Abstract
Proliferation-differentiation balance of epithelial cells is regulated by krüppel-like factors (KLF) 4 and 5, and the unbalanced expression relates to carcinoma progression. However, little is known about the expression and role in oral carcinomas. This study examined expression of KLF4 and KLF 5 in the carcinomas by immunohistochemistry (n = 67) and the involvement in proliferation and differentiation of carcinoma cells. KLF4 was detected in keratinizing carcinoma cells and KLF5 in non-keratinizing cells. KLF4 staining declined in the patient with lymph node metastasis (P < 0.05) and in parallel with the histological dedifferentiation (P = 0.09). Exogenous overexpression of KLF4 arranged cells in a cobble-like structure with desmosomes and KLF5 elongated cells like fibroblasts without desmosomes. KLF4 suppressed fibronectin expression, and KLF5 down-regulated and degraded E-cadherin. The proliferation was not affected by KLFs. Thus, down-regulation of KLF4 and up-regulation of KLF5 may stimulate oral carcinoma progression through the dedifferentiation of carcinoma cells.
Keywords: Differentiation, KLF, oral carcinoma, progression
Introduction
Over 50% of patients with oral squamous cell carcinomas (OSCCs) experience a relapse [1]. Phenotypic alterations of carcinoma cells progress carcinomas to the more advanced states [2], and the inhibition of differentiation and sustained proliferation are the most prominent features of the aggressive subset [3,4]. Carcinoma cells loose characteristics of epithelial cells and dedifferentiate into mesenchymal cell-like cells especially at the invasive front of carcinoma cells in the patients with worst prognosis [5-7].
Krüppel-like factors (KLFs) are developmentally regulated transcription factors that bind to GC-rich elements in the promoter of target genes and consist of 17 members in human. In contrast to the ubiquitous expression such as KLF6/10/11, KLF4 and KLF5 are preferentially expressed in epithelial cells. KLF4 is expressed in post-mitotic differentiating epithelial cells and KLF5 in less-differentiated proliferating epithelial cells, indicating that they coordinately regulate epithelial development and homeostasis in an opposite fashion [8]. Previous studies documented their aberrant expression contributes to progression of adenocarcinomas and transitional cell carcinomas, but it is obscure in squamous cell carcinomas [9,10]. Although KLF4 and KLF5 are strongly expressed in embryonic stem cells, genes regulated by KLF5 in embryonic stem cells and primary cultured keratinocytes are largely different [11], indicating the divergent role in different cell-types. In this study, we examined the expression in OSCCs and the role in carcinoma cell phenotypes focusing on proliferation and differentiation.
Materials and methods
OSCC cells and tissues
OSCC cell lines (Ca9.22, Ho1-u-1, HOC313, HSC2, HSC3, KOSC2, OSC10, SCCKN and TSU) and a normal keratinocyte cell line, HaCaT [12], were maintained in 10% fetal bovine serum-containing medium. OSCC tissues excised at Kanazawa University Hospital were used (n = 67). Mean age of the patients was 63.5 yrs (37-93 yrs), and the clinicopathological data were listed in Table 1. Histologically normal epithelium far distant from carcinomas was used as the normal control (n = 3). All tissues were obtained with the written consent of the patient and with approval by institutional review boards of Kanazawa University and Nippon Dental University.
Table 1.
