Abstract
Objective: To describe the expression profiles of FOXA1, DUSP6, and HA117 in different portions of the colon of patients diagnosed with Hirschsprung’s disease (HSCR). Methods: Colon specimens were collected from34 HSCR patients and grouped into 3 segments: proximal anastomosis, dilated segment and stenotic segment. Levels of FOXA1, DUSP6, and HA117 RNA were evaluated by real-time PCR. Levels of FOXA1 and DUSP6 protein were analyzed by immunohistochemistry and Western blotting. Results: The levels of FOXA1 and DUSP6 RNA were significantly lower in the stenotic segment compared to proximal anastomosis (P < 0.05). The level of HA117 RNA was significantly higher in the stenotic segment compared to proximal anastomosis (P < 0.05). In proximal anastomosis, FOXA1 and DUSP6 were both expressed at the protein level in ganglion cells and nerve fibers between the circular and longitudinal muscles. In the stenotic segments, positive staining for FOXA1 and DUSP6 was diminished. The levels of FOXA1 and DUSP6 protein were significantly lower in the stenotic segment compared to proximal anastomosis (P < 0.05). Conclusion: Suppression of the FOXA1/DUSP6 signaling pathway may contribute to the development of HSCR. LncRNA HA117 may have an anti-differentiation function, and play a pivotal role in the progression of HSCR.
Keywords: Hirschsprung’s disease, FOXA1, DUSP6, HA117
Introduction
Hirschsprung’s disease (HSCR) is a common congenital gastrointestinal malformation, with an incidence of 1 in 5,000 live births [1]. HSCR is characterized by the absence of parasympathetic intramural ganglion cells in the distal gastrointestinal tract. The enteric nervous system (ENS) is a network of neurons and glia within the wall of the bowel that controls essential functions of the gastrointestinal tract [2]. The ENS is derived from the neural crest (NC). HSCR occurs when enteric NC-derived cells (ENCDCS) fail to colonize the distal bowel [3].
A large number of chromosomal anomalies have been described in association with HSCR. In particular, molecular analyses have detected RET, endothelin receptor B (EDNRB), glial cell line-derived neurotrophic factor (GDNF), and endothelin 3 (EDN3) mutations in HSCR patients. Loss of function mutations in RET account for up to 50% of familial and 15-20% of sporadic HSCR cases [4,5]. EDNRB mutations may account for 5-15% of HSCR cases. Mutations in GDNF and EDN3 have also been described. Importantly, all these genes are essential for ENS survival, migration, differentiation, and growth [4,7,8].
Recent studies suggest that mutations in individual genes are not sufficient to cause HSCR. Rather, current opinion proposes that genes synergistically cooperate in generating a HSCR phenotype [4]. Additional genes that may be responsible for the development of HSCR include forkhead box A1 (FOXA1), dual-specificity phosphatase 6 (DUSP6), and HA117. FOXA1 is considered a critical transcription factor that can promote the survival, migration, differentiation, and growth of cells, tissues, and organs [9]. DUSP6 is a differentiation gene that plays a major role in embryonic development [10]. HA117 is an all-trans retinoic acid (ATRA)-induced long noncoding RNA (Lnc RNA) that may have an anti-differentiation or anti-maturation role in the genesis of HSCR [11]. To date, the expression and potential roles of FOXA1, DUSP6, and HA117 in HSCR have not been characterized.
The objective of this study was to describe the expression profiles of FOXA1, DUSP6, and HA117 in different portions of the HSCR colon. This may provide further insight into the etiology of HSCR.
Materials and methods
Study design
Surgically resected colon tissues were obtained intra- and post-operatively from 34 randomly selected HSCR patients (23 males and 11 females; age 21 days to 5 years) at the Children’s Hospital of Chongqing Medical University between June 2012 and April 2014. According to the resection location and size of the bowel, colon tissues were divided into 3 groups: 1) proximal anastomosis (proximal anastomosis of resected colon), corresponding to the normal segment (controls); 2) dilated segment, corresponding to the hypoganglionic segment; and 3) stenotic segment, corresponding to aganglionic segment of the resected colon [11]. All patients were recruited after obtaining written informed consent from their parents. The study was approved by the Ethics Committee at the Children’s Hospital of Chongqing Medical University.
