Abstract
We investigated the association between the clinical outcome and GSTP1 and XRCC1 gene polymorphisms in advanced NSCLC patients with cisplatin-based chemotherapy. We prospectively recruited 325 NSCLC patients between January 2010 and January 2014. Genotypes of GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln were conducted using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) assay. AG and GG genotypes of GSTP1 A313G were correlated with a higher CR + PR when compared with AA genotype. Furthermore, GA and AA genotypes of XRCC1 Arg399Gln were associated with more CR + PR when compared with GG genotype. In the Cox proportional hazards model, GG genotype of GSTP1 A313G was significantly correlated with a longer median survival time when compared with AA genotype, and it is associated with a heavy decreased risk of death from NSCLC. Moreover, GA and AA genotypes of XRCC1 Arg399Gln had a significantly longer median survival time, and GA and AA genotypes were significantly associated with a moderate reduced risk of death from NSCLC. GSTP1 A313G and XRCC1 Arg399Gln gene polymorphisms might influence the response to cisplatin-based chemotherapy and affect the clinical outcome of advanced NSCLC.
Keywords: GSTP1, XRCC1, polymorphism, non-small cell lung cancer, cisplatin-based chemotherapy
Introduction
Lung cancer is one of the most common health problems worldwide [1]. It is estimated that 80% of lung cancer is non-small cell lung cancer (NSCLC), and patients with NSCLC are usually diagnosed in an advanced stages, which leaves curable surgery impossible [2]. Although many clinical trials have demonstrated that patients with complete NSCLC resection also benefit from cisplatin-based adjuvant chemotherapy, the efficacy and toxicity are largely individual [3]. Numerous evidences revealed that treatment selections with tumor stage [4] or with pathology [5] can be affected by adjuvant therapeutic regimens, and expression of molecular biomarkers for selection of patients also plays a crucial role in individual differences of cisplatin-based chemotherapy [6]. Moreover, increasing evidences have reported that molecular markers do help physician to stratify the patients for treatment options and showed treatment benefit [7-10].
Glutathione S-transferase P1 (GSTP1) is one of the glutathione S-transferase class members, and is widely expressed in different human tissues including lung and liver. It has an important role in the phase II metabolism of xenobiotics, and also has a role in the key process of cisplatin biotransformation [11]. GSTP1 A313G mutant alleles, causing an Ile to Val at codon 105 of the substrate-binding site of GSTP1 protein, decrease the biotransformation activity of GST proteins, followed by increasing intracellular cisplatin-GSH-conjugates and inhibiting the metabolism of cisplatin [12,13].
X-ray repair cross-complementing group 1 (XRCC1) is a major member of base excision repair pathway, which plays a prominent role in both single-strand break repair and base excision repair [14]. XRCC1 protein interacts with ligase and poly (ADP-ribose) polymerase to efficiently repair DNA damage including cisplatin-induced damage [15].
Previous studies have reported the role of GSTP1 and XRCC1 gene polymorphisms have an important role in influencing the clinical outcome of NSCLC, but the results are inconsistent [16-19]. Therefore, we investigated the association between the clinical outcome and GSTP1 and XRCC1 gene polymorphisms in advanced NSCLC patients with cisplatin-based chemotherapy.
Materials and methods
Study population
In this study, we prospectively recruited 325 NSCLC patients from Affiliated Hospital of Inner Mongolia Medical University between January 2012 and December 2014. All patients did not receive any anticancer therapies before surgery and had adequate hematology, renal and liver function with the Eastern Cooperative Oncology Group (ECOG) performance status 0 or 1. During and after adjuvant chemotherapy, the patients were routinely followed up every four weeks until death or the end of this study.
Chemotherapy
All participants received one of the following cisplatin-based combination chemotherapy regimen: cisplatin plus gemcitabine (GP), cisplatin plus vinorelbine (NP), cisplatin plus paclitaxel (TP), and cisplatin plus docetaxel (DP). The patients received a physical examination, computed tomography, and magnetic resonance imaging before adjuvant chemotherapy. The chemotherapy was repeated at three-weekly intervals for up to six cycles unless unacceptable toxicity, disease progression or patients’ refusal to continue treatment.
Clinical evaluation
The patients were required to complete at least two cycles of chemotherapy to evaluate treatment response according to RECIST criteria [20]. The percentage of patients with complete response (CR) and partial response (PR) after treatment were considered as responders, and patients with stable disease (SD) and progressive disease (PD) to chemotherapy were regarded as non-responders. Overall survival (OS) was defined as the period between the date of chemotherapy and the data of confirmed death from any cause.