KLF immunostaining score and clinicopathological parameters (n = 67)
| Category | Subcategory (n) | KLF4 | KLF5 | ||
|---|---|---|---|---|---|
|
| |||||
| Mean ± SD | P † | Mean ± SD | P † | ||
| Age | < 65 yrs (38) | 4.55 ± 3.89 | 0.07 | 3.84 ± 3.18 | 0.87 |
| ≥ 65 yrs (29) | 2.79 ± 2.46 | 3.10 ± 2.88 | |||
| Gender | Female (27) | 4.75 ± 4.10 | < 0.05 | 3.40 ± 2.46 | 0.44 |
| Male (40) | 3.13 ± 2.69 | 3.89 ± 3.53 | |||
| T stage* | T1 (13) | 6.46 ± 3.84 | 0.13 | 4.56 ± 3.43 | 0.83 |
| T2 (33) | 2.97 ± 3.30 | 3.77 ± 3.34 | |||
| T3 (7) | 2.29 ± 1.98 | 1.50 ± 1.52 | |||
| T4 (14) | 4.07 ± 2.59 | 4.00 ± 2.79 | |||
| N stage* | N0 (39) | 4.13 ± 4.69 | < 0.05 | 3.15 ± 3.15 | 0.81 |
| N1 (16) | 3.56 ± 3.37 | 4.86 ± 2.85 | |||
| N2 (11) | 3.18 ± 2.44 | 2.71 ± 2.87 | |||
| N3 (1) | 1.00 | 8.00 | |||
| Clinical stage* | Stage 1 (13) | 6.23 ± 3.77 | 0.12 | 4.56 ± 3.43 | 0.74 |
| Stage 2 (23) | 2.71 ± 3.10 | 2.93 ± 3.24 | |||
| Stage 3 (12) | 4.00 ± 3.67 | 4.27 ± 3.07 | |||
| Stage 4 (19) | 3.37 ± 2.69 | 3.46 ± 2.86 | |||
| Differentiation | Well (29) | 4.93 ± 3.35 | 0.09 | 3.95 ± 3.33 | 0.66 |
| Moderate (23) | 3.40 ± 3.72 | 4.13 ± 3.40 | |||
| Poor (15) | 1.67 ± 1.67 | 3.25 ± 2.86 | |||
Patients were categorized by tumor size (T stage) and clinical stage according to the UICC WHO grading system and by the stage of lymph node metastasis (N stage).
Pearson’s chi-square test.
Immunostaining
Unstained tissue sections microwaved in 0.01 M sodium citrate buffer, pH 6.0, were incubated with primary antibodies against KLF4 (R&D Biosystems, Minneapolis, USA) and KLF5 (Sigma-Aldrich, St. Louis, USA). The nuclear immunostaining was evaluated as previously described [13]: percentage of staining was scored by on a scale of 0-4; 0, totally negative; 1, < 10%; 2, ≥ 10- < 40%; 3, ≥ 40- < 60%; and 4, ≥ 60%. Positive staining was classified as: 1, weak; 2, moderate; and 3, strong at the area of strongest staining due to the variable staining intensity. The staining score was calculated by multiplying the percentage and intensity scores to give a value of between 0-12.
Real-time PCR
Total RNA extracted from cell lines was reverse-transcribed into cDNA and subjected to real-time PCR using StepOne Real-time PCR system and TaqMan probes for KLF4 (Hs00358836_m1) or KLF5 (Hs00156145_m1; Applied Biosystems, Foster City, USA). Expression levels were normalized against ACTB (TaqMan Endogenous Control Human ACTB, Applied Biosystems). The expression levels were calculated by the standard curve method (2-∆∆Ct), and the relative fold of expression compared with that in HaCaT cells was determined.
Immunoblot
After transfection of HA-tagged KLF4 and KLF5 cDNA or the siRNA (50 nM; KLF4, #s17793; KLF5, #s2116; negative control, Silencer Negative Control #1; Ambion, Austin, USA) for 48 h, cells lysed in a buffer containing protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) were applied for the immunoblot using antibodies to KLF4, E-cadherin (R&D Biosystems), KLF5, cyclin D1, p21Cip1/Waf1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), HA (Cell Signaling Technology, Danvers, USA), fibronectin, involucrin and β-actin (Sigma-Aldrich).
Electron microscopy
Cells transfected with KLF cDNA or vector alone were cultured for 48 h and fixed in 2.5% glutaraldehyde and 1% osmium tetroxide, and embedded in Epon. Ultra-thin sections stained with uranyl acetate and lead citrate were observed using the transmission electron microscope (H-7500, 80 kV; Hitachi High-Tech, Tokyo, Japan).
Statistical analysis
Correlation of KLF staining score and clinicopathological parameters of carcinomas was analyzed by Pearson’s chi-square.