Resected colon tissues were immediately frozen in liquid nitrogen and stored at -80°C until use for quantitative real time-polymerase chain reaction (Q-PCR), immunohistochemical staining, and western blotting analyses.
Quantitative real time-PCR analysis
Total RNA was isolated from tissues using TRIzol reagent according to the manufacturer’s instructions (Life Technologies, Grand Island, NY, USA). Total RNA was extracted with chloroform, precipitated with isopropyl alcohol, and dissolved in diethyl pyrocarbonate (DEPC)-treated water. RNA was reverse transcribed to cDNA using the PrimeScript™ RT reagent Kit according to the manufacturer’s instructions (RR037A, Takara, Otsu, Japan). The levels of FOXA1, DUSP6, HA117 and β-actin were detected using the CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, USA) with SYBR Premix Ex Taq™ II (Takara, Otsu, Japan). Primer sequences (Beijing Genomics Institute, China) were: FOXA1, forward: 5’-AAGGGCATGAAACCAGCGAC-3’, reverse: 5’-GCCTGAGTTCATGTTGCTGAC-3’; DUSP6, forward: 5’-GCAGCGACTGGAACGAGAATA-3’, reverse: 5’-GGAGAACTCGGCTTGGAACTT-3’; HA117, forward: 5’-CAGAGTCAGGGACTTCAGCCTTAT-3’, reverse: 5’-CTGTTTCCTTCTCACTCCCAACCA-3’; β-actin, forward: 5’-AGACCTGTACGCCAACACAG-3’, reverse: 5’-GTACTTGCGCTCAGGAGGAG-3’. cDNA amplification was performed at: 95°C for 30 sec, 95°C for 5 sec, and 59°C for 30 sec, followed by 40 cycles at 60°C for 30 sec, and 95°C for 15 sec.
Immunohistochemical staining
Tissues were fixed in 10% formaldehyde and embedded in paraffin. Consecutive 5 μm sections were cut with a microtome. Sections were dewaxed, rehydrated, and washed in PBS for 5 min. After antigen retrieval (citrate buffer, pH 6.0; 95°C; 20 min), slides were allowed to cool for 30 min at room temperature (RT) and were washed again in PBS for 5 min. Quenching of endogenous peroxidase activity was performed in 3% H2O2 in PBS. Nonspecific sites were blocked by incubating slides with 10% goat serum/PBS for 20 min. Calretinin (CR), a crucial and hypersensitive protein in the development of the ENS, served as a positive control [12]. Incubation with primary antibodies (rabbit anti-FOXA1, monoclonal, 1:100 Abcom; rabbit anti-DUSP6, monoclonal, 1:100 Abcom; rat anti-calretinin, monoclonal, 1:100 Santa) was carried out in a humid chamber overnight at 4°C. Horseradish peroxidase (HRP) labeled secondary antibody included in the MaxVision™ HRP-Polymer anti- mouse/rabbit IHC kit (MaxVision, China) was applied for 30 minutes at RT, followed by 1-2 minutes incubation with DAB (Zsbio, China) for color development. Slides were counterstained with hematoxylin (Harleco®), washed, and dehydrated with alcohol and xylene. PBS was used instead of the primary or secondary antibodies as the negative control. The appearance of tan/brown granules in the tissues was considered a positive result.
