Abstract
Epidemiological and histopathological studies have indicated that proliferative inflammatory atrophy (PIA) of the prostate is closely associated with the onset and development of prostate cancer (PCa). However, accurate isolation of PIA still remains a difficult matter, as well as high-quality RNA extraction from isolated PIA. These issues generated a lack of molecular evidence to support the mechanistic explanation proposed for the progression of PIA to PCa. Therefore, the isolation of PIA and the extraction of high-quality RNA from isolated PIA are of great importance to further demonstrate the correlation between PIA and the development of PCa at a molecular level. In this study, clinical samples from radical prostatectomy were stored in liquid nitrogen, PIA was identified by H&E staining of cryosections, PIA clusters were isolated by manual microdissection, total RNA was extracted from the PIA clusters by Trizol, and RNA quality was determined using the Agilent 2100 Bioanalyzer. Our results showed that PIA might be isolated by manual microdissection of cryosections stored in liquid nitrogen from clinical radical prostatectomy and used for extracting high-quality RNA (RIN > 7.5) by Trizol. Therefore, the present study established a valid method to discover molecular evidence in support of the correlation between PIA and the development of PCa.
Keywords: Proliferative inflammatory atrophy (PIA), prostate cancer (PCa), cryosection
Introduction
Prostate cancer (PCa) is one of the most common tumors of the urinary tract. According to the latest statistics issued by the National Institute of Health (NIH), the morbidity and mortality rates of prostate cancer in the US in 2013 are 238,590 with an average of 29,720 per year [1]. Although there is no clear statistical evidence, morbidity and mortality rates of prostate cancer is also rising in China due to the improvements in living standards, diet and lifestyle adjustments, and advances in diagnostic technologies [2]. Many studies have found that chronic inflammation contribute to the development of different types of cancer such as esophageal cancer, bladder cancer, liver cancer, stomach cancer, and colorectal cancer, through a variety of mechanisms [3,4]. However, the relationship between chronic prostatitis and PCa is still unclear, although many evidences indicate that chronic prostatitis, especially nonresolving prostatitis, may very likely lead to PCa.
Centesis and clinical histopathological assessments of PCa patients showed that PCa is often found in association with proliferative inflammatory atrophy (PIA) and prostatic intraepithelial neoplasia (PIN) [3,5-7]. Karakiewicz et al. found that the majority of PCa patients have a history of prostatitis [6,8-12]. Studies on the relationship between PCa and prostatitis show that the abnormal expression of scavenger receptor 1 (MSR1), macrophage inhibitory cytokine-1 (MIC-1), interleukin-8 (IL-8), interleukin-10 (IL-10), vascular endothelial growth factor (VEGF), intercellular adhesion molecule (ICAM), dendritic receptors, NF-κB, cyclooxygenase 2, and other inflammatory signaling genes is closely associated with the onset and development of PCa [5,13-15]. Therefore, a pathological model of PIA to PCa progression (PIA-PCa model) has been proposed. However, the difficulty in isolating PIA or extract high-quality RNA from isolated PIA, is precluding additional studies to discover molecular evidence in support of the PIA-PCa model.
The isolation of PIA and the extraction of high-quality total RNA from isolated PIA are essential requirements for the molecular biology analysis of the mechanisms of the PIA-PCa model. A number of procedures such as RNA sequence analysis, cDNA library construction, in vitro translation, and blotting assays require enough purity and integrity of RNA. Although RNA extraction is already a mature technique, the successful extraction of high-quality RNA from isolated PIA is still difficult for most skilled researchers due to the relatively small amount of PIA and PCa cells and their close proximity to each other. To address this issue, in this study we combined cryosection with manual microdissection to isolate PIA, improving the experimental conditions at all stages of RNA extraction, and analyzing the relationship between tissue preparation and the quality of extracted RNA. Hence, our study enables to achieve a high quality of both lncRNA expression profiling and PIA mRNA content, establishing a valid method to discover molecular evidences in support of the PIA-PCa model.
Materials and methods
Clinical tissue specimens
The prostate cancer tissue samples were collected from The First Affiliated Hospital of Nanchang University in 2013. The samples were immediately extracted from the abdominal cavity soon after excision, and washed with saline three times. Next, the samples were cut into 3 × 3 cm squares and immediately frozen in liquid nitrogen after removing necrotic, connective, and adipose tissues. The collection and study of tissue samples from all patients were both approved by the Medical Research Ethics Committee of The First Affiliated Hospital of Nanchang University. All tissue samples that were included in this study meet the following two criteria: 1) they belong to patients that have never received chemotherapy and radiotherapy, and 2) they were all pathologically confirmed after surgical resection to be PCa associated with PIA.