Results
Expression of KLFs
In normal oral epithelium, KLF4 was localized in the nucleus of suprabasal cells and KLF5 preferentially in the basal cells (Figure 1). KLF4 staining intensity was increased in the epithelium near carcinoma cells. Although the intensity and percentage of the staining was variable, 91.2% and 87.5% of the cases were positively stained for KLF4 and KLF5, respectively. KLF4 was primarily detected in carcinoma cells near the center of the carcinoma cell nests that form keratin pearls, and rapidly declined at the periphery of the nests. KLF5 was detected in the carcinoma cells at the periphery and decreased near the center. In non-keratinized carcinoma cell nests, KLF5 localized in carcinoma cells throughout the nests, and KLF4 was minimally stained (data not shown). The cytoplasmic staining was negligible.
Figure 1.
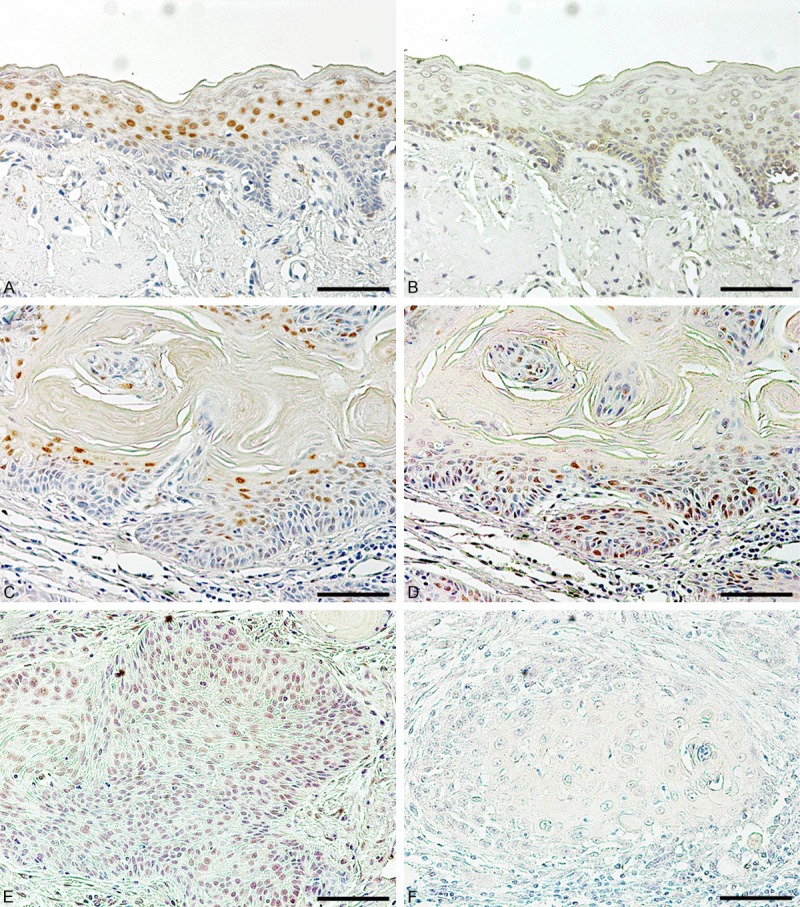
Immunohistochemical staining of KLF4 and KLF5 in oral epithelium and carcinomas. Expression of KLF4 (A, C) and KLF5 (B, D, E) in normal oral epithelium adjacent to carcinoma cells (A, B) and in oral carcinoma tissues with (C, D) or without keratin pearls (E) was examined by immunostaining. Non-immune IgG instead of primary antibody was used as a negative control (F). Bar = 40 µm.
Implications in carcinoma pathology
KLF staining scores with respect to the pathological parameters were summarized in Table 1. KLF4 staining was significantly decreased in the tumors of advanced N-stage, suggesting an association with the patient prognosis. The patient survival in the positive group (staining scores 6-12, n = 20) and the negative group (staining scores 0 and 1, n = 26) excluding patients who died of other diseases (n = 7) was analyzed. Although the negative group lowered the long-term survival (P < 0.05, log-rank test), multivariate hazard analysis showed no significant difference (P = 0.40; risk ratio, -0.366; 95% confidence interval, -1.246- + 0.482), suggesting an association of KLF4 staining with other parameters. The staining score tended to decline with the differentiation, and both the percentage and intensity of the staining decreased with dedifferentiation (percentage, P = 0.07; intensity, P < 0.05). The differentiation state (well- vs. moderately-/poorly-differentiated carcinomas) was an independent prognostic determinant (P < 0.005; risk ratio, 0.652; 95% confidence interval, 0.225-1.117). The role of the significant decrease in males was unknown since the gender equally distributed in each parameter (data not shown). KLF5 staining did not associate with any parameters (Table 1).