Western blot analysis
Total protein was extracted from pulverized samples by ultrasonication. CR represented a positive control. β-actin was considered the loading control. 100 µg total protein was subjected to SDS gel electrophoresis (10% polyacrylamide). Proteins were blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA), nonspecific binding sites were blocked with 1% skimmed milk diluted with TBS for 2 h at room temperature (RT), and membranes were incubated with primary antibodies (rabbit anti-FOXA1, monoclonal, 1:1000 Abcom; rabbit anti-DUSP6, monoclonal, 1:1000 Abcom; rat anti-Calretinin, monoclonal, 1:500 Santa; rabbit anti-β-actin, monoclonal, 1:3000 Abcom) at 4°C overnight. The next day, membranes were washed four times with TBST for 10 min and incubated with a secondary antibody (1:2000) for 90 min at RT. Membranes were washed and a developing reagent (Bio-Rad, USA) was added for band detection.
Statistical analysis
GraphPad Prism 5 (GraphPad Software, Inc, USA) was used for statistical analysis. Data are presented as means ± SEM. Levels of FOXA1, DUSP6, and HA117 RNA and protein in anastomotic, dilated, and stenotic segments were compared using one-way analysis of variance (ANOVA). Differences between two groups were assessed by Student’s t test. P < 0.05 was considered statistically significant.
Results
Levels of FOXA1, DUSP6 and HA117 RNA
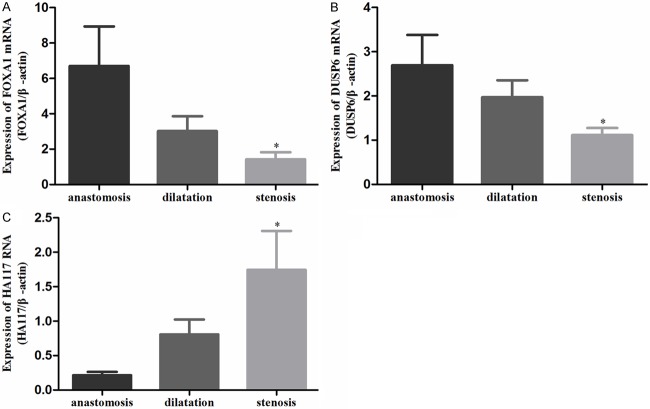
In proximal anastomosis, the dilated segment, and the stenotic segment of resected HSCR colons, the levels of FOXA1 RNA were 6.703 ± 2.235, 3.017 ± 0.848, and 1.432 ± 0.396 (Figure 1A); the levels of DUSP6 RNA were 2.694 ± 0.683, 1.971 ± 0.385, and 1.113 ± 0.162 (Figure 1B); and the levels of HA117 RNA were 0.214 ± 0.049, 0.807 ± 0.217, and 1.744 ± 0.565 (Figure 1C), respectively. The levels of FOXA1 and DUSP6 RNA were significantly lower in the stenotic segment compared to proximal anastomosis (P < 0.05). The level of HA117 RNA was significantly higher in the stenotic segment compared to proximal anastomosis (P < 0.05).
Figure 1.
FOXA1 RNA (A), DUSP6 RNA (B), and HA117 RNA (C) in different segments of HSRC detected by real-time PCR.
Immunohistochemical staining
In proximal anastomosis, FOXA1, DUSP6, and CR protein were positively identified in ganglion cells and nerve fibers between circular and longitudinal muscle (Figure 2). In stenotic segments, positive staining for FOXA1, DUSP6, and CR were diminished. No positive staining was observed in the blank controls.
Figure 2.