Identification of PIA by H&E staining of cryosections
The tissue samples were embedded in OCT cryomold (Sakura Finetek, Torrance, CA, USA) and placed in Leica CM1900 cryostat (Leica, Germany) at -28°C for 2 h. OCT embedding medium was removed from the samples using RNase-free water treated surgical blade, the samples were cut into to 6 μm-thick slices, and immediately fixed in 95% ethanol for 30 s to 1 min. H&E staining procedure was applied, the sections were dehydrated by passing through increasing concentrations of ethanol (70%, 80%, 90%, and 100%), cleared with xylene, mounted on neutral resin, and their tissue structure was examined under a microscope. The above steps were repeated until PIA was found.
Isolation of PIA by manual microdissection, extraction of total RNA, and determination of RNA quality
Once PIA was found following the procedure in Section 2.2, the approximate area of the PIA cluster was outlined with a marker under the microscope. The comparison with the sliced sections allowed us to locate and remove the tissue in excess using a surgical blade treated with RNase-free water (the removed area was 0.2-0.5 cm into the boundary of the marked area). The remaining tissue was cut into of 6 μm-thick slices, H&E staining was performed, and the slices were examined under a microscope to identify PIA. The above steps were repeated until PIA was the only remaining tissue that could be seen under the microscope. A total of 30-60 6 μm-thick slices were cut out; the number of slices was depending by the size of the PIA cluster. The completeness of the PIA cluster was determined in the middle and last sections. The slices that were not undergoing H&E staining were placed directly into Trizol reagent for RNA extraction. Agilent 2001 Bioanalyzer was used to estimate the quality of the extracted RNA.
Results
Identification of PIA by H&E staining
According to the criteria established by the 2006 International Working Group Classification System for Focal Prostate Atrophy Lesions, both simple atrophy (SA) and postatrophic hyperplasia (PAH) are classified as PIA. Epithelial cells of atrophic lesions include the basal cell layer and the luminal cell layer. While the luminal cells in the atrophic lesions displayed a basophilic cytoplasm which was dark staining under microscope of the relatively number of. The luminal cells were flat with no clear cytoplasm. There were a few cytoplasms in the basal layer and many gaps (Figure 1).
Figure 1.
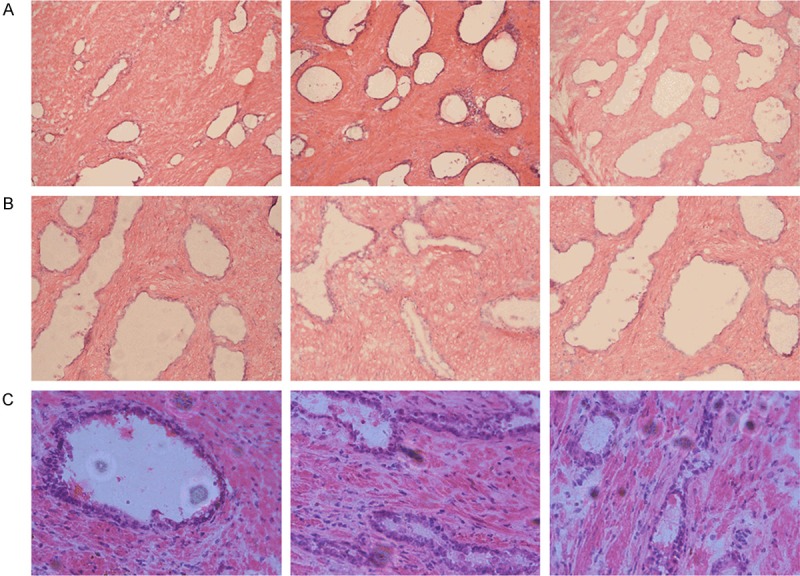
Microscope images of the PIA cluster, A: 100×, B: 200×, C: 400×.