Involvement in carcinoma differentiation
Differentiation of epithelial cells is intertwined with the cell shape. Differentiated squamous epithelial cells flatten cell bodies and form desmosome-mediated cobble-like structures, and the dedifferentiated cells elongate like fibroblasts without desmosome. Therefore we examined the cell shape after the transfection. KLF4 flattened almost all carcinoma cells at a single cell level and arranged in a cobble-like structure at the high-density area, and KLF5 elongated cell bodies with a front-rear polarity even in the high-density area (Figure 2). In electron micrograms KLF4-trasfected cells displayed well-developed desmosomes in contrast to no apparent cell-cell adhesion structure in KLF5-transfected cells (Figure 3).
Figure 2.
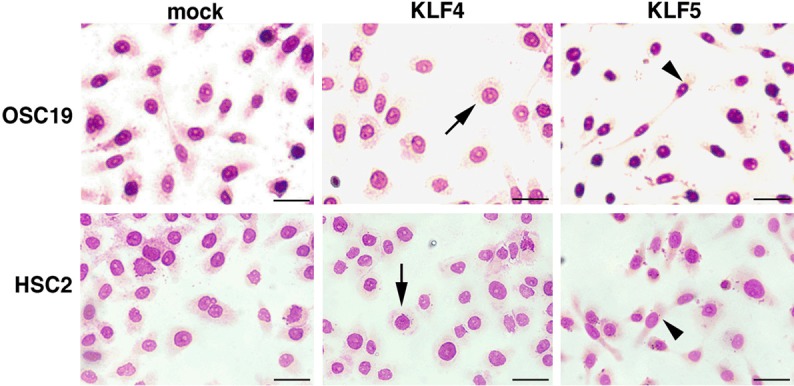
Morphological alterations of KLF-transfected cells. Morphological changes of carcinoma cells transfected vector alone (mock), KLF4 or KLF5 cDNA. Cells were visualized by Diff-Quik staining. KLF4 flattened cells at a single cell level (arrows) and arranged in a cobble-like structure at the high-density area, and KLF5 elongated cells with a front-rear polarity (arrow heads). Bar = 20 µm.
Figure 3.
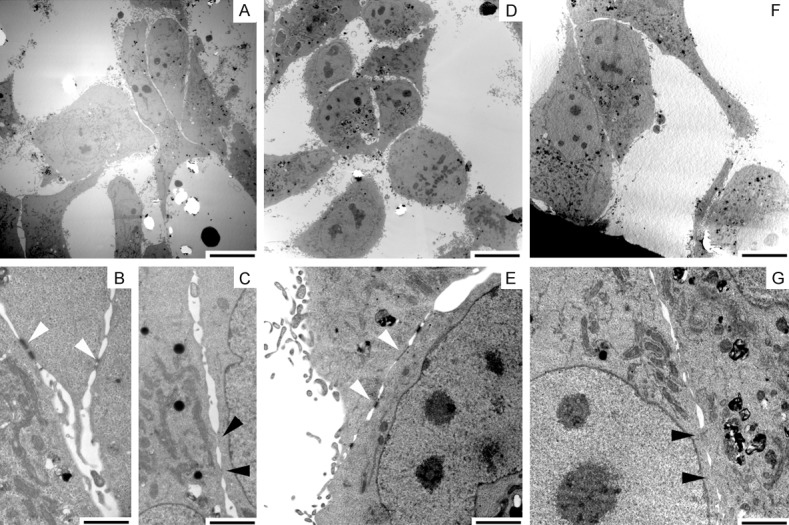
Ultrastructure of KLF-transfected cells. OSC19 cells transfected with KLF4 cDNA (D, E), KLF5 cDNA (F, G) or vector alone (A-C) were observed by transmission electron microscope. KLF4-trasfected cells developed desmosomes (open arrowheads), and KLF5-transfected cells had no apparent cell-cell adhesion structures (closed arrowheads). Cells transfected vector contacted with and without desmosomes. Bars = 10 µm (A, D, F) and 2 µm (B, C, E, G).