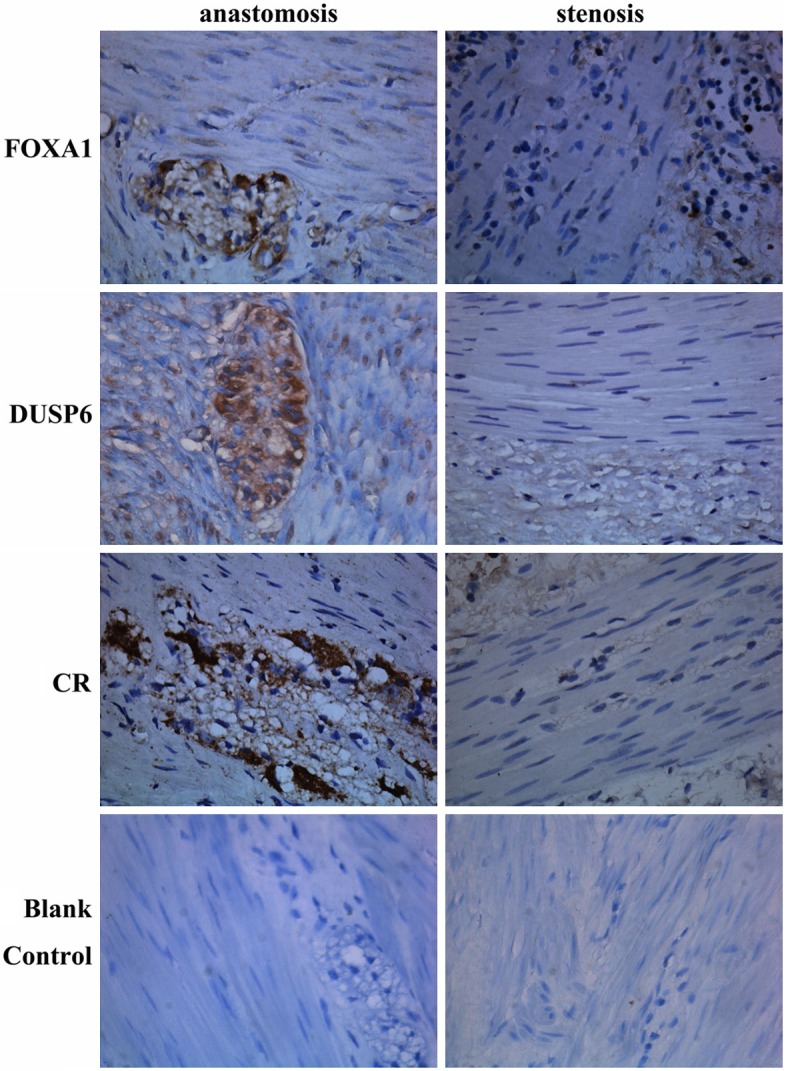
FOXA1, DUSP6, and CR protein expression in different segments of HCSR by immunohistochemical staining (400×).
Levels of FOXA1 and DUSP6 protein expression
In proximal anastomosis, the dilated segment, and the stenotic segment of resected HSCR colons, the levels of FOXA1 protein were 1.416 ± 0.153, 0.830 ± 0.060, and 0.821 ± 0.072 (Figure 3A, 3B); the levels of DUSP6 protein were 1.187 ± 0.090, 1.067 ± 0.113, and 0.809 ± 0.040 (Figure 3A, 3C), and the levels of CR protein were 1.266 ± 0.120, 0.887 ± 0.085, and 0.897 ± 0.051 (Figure 3A, 3D), respectively. The level of FOXA1 protein was significantly lower in the dilated and stenotic segments compared to proximal anastomosis (P < 0.05). The levels of DUSP6 and CR protein were significantly lower in the stenotic segment compared to proximal anastomosis (P < 0.05).
Figure 3.
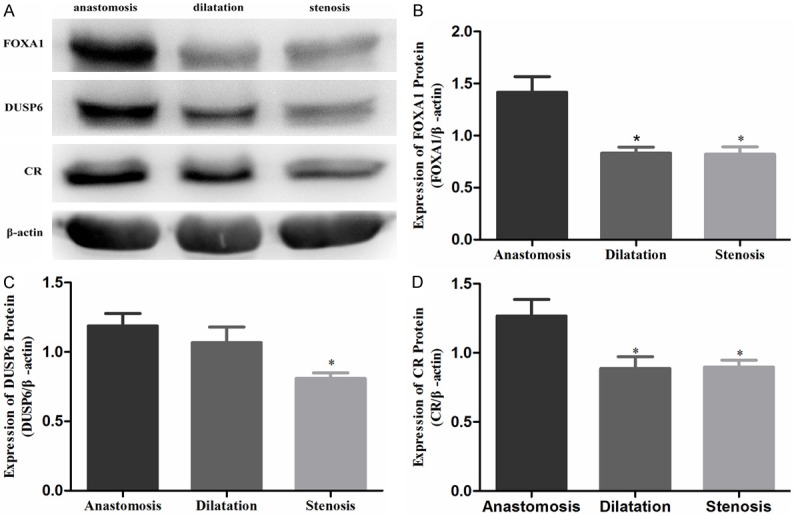
FOXA1 (A, B), DUSP6 (A, C), and CR (A, D) protein in different segments of HSRC detected by Western blotting.