Isolation of PIA by manual microdissection
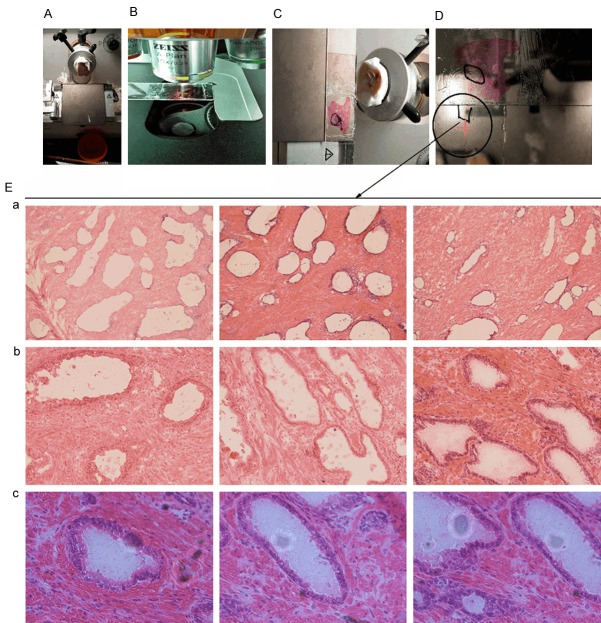
The size of the selected tissue sections was 2 × 2.5 cm. The PIA clusters were identified under the microscope and isolated by manual microdissection. The size of the PIA clusters was approximately 0.2 × 0.15 cm. The PIA clusters were observed under the microscope in a full field-of-view to ensure that there were no normal and cancerous prostate tissues in the first, middle, and last slices (Figure 2).
Figure 2.
A schematic diagram showing the isolation of PIA by manual microdissection. A: Tissue section size before cutting, B: H&E staining of 6 μm-thick slices to locate PIA, C: The identification of PIA after H&E staining, D: Scheme of manual isolation of PIA clusters, E: Microscope images of PIA clusters after isolation. a: 100×, b: 200×, c: 400×.
RNA extraction (RIN > 7) by Trizol
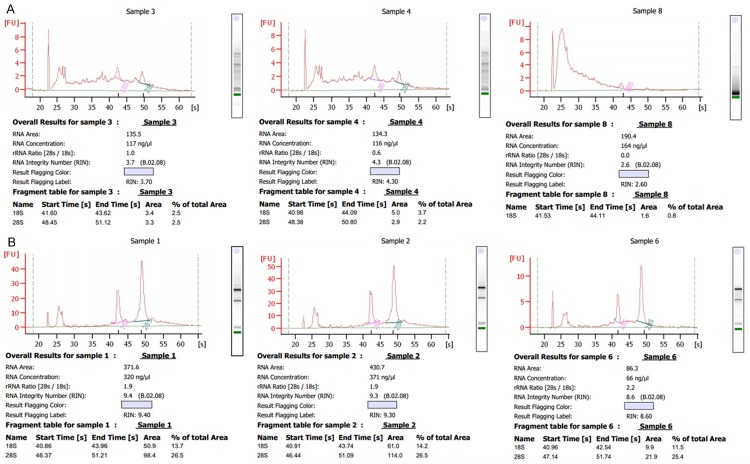
The quality and concentration of the extracted RNA was evaluated using Agilent 2100 Bioanalyzer. The RIN values of the RNA from triplicate experiments subjected to H&E staining were 3.7, 4.3, and 2.6; the concentrations were 117, 116, and 164 ng/μl. The RIN values from samples that did not undergo H&E staining were 9.4, 9.3, and 8.6; the concentrations were 320, 371, and 66 ng/μl (Figure 3).
Figure 3.
Evaluation of RNA quality and concentration using Agilent 2100 Bioanalyzer. A: RNA quality after H&E staining, B: RNA quality without H&E staining.
Discussion
Recent clinical and epidemiological studies strongly suggest that inflammation is closely associated with the onset, development, and metastasis of PCa [16]. PIA is a recently proposed concept, and is defined by De Marzo as the proliferation of epithelial cells on the basis of focal prostatic atrophy [17]. A number of laboratory and clinical data indicate that the onset and development of PCa follow a progression from PIA to PIN, followed by the malignant transformation into PCa, or even directly from PIA to PCa [18-20]. However, the existence of PIA in PCa is limited, and it is difficult to isolate PIA or to obtain high-quality total RNA and proteins from isolated PIA. Therefore, the pathological model of PIA to PCa progression could not be sufficiently validated.