As a preliminary examination at the protein level, the endogenous KLF expression was quantified at the mRNA level (Figure 4A). Compared to normal keratinocytes HaCaT cells, KLF4 expression was comparable in SCCKN cells and Ca9.22 cells, but very small or negligible in other cell lines. KLF5 was expressed 2.29-fold higher in HSC2 cells and lower in other cell lines. We then transfected the cDNA into cells expressing KLFs at various levels (HSC3, Ho1-u-1, TSU, and OSC19 cells). After 48 h transfection, E-cadherin was down-regulated and/or reduced in size to 100 kDa in KLF5-transfected cells (Figure 4B), and KLF5 siRNA did not change the expression of intact E-cadherin (Figure 4C). Involucrin was not affected by KLF5 cDNA, but increased by the siRNA, and fibronectin was not affected. KLF4 cDNA apparently reduced fibronectin expression but did not affect expression of the differentiation markers. KLF4 siRNA increased fibronectin slightly.
Figure 4.
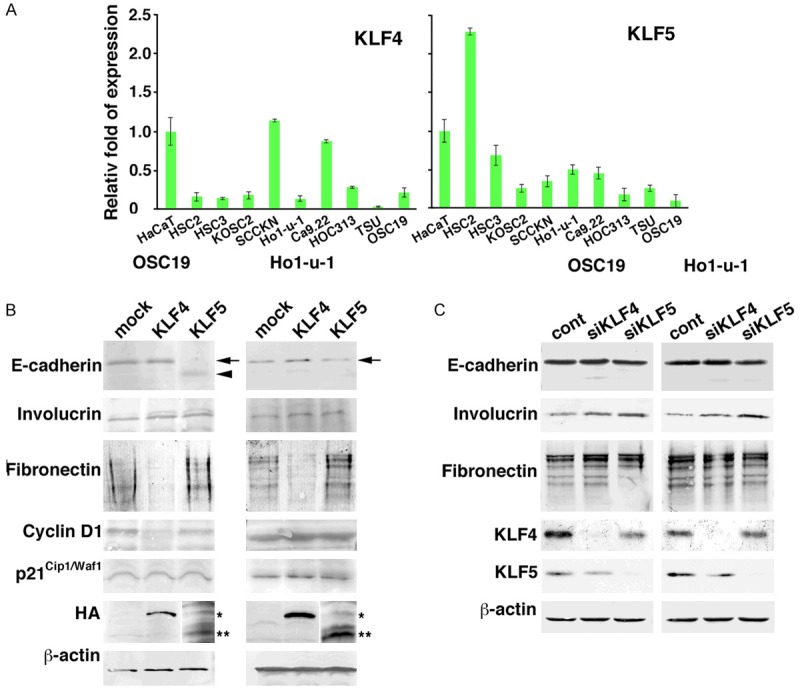
Phenotypic alterations of carcinoma cells by KLFs. A: Quantitative analysis of KLF4 and KLF5 mRNA in oral carcinoma cell lines and normal keratinocytes (HaCaT). The relative fold of expression against HaCaT cells was depicted (n = 4). Bars = mean ± S.D. B: Expression of protein involved in the differentiation (E-cadherin, Involucrin), dedifferentiation (Fibronectin) and proliferation (Cyclin D1, p21Cip1/Waf1). KLF4 (*) and KLF5 (**) expression were probed by anti-HA antibody. C: Expression of E-cadherin, involucrin and fibronectin upon transfection of siRNA against KLF4 (siKLF4) and KLF5 (siKLF5). Endogenous KLF4 and KLF5 expression was probed with anti-KLF4 and -KLF5 antibodies. b-actin was used as an internal control.
KLFs and proliferation
Cyclin D1 was down-regulated by KLF4 in OSC19 cells but not in other cell lines (Figure 4B). Expression of a proliferation negative regulator, p21Cip1/Waf1, was not affected by KLFs in any of the carcinoma cell lines examined. To analyze the involvement of KLFs in proliferation more directly, a proliferation assay was performed. Excluding OSC19 cells, carcinoma cells were not affected by KLF4 and KLF5 (Figure 5).
Figure 5.