Discussion
The present study investigated the RNA and protein expression profiles of FOXA1, DUSP6, and HA117 in proximal anastomosis, dilated, and stenotic colonic segments from HSCR patients. FOXA1 and DUSP6 protein were localized to the ganglion cells and nerve fibers in proximal anastomosis. Levels of FOXA1 and DUSP6 RNA and protein were significantly decreased in stenotic segments compared to proximal anastomosis. These data suggest that FOXA1 and DUSP6 may act synergistically to promote development of the ENS, and that suppression of FOXA1 and DUSP6 expression may contribute to ganglion cell hypogenesis. Conversely, the level of HA117 RNA was significantly increased in stenotic segments compared to proximal anastomosis. This observation confirms the results of a recent study that used real time-PCR, fluorescence in situ hybridization, and immunohistochemistry to show that HA117 expression was higher in the stenotic segments compared to the dilated segments and proximal anastomosis in colons from HSCR patients [11].
HSCR is based on a defect of craniocaudal migration of neuroblasts originating from the NC that normally reach the small intestine in week 7 of gestation and the rectum in week 12 [13]. The pathology of HSCR involves non-Mendelian inheritance, gender-dependent penetrance, variable expression, and the involvement of one or more gene(s) [13]. Genetic mapping in multiplex families and analysis of candidate genes have identified mutations in eight partially-interdependent genes in HCSR, with mutations occurring primarily in the RET gene [7]. RET is a tyrosine kinase receptor that is widely expressed in ganglion cells. It is the receptor for members of the GDNF family of extracellular signaling molecules. RET mutations are thought to cause defective migration, proliferation, differentiation, and survival of the enteric neuroblasts that form the enteric ganglia [1]. Dominant loss-of-function mutations in RET have been identified in familial and sporadic HCSR cases. RET has also been shown to act as a modifier gene in other syndromic forms of Hirschsprung Disease [14,15]. However, the overall frequency of RET mutations detected in sporadic HSCR is low, which suggests that other, previously unidentified genetic mechanisms, may be responsible for the development of HSCR. Since RET is considered to be the major HSCR susceptibility gene [4], it is likely that additional HSCR-associated genetic abnormalities occur at multiple nodes in the RET signaling cascade.
FOXA1 is an important transcription factor that can regulate RET expression [16]. FOXA1 is the founding member of the FOX family of transcription factors. FOXAs are required for postnatal survival due to their essential roles in controlling pancreatic and renal function and regulating many genes involved in development of the liver, lung, brain, prostate, and mammary gland [17]. DUSP6 is a downstream target of RET signaling [18]. DUSP6 is a specific negative feedback regulator of phosphorylated extracellular signal-regulated kinase, and is associated with cellular proliferation and differentiation [19]. DUSP6 expression can be altered by RET [18,19]. The increased expression of FOXA1 and DUSP6 in stenotic segments compared to proximal anastomosis in resected HSCR colons supports a role for these factors in the etiology of HCSR. Aberrant expression of FOXA1 and DUSP6 may prevent ENS differentiation and development.
