerative Medicine Grant GC1R-06673, Center of Excellence for Stem Cell Genomics. Y.Z. was supported by the Walter V. and Idun Berry Fellowship. Y.Z. was supported by NIH Grant K99NS089780. S.A.S. was supported by NIH Grants T32GM007365 and F30MH106261, and a Bio-X Bowes Fellowship from Stanford University. S.D. was supported by a fellowship from Svenska Sällskapet för Medicinsk Forskning.
Footnotes
S.R.Q. is a founder of and consultant for Fluidigm.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE67835).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507125112/-/DCSupplemental.
References
- 1.Ke R, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10(9):857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 2.Lee JH, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10(3):442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297(5582):836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 4.Park HY, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343(6169):422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capodieci P, et al. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2(9):663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunskill EW, et al. Single cell dissection of early kidney development: Multilineage priming. Development. 2014;141(15):3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 8.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 10.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32(10):1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Marinovich A, Barres BA. Purification and characterization of adult oligodendrocyte precursor cells from the rat optic nerve. J Neurosci. 1998;18(12):4627–4636. doi: 10.1523/JNEUROSCI.18-12-04627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodama T, et al. Neuronal classification and marker gene identification via single-cell expression profiling of brainstem vestibular neurons subserving cerebellar learning. J Neurosci. 2012;32(23):7819–7831. doi: 10.1523/JNEUROSCI.0543-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista-Brito R, et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63(4):466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sultan KT, Brown KN, Shi S-H. Production and organization of neocortical interneurons. Front Cell Neurosci. 2013;7:221. doi: 10.3389/fncel.2013.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sloviter RS, Sollas AL, Barbaro NM, Laxer KD. Calcium-binding protein (calbindin-D28K) and parvalbumin immunocytochemistry in the normal and epileptic human hippocampus. J Comp Neurol. 1991;308(3):381–396. doi: 10.1002/cne.903080306. [DOI] [PubMed] [Google Scholar]
- 18.Wittner L, et al. Preservation of perisomatic inhibitory input of granule cells in the epileptic human dentate gyrus. Neuroscience. 2001;108(4):587–600. doi: 10.1016/s0306-4522(01)00446-8. [DOI] [PubMed] [Google Scholar]
- 19.Zeng H, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149(2):483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deans MR, et al. Control of neuronal morphology by the atypical cadherin Fat3. Neuron. 2011;71(5):820–832. doi: 10.1016/j.neuron.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1) J Neurosci. 2006;26(5):1624–1634. doi: 10.1523/JNEUROSCI.4199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: Hiding in plain sight. Immunol Rev. 2006;213:48–65. doi: 10.1111/j.1600-065X.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci USA. 2007;104(16):6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21(3):505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 25.Chacon MA, Boulanger LM. MHC class I protein is expressed by neurons and neural progenitors in mid-gestation mouse brain. Mol Cell Neurosci. 2013;52:117–127. doi: 10.1016/j.mcn.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Shatz CJ. MHC class I: An unexpected role in neuronal plasticity. Neuron. 2009;64(1):40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cebrián C, Loike JD, Sulzer D. Neuronal MHC-I expression and its implications in synaptic function, axonal regeneration and Parkinson’s and other brain diseases. Front Neuroanat. 2014;8:114. doi: 10.3389/fnana.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71(1):45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.