DNA extraction and genotyping
Each patient with NSCLC was asked to provide 5 ml peripheral blood, and the blood was kept at -70°C until use. Genomic DNA was isolated from peripheral blood lymphocytes using the TIANamp Blood DNA Kit (Tiangen, Beijing, China) by the manufacturer’s protocol. Genotypes of GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln were conducted using Polymerase Chain Reaction Restriction Fragment Length Polymorphism (PCR-RFLP) assay (Applied Biosystems, Foster City, CA, USA). The positive and reverse primer sequences of GSTP1 A313G were 5’-ACCCCAGGGCTCTATGGGAA-3’ and 5’-TGAGGGCACAAGAAGCCCCT-3’, respectively; for XRCC1 Arg194Trp, the positive and reverse primer sequences were 5’-GCCAGGGCCCCTCCTTCAA-3’ and 5’-TACCCTCAGACCCACGAGT-3’, respectively; for XRCC1 Arg280His, the positive and reverse primer sequences were 5’-CAGTGGTGCTAACCTAATC-3’ and 5’-AGTAGTCTGCTGGCTCTGG-3’, respectively; for XRCC1 Arg399Gln, the positive and reverse primer sequences were 5’-CAGTGGTGCTAACCTAATC-3’ and 5’-AGTAGTCTGCTGGCTCTGGG-3’, respectively. The PCR reaction was performed in a 25 μL reaction solution, and it included 25 mM MgCl2, 200 μM primers, 2 mM 4×dNTP, 20 μM Primer and 5 U/μl Taq DNA. PCR reaction was performed using the following conditions: beginning at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 25 s, and annealing at 53°C for 40 s, with a final extension at 72°C for 6 min.
Statistical analysis
The clinical information on efficacy and toxicity were compared across genotype, using χ2 tests for categorical variables and one-way analysis of variance for continuous variables. Survival curves were analyzed by the Kaplan-Meier method, and the impact of the SNPs on OS was assessed using the log-rank test. Meanwhile, baseline characteristics were adjusted in order to avoid potential confounding effects. The association between response to chemotherapy and GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms were described as odds ratio (ORs) and 95% confidence interval (CI) in unconditioned Logistic regression. The prognostic value of GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms for the OS was estimated by multivariate analysis using the Cox proportional hazards models, describing as the hazard ratio (HR) and 95% CI. P values < 0.05 with two-sided were considered statistical differences. Data were performed by the statistical software SPSS Statistics (version 19.0, SPSS Inc., Chicago, IL, USA).
Results
In this present study, the mean age of 325 NSCLC patients were 57.6±12.4 years old, and 116 (35.69%) patients were males and 209 (64.31%) were females (Table 1). 222 (68.31%) patients were current or former smokers, and 138 (42.46%) were at the TNM stage, 176 (54.15%) were adenocarcinoma NSCLC and 124 (38.15%) were squamous cell carcinoma (38.15%). There were 228 (70.15%) patients showed CR + PR to chemotherapy, and 97 (29.85%) showed SD + PD to chemotherapy.
Table 1.
Demographic and clinical characteristics of included NSCLC patients
| Characteristics | Number of patients | Percent (%) |
|---|---|---|
| Age, years | ||
| < 60 | 173 | 53.23 |
| ≥ 60 | 152 | 46.77 |
| Gender | ||
| Male | 116 | 35.69 |
| Female | 209 | 64.31 |
| Tobacco smoking | ||
| Never | 103 | 31.69 |
| Current or former | 222 | 68.31 |
| TNM stage | ||
| IIIB | 138 | 42.46 |
| IV | 187 | 57.54 |
| Histology | ||
| Adenocarcinoma | 176 | 54.15 |
| Squamous cell carcinoma | 124 | 38.15 |
| Other | 25 | 7.70 |
| Response to chemotherapy | ||
| CR + PR | 228 | 70.15 |
| SD + PD | 97 | 29.85 |
Genotype frequencies of GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms were found to be in Hardy-Weinberg equilibrium (P > 0.05). By unconditioned logistic regression, we found that GSTP1 A313G polymorphism had a significant association with response to chemotherapy, and AG and GG genotypes were correlated with a higher CR + PR when compared with AA genotype, and the Ors (95% CI) were 2.31 (1.35-3.95) and 5.68 (1.61-30.46), respectively (Table 2). Furthermore, GA and AA genotypes of XRCC1 Arg399Gln were associated with more CR + PR when compared with GG genotype, and the Ors (95% CI) were 1.97 (1.13-3.43) and 3.76 (1.52-10.53), respectively.