HA117 is a highly expressed sequence in a multidrug resistant ATRA-treated leukemia cell line. HA117 is located on chromosome 14 [11]. Its adjacent upstream gene, DPF3 (D4, zinc and double PHD fingers, family 3), plays a role in neuronal differentiation and skeletal muscle and heart development [20]. HA117 expression levels are negatively associated with DPF3 in HSCR colonic segments. Therefore, HA117 may contribute to the genesis and progression of HSCR by exerting an anti-differentiation or anti-maturation action [11]. This process may be mediated by ATRA and RET, as retinoic acid upregulates RET and is involved in ENS development [21].
The recent expansion of molecular genetics has identified other HSCR-related genes. The most common chromosomal abnormality associated with HSCR is Down syndrome (trisomy 21), which occurs in 2-10% of all individuals with HSCR. Although individuals with Down syndrome are at a hundred-fold higher risk for HSCR than the general population, none of the established HSCR genes reside on chromosome 21; therefore, the relationship between HSCR and Down syndrome remains to be elucidated [6]. Other HSCR-associated genes include laminin, sex determining region Y-box 10 (SOX10), nitric oxide synthase (NOS), heme oxygenase-2 (HO-2), zinc finger homeobox protein 1b (ZFHX1B), endothelin converting enzyme 1 (ECE1), C-fos, Bcl-2, nerve growth factor (NGF), NGF receptor (NGFR), and CXCR4 [22]. Altered expression of each of these genes individually may not be sufficient to cause disease, but they may synergistically cooperate with the RET signaling pathway and other genetic aberrations to generate the HSCR phenotype. Future research is required to clearly define the role of all these genes in the pathogenesis of HSCR.
This study was associated with a few limitations. First, the sample size was small. Second, clinical details other than a diagnosis of HSCR were not considered. Finally, the FOXA1, DUSP6, and HA117 genes were not sequenced so the cause of the alterations in RNA and protein expression could not be identified. Further studies are necessary to fully understand the role of FOXA1, DUSP6, and HA117 in HSCR.
In summary, FOXA1 and DUSP6 may promote ENS differentiation and development, most likely as upstream and downstream factors, respectively, in the RET signaling cascade. The suppression of the FOXA1/DUSP6 signaling pathway may contribute to the development of HSCR. LncRNA HA117 may have an anti-differentiation function in the development of the ENS, and play a pivotal role in the progression of HSCR.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81370474).
Disclosure of conflict of interest
None.
References
- 1.Martucciello G, Pini Prato A, Puri P, Holschneider AM, Meier-Ruge W, Jasonni V, Tovar JA, Grosfeld JL. Controversies concerning diagnostic guidelines for anomalies of the enteric nervous system: a report from the fourth International Symposium on Hirschsprung’s disease and related neurocristopathies. J Pediatr Surg. 2005;40:1527–1531. doi: 10.1016/j.jpedsurg.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 2.Furness JB. The Enteric Nervous System. Malden, MA: Blackwell; 2006. [Google Scholar]
- 3.Iwashita T, Kruger GM, Pardal R, Kiel MJ, Morrison SJ. Hirschsprung disease is linked to defects in neural crest stem cell function. Science. 2003;301:972–976. doi: 10.1126/science.1085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gath R, Goessling A, Keller KM, Koletzko S, Coerdt W, Müntefering H, Wirth S, Hofstra RM, Mulligan L, Eng C, von Deimling A. Analysis of the RET, GDNF, EDN3, and EDNRB genes in patients with intestinal neuronal dysplasia and Hirschsprung disease. Gut. 2001;48:671–675. doi: 10.1136/gut.48.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanchuk SM, Myers SM, Eng C, Lapchak PA, Chakravarti A. De novo mutation of GDNF, ligand for the RET/GDNFR-alpha receptor complex, in Hirschsprung disease. Hum Mol Genet. 1996;5:2023–2026. doi: 10.1093/hmg/5.12.2023. [DOI] [PubMed] [Google Scholar]