Technological progresses in high-throughput microarrays and transcriptome sequencing have provided the basis for elucidating the mechanisms behind the onset and development of diseases [21]. However, high-throughput microarrays and transcriptome sequencing required a high quality of RNA, especially regarding the integrity and amount of RNA. For example, Agilent lncRNA plus mRNA Human Gene Expression Microarray V3.0 requires a total amount of RNA to be more than 1 μg with RIN greater than 7; TruSeq trace transcriptome and lncRNA sequencing requires a total amount of RNA more than 2 μg with RIN more than 7 [22-25]. Therefore, this restriction of high-throughput microarrays and transcriptome sequencing limits the high-throughput analysis of trace substances (such as PIA), thus preventing the analysis of pathological mechanisms through transcriptome data.
The aim of the present study was to discover a valid method for isolating PIA, for extracting high-quality RNA, and for the expression profiling of mRNA and lncRNA content of PIA to validate PIA-PCa model from a molecular perspective. This study used manual microdissection to isolate tissue regions with PIA, which were define as PIA clusters. PIA clusters include cells within the atrophic lesion, such as basal cells, luminal cells, inflammatory cells, and interstitial cells between prostate glands. In order to ensure the purity of PIA clusters during their isolation by cryosection, the tissue specimens were sliced into 6 μm-thick slices and H&E staining was performed on the first, middle, and last slices to observe the purity of PIA clusters under a microscope. The analysis of the RNA quality and quantity revealed that RNA extracted from the PIA clusters using this method met the standards for high-throughput microarray and transcriptome sequencing. However, RNA extracted from H&E stained samples did not meet these standards. In conclusion, this study established a valid method for further transcriptome analyses to discover molecular evidence in support of the pathological model of PIA to PCa progression, as well as provided a method for the high-throughput characterization of other materials present in traces within tissues.
Acknowledgements
This project was supported by the Sciences Foundation Monumental Project of China (Grant No. 91229119) and the National Natural Science Foundation of China (Grant No. 81460389).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Sun YH. Current diagnosis and treatment of prostate carcinoma. Shanghai Medical Journal. 2011;7:487–488. (In Chinese) [Google Scholar]
- 3.Goldstraw MA, Fitzpatrick JM, Kirby RS. What is the role of inflammation in the pathogenesis of prostate cancer? BJU Int. 2007;99:966–968. doi: 10.1111/j.1464-410X.2007.06879.x. [DOI] [PubMed] [Google Scholar]
- 4.Platz EA, De Marzo AM. Epidemiology of inflammation and prostate cancer. J Urol. 2004;171:S36–40. doi: 10.1097/01.ju.0000108131.43160.77. [DOI] [PubMed] [Google Scholar]
- 5.Sun J, Turner A, Xu J, Grönberg H, Isaacs W. Genetic variability in inflammation pathways and prostate cancer risk. Urol Oncol. 2007;25:250–259. doi: 10.1016/j.urolonc.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Klein EA, Silverman R. Inflammation, infection, and prostate cancer. Curr Opin Urol. 2008;18:315–319. doi: 10.1097/MOU.0b013e3282f9b3b7. [DOI] [PubMed] [Google Scholar]
- 7.De Nunzio C, Kramer G, Marberger M, Montironi R, Nelson W, Schröder F, Sciarra A, Tubaro A. The controversial relationship between benign prostatic hyperplasia and prostate cancer: the role of inflammation. Eur Urol. 2011;60:106–117. doi: 10.1016/j.eururo.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 8.Karakiewicz PI, Benayoun S, Bégin LR, Duclos A, Valiquette L, McCormack M, Bénard F, Saad F, Perrotte P. Chronic inflammation is negatively associated with prostate cancer and high-grade prostatic intraepithelial neoplasia on needle biopsy. Int J Clin Pract. 2007;61:425–430. doi: 10.1111/j.1742-1241.2006.00905.x. [DOI] [PubMed] [Google Scholar]