Table 2.
Association of GSTP1 and XRCC1 gene polymorphisms with response to cisplatin-based chemotherapy in NSCLC patients
| Genotypes | Total number | Percent (%) | CR + PR N = 228 | Percent (%) | SD + PD N = 97 | Percent (%) | OR (95% CI)1 | P value |
|---|---|---|---|---|---|---|---|---|
| GSTP1 A313G | ||||||||
| AA | 148 | 45.54 | 88 | 38.60 | 60 | 61.86 | 1.0 (Ref.) | - |
| AG | 149 | 45.85 | 115 | 50.44 | 34 | 35.05 | 2.31 (1.35-3.95) | < 0.05 |
| GG | 28 | 8.62 | 25 | 10.96 | 3 | 3.09 | 5.68 (1.61-30.46) | < 0.05 |
| XRCC1 Arg194Trp | ||||||||
| CC | 169 | 52.00 | 115 | 50.44 | 54 | 55.67 | 1.0 (Ref.) | - |
| CT | 116 | 35.69 | 83 | 36.40 | 33 | 34.02 | 1.18 (0.68-2.05) | 0.53 |
| TT | 40 | 12.31 | 30 | 13.16 | 10 | 10.31 | 1.41 (0.61-3.47) | 0.39 |
| XRCC1 Arg280His | ||||||||
| AA | 211 | 64.92 | 146 | 64.04 | 65 | 67.01 | 1.0 (Ref.) | - |
| AG | 87 | 26.77 | 62 | 27.19 | 25 | 25.77 | 1.10 (0.62-2.00) | 0.72 |
| GG | 27 | 8.31 | 20 | 8.77 | 7 | 7.22 | 1.27 (0.49-3.74) | 0.6 |
| XRCC1 Arg399Gln | ||||||||
| GG | 151 | 46.46 | 92 | 40.35 | 59 | 60.82 | 1.0 (Ref.) | - |
| GA | 126 | 38.77 | 95 | 41.67 | 31 | 31.96 | 1.97 (1.13-3.43) | < 0.05 |
| AA | 48 | 14.77 | 41 | 17.98 | 7 | 7.22 | 3.76 (1.52-10.53) | < 0.05 |
Adjusted for age, gender, tobacco smoking, TNM stage and histology.
At the end of follow-up, 125 patients died from all causes, and the five-year survival rate was 38.46%. In the Cox proportional hazards model, GG genotype (34.50 months) of GSTP1 A313G was significantly correlated with a longer median survival time when compared with AA genotype (22.20 months), and it is associated with a heavy decreased risk of death from NSCLC (HR = 0.36, 95% CI = 0.11-0.98) (Table 3; Figure 1). Moreover, GA (26.50 months) and AA genotypes (23.80 months) of XRCC1 Arg399Gln had a significantly longer median survival time when compared with GG genotype (35.70 months), and GA and AA genotypes were significantly associated with a moderate reduced risk of death from NSCLC (for GA genotype, HR = 0.49, 95% CI = 0.29-0.83; for AA genotype, HR = 0.17, 95% CI = 0.06-0.41) (Table 3; Figure 2).
Table 3.
Association of GSTP1 and XRCC1 gene polymorphisms with overall survival of NSCLC patients
| Genotypes | Total Frequencies | Percent (%) | Death N = 125 | Percent (%) | Median survival time (months) | Log-rank P value | Adjusted HR (95% CI)1 | P value |
|---|---|---|---|---|---|---|---|---|
| GSTP1 A313G | ||||||||
| AA | 148 | 45.54 | 64 | 51.20 | 22.20 | < 0.05 | 1.0 (Ref.) | |
| AG | 149 | 45.85 | 54 | 43.20 | 27.10 | 0.75 (0.46-1.22) | 0.22 | |
| GG | 28 | 8.62 | 7 | 5.60 | 34.50 | 0.36 (0.11-0.98) | 0.03 | |
| XRCC1 Arg194Trp | ||||||||
| CC | 169 | 52.00 | 71 | 56.80 | 29.20 | 0.35 | 1.0 (Ref.) | |
| CT | 116 | 35.69 | 41 | 32.80 | 31.40 | 0.75 (0.45-1.26) | 0.26 | |
| TT | 40 | 12.31 | 13 | 10.40 | 32.50 | 0.74 (0.33-1.60) | 0.42 | |
| XRCC1 Arg280His | ||||||||
| AA | 211 | 64.92 | 87 | 69.60 | 33.10 | 0.22 | 1.0 (Ref.) | |
| AG | 87 | 26.77 | 31 | 24.80 | 31.70 | 0.79 (0.45-1.36) | 0.37 | |
| GG | 27 | 8.31 | 7 | 5.60 | 30.40 | 0.50 (0.17-1.30) | 0.13 | |
| XRCC1 Arg399Gln | ||||||||
| GG | 151 | 46.46 | 76 | 60.80 | 35.70 | < 0.05 | 1.0 (Ref.) | |
| GA | 126 | 38.77 | 42 | 33.60 | 26.50 | 0.49 (0.29-0.83) | < 0.05 | |
| AA | 48 | 14.77 | 7 | 5.60 | 23.80 | 0.17 (0.06-0.41) | < 0.05 | |
Adjusted for age, gender, tobacco smoking, TNM stage and histology.