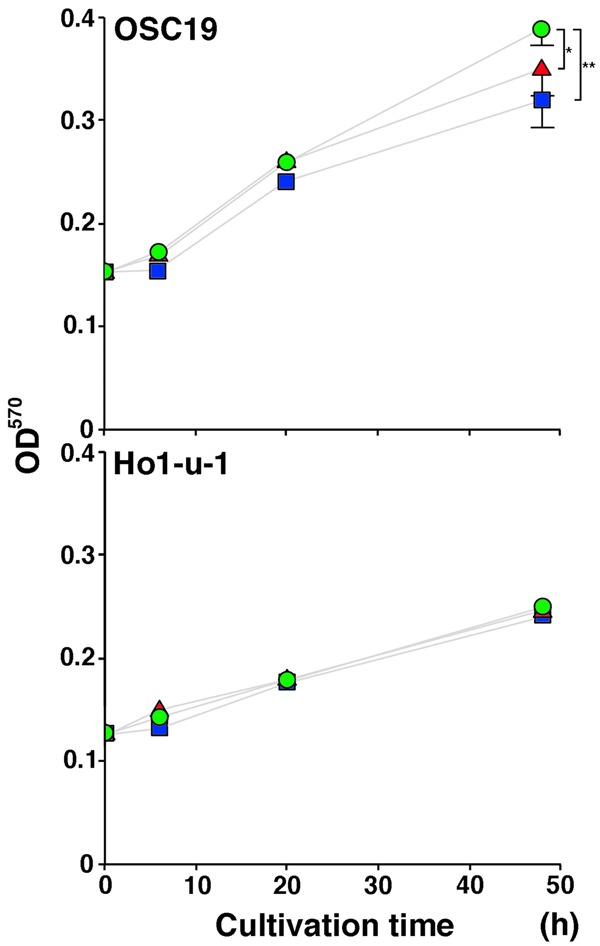
Carcinoma cell proliferation by KLF transfection. Proliferation of carcinoma cells was monitored by the MTT assay after transfection of vector alone (circle), KLF4 (square) or KLF5 (triangle). *P < .05, **P < .01 (t-test).
Discussion
Carcinomas frequently unbalance its proliferation and differentiation [8], and aggressive OSCCs inactivate expression of mucosa-associated lymphoid tissue 1 that differentially regulates KLF4 and KLF5 [7,14], suggesting an impact of the KLFs on OSCC progression. It prompts us to examine the expression in OSCCs and its involvement in the proliferation and differentiation.
KLF4 and KLF5 differentially localized in normal epithelium covering the oral cavity, KLF4 was found in the suprabasal cells and KLF5 in the basal cells. It confirmed the localization in the skin [8]. The basal cells maintains a less-differentiated state and supply newly divided cells into the suprabasal cell layer undergoing a terminal differentiation program, and the terminally differentiating cells cannot re-enter the proliferation state [15]. Carcinoma cells basically retain the characteristics of epithelial cells, and keratinization is a hallmark of this differentiation. Irregular masses composed of carcinoma cell nests invade into the underlying tissues, and keratinization takes place in the form of keratin pearls that are typically observed at the center of the nests. Thus, carcinoma cells near the center exhibit a more differentiated phenotype than the cells at the periphery [16].
KLF4 was predominantly stained in the keratinizing carcinoma cells in agreement with a previous study [10]. As the staining score was decreased with histological dedifferentiation of OSCCs, KLF4-transfected carcinoma cells flattened and aligned in a cobble-like structure with well-developed desmosomes. These morphological changes support an association of KLF4 with the differentiation. However, at the protein level, KLF4 did not affect the expression of differentiation markers in contrast to previous studies [17,18]. Instead, it repressed fibronectin expression that are regulated by KLF17 binding on the gene promoter [19] and increased in an aggressive subset of carcinomas [20,21]. These data indicate that KLF4 suppresses dedifferentiation of the carcinoma cells rather than the initiation of differentiation.