- 9.Molimard B, Camparo P, Desfemmes FR, Durand X, Haus R, Deligne E, Houlgatte A, Compérat E. Predictive value of the asymptomatic prostatic inflammation in the development of prostate cancer. Prog Urol. 2010;20:508–514. doi: 10.1016/j.purol.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Perletti G, Montanari E, Vral A, Gazzano G, Marras E, Mione S, Magri V. Inflammation, prostatitis, proliferative inflammatory atrophy: ‘Fertile ground’ for prostate cancer development? Mol Med Rep. 2010;3:3–12. doi: 10.3892/mmr_00000211. [DOI] [PubMed] [Google Scholar]
- 11.Sutcliffe S, Platz EA. Inflammation in the etiology of prostate cancer: an epidemiologic perspective. Urol Oncol. 2007;25:242–249. doi: 10.1016/j.urolonc.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Bastian PJ, Nuhn P, Stadler TC, Roosen A, Stief CG. Prostatic inflammation and prostate cancer. Urologe A. 2010;49:636–638. doi: 10.1007/s00120-010-2254-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim BH, Kim CI, Chang HS, Choe MS, Jung HR, Kim DY, Park CH. Cyclooxygenase-2 overexpression in chronic inflammation associated with benign prostatic hyperplasia: is it related to apoptosis and angiogenesis of prostate cancer? Korean J Urol. 2011;52:253–259. doi: 10.4111/kju.2011.52.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shetty AV, Thirugnanam S, Dakshinamoorthy G, Samykutty A, Zheng G, Chen A, Bosland MC, Kajdacsy-Balla A, Gnanasekar M. 18alpha-glycyrrhetinic acid targets prostate cancer cells by down-regulating inflammation-related genes. Int J Oncol. 2011;39:635–640. doi: 10.3892/ijo.2011.1061. [DOI] [PubMed] [Google Scholar]
- 15.Haverkamp J, Charbonneau B, Ratliff TL. Prostate inflammation and its potential impact on prostate cancer: a current review. J Cell Biochem. 2008;103:1344–1353. doi: 10.1002/jcb.21536. [DOI] [PubMed] [Google Scholar]
- 16.Sugar LM. Inflammation and prostate cancer. Can J Urol. 2006;13(Suppl 1):46–47. [PubMed] [Google Scholar]
- 17.De Marzo AM, Marchi VL, Epstein JI, Nelson WG. Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis. Am J Pathol. 1999;155:1985–1992. doi: 10.1016/S0002-9440(10)65517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Leenders GJ, Gage WR, Hicks JL, van Balken B, Aalders TW, Schalken JA, De Marzo AM. Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am J Pathol. 2003;162:1529–1537. doi: 10.1016/S0002-9440(10)64286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putzi MJ, De Marzo AM. Morphologic transitions between proliferative inflammatory atrophy and high-grade prostatic intraepithelial neoplasia. Urology. 2000;56:828–832. doi: 10.1016/s0090-4295(00)00776-7. [DOI] [PubMed] [Google Scholar]
- 20.Nelson WG, De Marzo AM, Isaacs WB. Inflammation, proliferative inflammatory atrophy, glutathione S-transferase Pi, and prostate cancer prevention. Cancer Epidemiology Biomarkers Prevention. 2003;12:1362S–1362S. [Google Scholar]
- 21.Yang L, Wei G, Tang K, Nardini C, Han JD. Understanding human diseases with highthroughput quantitative measurement and analysis of molecular signatures. Sci China Life Sci. 2013;56:213–219. doi: 10.1007/s11427-013-4445-9. [DOI] [PubMed] [Google Scholar]
- 22.Walter MA, Seboek D, Demougin P, Bubendorf L, Oberholzer M, Müller-Brand J, Müller B. Extraction of high-integrity RNA suitable for microarray gene expression analysis from long-term stored human thyroid tissues. Pathology. 2006;38:249–253. doi: 10.1080/00313020600696272. [DOI] [PubMed] [Google Scholar]
- 23.Warshauer DH, Davis CP, Holt C, Han Y, Walichiewicz P, Richardson T, Stephens K, Jager A, King J, Budowle B. Massively parallel sequencing of forensically relevant single nucleotide polymorphisms using TruSeq forensic amplicon. Int J Legal Med. 2015;129:31–36. doi: 10.1007/s00414-014-1108-8. [DOI] [PubMed] [Google Scholar]
- 24.Van Nieuwerburgh F, Soetaert S, Podshivalova K, Ay-Lin Wang E, Schaffer L, Deforce D, Salomon DR, Head SR, Ordoukhanian P. Quantitative bias in Illumina TruSeq and a novel post amplification barcoding strategy for multiplexed DNA and small RNA deep sequencing. PLoS One. 2011;6:e26969. doi: 10.1371/journal.pone.0026969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sultan M, Dökel S, Amstislavskiy V, Wuttig D, Sültmann H, Lehrach H, Yaspo ML. A simple strand-specific RNA-Seq library preparation protocol combining the Illumina TruSeq RNA and the dUTP methods. Biochem Biophys Res Commun. 2012;422:643–646. doi: 10.1016/j.bbrc.2012.05.043. [DOI] [PubMed] [Google Scholar]