- 6.Angrist M, Bolk S, Halushka M, Lapchak PA, Chakravarti A. Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet. 1996;14:341–344. doi: 10.1038/ng1196-341. [DOI] [PubMed] [Google Scholar]
- 7.Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, Brooks AS, Antinolo G, de Pontual L, Clement-Ziza M, Munnich A, Kashuk C, West K, Wong KK, Lyonnet S, Chakravarti A, Tam PK, Ceccherini I, Hofstra RM, Fernandez R. Hirschsprung Disease Consortium. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. Requirement of signaling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- 9.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–40. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bermudez O, Pagès G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Luo Y, Li S, Wang S, Wang N, Jin X. Expression profiles of HA117 and its neighboring gene DPF3 in different colon segments of Hirschsprung’s disease. Int J Clin Exp Pathol. 2014;7:3966–3974. [PMC free article] [PubMed] [Google Scholar]
- 12.Guinard-Samuel V, Bonnard A, De Lagausie P, Philippe-Chomette P, Alberti C, El Ghoneimi A, Peuchmaur M, Berrebi-Binczak D. Calretinin immunohistochemistry: a simple and efficient tool to diagnose Hirschsprung disease. Mod Pathol. 2009;22:1379–1384. doi: 10.1038/modpathol.2009.110. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez RM, Bleda M, Luzon-Toro B, García-Alonso L, Arnold S, Sribudiani Y, Besmond C, Lantieri F, Doan B, Ceccherini I, Lyonnet S, Hofstra RM, Chakravarti A, Antiñolo G, Dopazo J, Borrego S. Pathways systematically associated to Hirschsprung’s disease. Orphanet J Rare Dis. 2013;8:187. doi: 10.1186/1750-1172-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam PK, Garcia-Barceló M. Genetic basis of Hirschsprung’s disease. Pediatr Surg Int. 2009;25:543–558. doi: 10.1007/s00383-009-2402-2. [DOI] [PubMed] [Google Scholar]
- 15.Pontual L, Jaubert F, Espinosa-Parrilla Y, Munnich A, Brunet JF, Goridis C, Brunet JF, Goridis C, Feingold J, Lyonnet S, Amiel J. Mutations of the RET gene in isolated and syndromic Hirschsprung’s disease in human disclose major and modifier alleles at a single locus. J Med Genet. 2006;43:419–423. doi: 10.1136/jmg.2005.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanheimer PM, Woodfield W, Cyr AR, Kulak MV, White-Baer LS, Bair TB, Weigel RJ. Expression of the RET proto-oncogene is regulated by TFAP2C in breast cancer independent of the estrogen receptor. Ann Surg Oncol. 2013;20:2204–2212. doi: 10.1245/s10434-012-2570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–30. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degl’Innocenti D, Romeo P, Tarantino E, Sensi M, Cassinelli G, Catalano V, Perrone F, Pilotti S, Seregni E, Pierotti MA, Greco A, Borrello MG. DUSP6/MKP3 is overexpressed in papillary and poorly differentiated thyroid carcinoma and contributes to neoplastic properties of thyroid cancer cells. Endocr Relat Cancer. 2013;20:23–37. doi: 10.1530/ERC-12-0078. [DOI] [PubMed] [Google Scholar]
- 19.Ma J, Yu X, Guo L, Lu SH. DUSP6, a tumor suppressor, is involved in differentiation and apoptosis in esophageal squamous cell carcinoma. Oncol Lett. 2013;6:1624–1630. doi: 10.3892/ol.2013.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange M, Kaynak B, Forster UB, Tönjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling SS. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simkin JE, Zhang D, Rollo BN, Newgreen DF. Retinoic acid upregulates ret and induces chain migration and population expansion in vagal neural crest cells to colonise the embryonic gut. PLoS One. 2013;8:e64077. doi: 10.1371/journal.pone.0064077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrego S, Ruiz-Ferrer M, Fernandez RM, Antinolo G. Hirschsprung’s disease as a model of complex genetic etiology. Histol Histopathol. 2013;28:1117–1136. doi: 10.14670/HH-28.1117. [DOI] [PubMed] [Google Scholar]