In contrast, KLF5 elongated the cells with a front-rear polarity and impaired desmosome formation which are a signature of the dedifferentiation [22]. Although KLF5 did not affect fibronectin expression, it decreased E-cadherin expression and processed it into a smaller species if not all carcinoma cells. The processing into a 100 kDa species is worth noting. E-cadherin establishes epithelial cell-cell adhesion through the homophilic binding of extracellular domain, and catenins bound to the cytoplasmic domain stabilizes E-cadherin at cell surface [23]. Expression of E-cadherin and catenins closely correlates with each other and rapidly declines in less-differentiated and diffusely invasive OSCCs [24,25]. Processing into the 100 kDa species is consistent with that by calpain, and calpain cleavage of the catenin-binding site destabilizes E-cadherin-mediated cell-cell adhesion [26]. In addition, KLF5 siRNA increased involculin expression. Although further investigations are required to define an exact role of KLF5 in oral carcinoma cells, it appears likely that KLF5 negatively regulates the differentiation that eventually dedifferentiates the cells.
Previous studies documented that KLF4 and KLF5 decelerate and accelerate proliferation of carcinoma cells, respectively [27,28], and that they regulate expression of cyclin D1 and p21Cip1/Waf1 contrary [29-31]. However the current study showed that KLFs did not change the proliferation and the cell cycle regulator expression except in OSC19 cells. Additional factors may be required to regulate the proliferation as exemplified by the fact that KLF5 oppositely regulates p15INK4b expression in the presence or absence of TGF-β [32].
KLF4 stimulates its own transcription through the binding to the promoter, and KLF5 represses KLF4 expression by competing with the binding [33], indicating KLF5 as an up-stream repressor of KLF4. The present study demonstrated that KLF4 and KLF5 suppress dedifferentiation and differentiation of oral carcinoma cells, respectively. Sequential loss of KLF4 and gain of KLF5 may strongly induce phenotypic alterations of the cells with aggressive behaviors such as the epithelial-mesenchymal transition [3,4]. Since the initiation of differentiation and the inhibition of dedifferentiation are expected as a novel strategy for cancer therapy [2,34], future investigations on the regulation of KLF4 and KLF5 expression are necessary to unveil a mechanism of OSCC progression.
Acknowledgements
We would like to thank Drs. Inoue and Yuasa for generous providing of KLF cDNAs, and Drs. D’Armiento and Woode for critical reading of the manuscript. This work was supported by JSPS KAKENHI Grant Numbers 22592103, 26462859, and based on a thesis submitted to Graduate School of Life Dentistry at Tokyo, The Nippon Dental University, in partial fulfillment of the requirements for the Doctor of Dental Surgery degree.
Disclosure of conflict of interest
None.
References
- 1.Choi S, Myers JM. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 2.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Meledez-Zajga J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma M, Sah P, Sharma SS, Radhakrishnan R. Molecular changes in invasive front of oral cancer. J Oral Maxillofac Pathol. 2013;17:240–247. doi: 10.4103/0973-029X.119740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 7.Chiba T, Maeda G, Kawashiri S, Kato K, Imai K. Epigenetic loss of mucosa-associated lymphoid tissue 1 expression in patients with oral carcinomas. Cancer Res. 2009;69:7216–7223. doi: 10.1158/0008-5472.CAN-09-1140. [DOI] [PubMed] [Google Scholar]
- 8.McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abrigo M, Alvarez R, Paparella ML, Calb DE, Bal de Kier Joffe E, Raimondi AR. Impairing squamous differentiation by Klf4 deletion is sufficient to initiate tongue carcinoma development upon K-Ras activation in mice. Carcinogenesis. 2014;35:662–669. doi: 10.1093/carcin/bgt349. [DOI] [PubMed] [Google Scholar]
- 10.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, Grizzle WE, Ruppert JM. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 11.Parisi S, Cozzuto L, Tarantino C, Passaro F, Ciriello S, Aloia L, Antonini D, De Simone V, Pastore L, Russo T. Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state. BMC Biol. 2010;8:128. doi: 10.1186/1741-7007-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploidy human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohyama Y, Kawamoto Y, Chiba T, Kikuchi K, Sakashita H, Imai K. Differential expression of fatty acid-binding proteins and pathological implications in the progression of tongue carcinoma. Mol Clin Oncol. 2014;2:19–25. doi: 10.3892/mco.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohyama Y, Kawamoto Y, Chiba T, Maeda G, Sakashita H, Imai K. Inhibition of TGF-β and EGF pathway gene expression and migration of oral carcinoma cells by mucosa-associated lymphoid tissue 1. Br J Cancer. 2013;109:207–214. doi: 10.1038/bjc.2013.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandarillas A. The mysterious human epidermal cell cycle, or an oncogene-induced differentiation checkpoint. Cell Cycle. 2012;11:4507–4516. doi: 10.4161/cc.22529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lever WF, Schaumburg-Lever G. Histopathology of the Skin. 7th edition. Philadelphia: Lippincott; 1999. [Google Scholar]
- 17.Chew YC, Adhikary G, Xu W, Wilson GM, Eckert RL. Protein kinase C∂ increase Kruppel-like factor 4 protein, which drives involucirn gene transcription in differentiating keratinocytes. J Biol Chem. 2013;288:17759–17768. doi: 10.1074/jbc.M113.477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yori JL, Johnson E, Zhou G, Jain MK, Keri RA. Krüppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J Biol Chem. 2010;285:16854–16863. doi: 10.1074/jbc.M110.114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z, Han Q, Zhou N, Wang S, Lu S, Bai C, Zhao RC. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7:884–894. doi: 10.1016/j.molonc.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Garcia B, Eiró N, Marin L, Bonzález-Reyes S, González LO, Lamelas ML, Vizoso FJ. Expression and prognostic significance of fibronectin and matrix metalloproteinases in breast cancer metastasis. Histopathology. 2014;64:512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- 21.Ibbetson SJ, Pyne NT, Pollard AN, Olson MF, Samuel MS. Mechanotransduction of pathways promoting tumor progression are activated in invasive human squamous cell carcinoma. Am J Pathol. 2013;183:930–937. doi: 10.1016/j.ajpath.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Godde NJ, Galea RC, Elsum IA, Humbert PO. Cell polarity in motion: redefining mammary tissue organization through EMT and cell polarity transitions. J Mammary Gland Biol Neoplasia. 2010;15:149–168. doi: 10.1007/s10911-010-9180-2. [DOI] [PubMed] [Google Scholar]
- 23.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto T, Soeno Y, Maeda G, Taya Y, Aoba T, Nasu M, Kawashiri S, Imai K. Progression of oral squamous cell carcinoma accompanied with reduced E-cadherin expression but not cadherin switch. PLoS One. 2012;7:e47899. doi: 10.1371/journal.pone.0047899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaya K, Sudo H, Maeda G, Kawashiri S, Imai K. Concomitant loss of p120-catenin and β-catenin membrane expression and oral carcinoma progression with E-cadherin reduction. PLoS One. 2013;8:e69777. doi: 10.1371/journal.pone.0069777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rios-Doria J, Day KC, Kuefer R, Rashid MG, Chinnaiyan AM, Rubin MA, Day ML. The role of calpain in the proteolytic cleavage of E-cadherin in prostate and mammary epithelial cells. J Biol Chem. 2003;278:1372–1379. doi: 10.1074/jbc.M208772200. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Krüppel-like factor 4 (gut-enriched Krüppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng HQ, Zhou Z, Huang J, Chaudhury L, Dong JT, Chen C. Krüppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene. 2009;28:3702–3713. doi: 10.1038/onc.2009.235. [DOI] [PubMed] [Google Scholar]
- 29.Ghaleb AM, Katzu JP, Kaester KH, Du JX, Yang VW. Krüppel-like factor 4 exhibits antiapoptotic activity following-irradiation-induced DNA damage. Oncogene. 2007;26:2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He M, Han M, Zheng B, Shu YN, Wen JK. Angiotensin II stimulates KLF5 phosphorylation and its interaction with c-Jun leading to suppression of p21 expression in vascular smooth muscle cells. J Biochem. 2009;146:683–691. doi: 10.1093/jb/mvp115. [DOI] [PubMed] [Google Scholar]
- 31.Yoon F, Yang VW. Requirement of Krüppel-liker factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem. 2004;279:5035–5041. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J Biol Chem. 2009;284:6071–6078. doi: 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang WW. Opposing effects of Krüppel-like factor 4 (gut-enriched Krüppel-like factor) and Krüppel-like factor 5 (intestinal-enriched Krüppel-like factor) on the promoter of the Krüpple-like factor 4 gene. Nucleic Acid Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–21. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]