Figure 1.
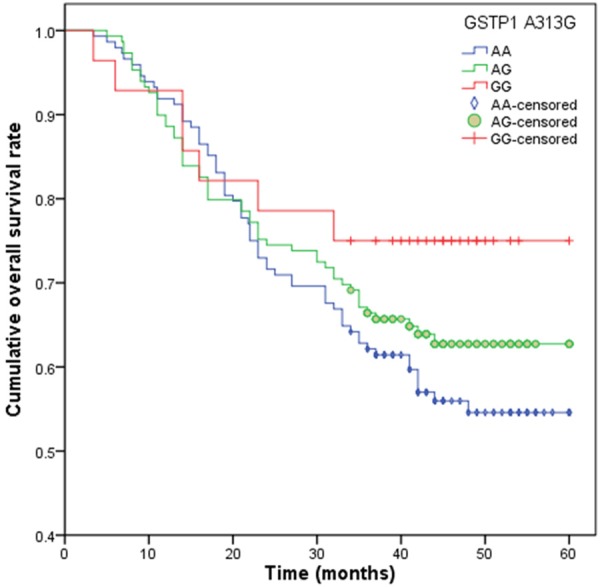
Overall survival time (months) in advanced NSCLC patients with GSTP1 A313G.
Figure 2.
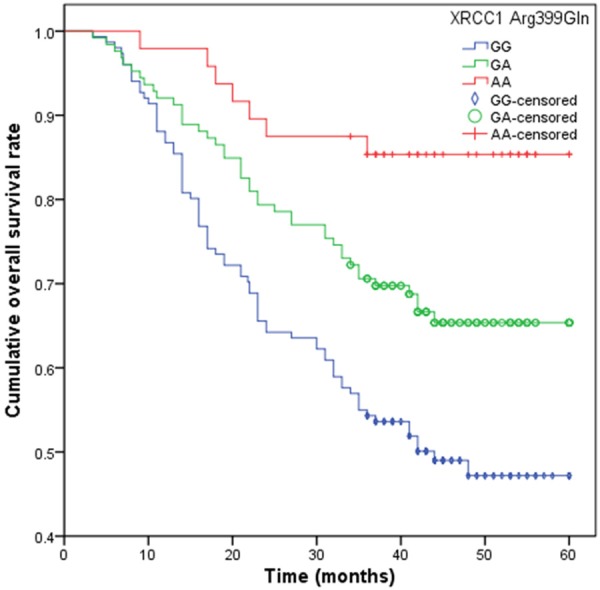
Overall survival time (months) in advanced NSCLC patients with XRCC1 Arg399Gln.
Discussion
It is well known that patients with advanced NSCLC usually show an individual response to cisplatin-based chemotherapy, and increasing evidences suggest that genetic factors maybe play an important role in the individual response [21,22]. Previous studies have reported that response to chemotherapy can be influenced by hereditary factors through increasing the cell activity of biotransformation, accumulation of intracellular cisplatin and the weakened capacity of DNA repairing [23-25]. In the present study, we investigated whether polymorphisms of GSTP1 A313G, XRCC1 Arg194Trp, Arg280His and Arg399Gln could influence the response to chemotherapy and clinical outcome of NSCLC, and we found that GG genotype of GSTP1 A313G and AA genotype of XRCC1 Arg399Gln were associated with better CR + PR to chemotherapy when compared with wide-type genotypes, and they are also associated with longer median survival time and an reduced risk of death from NSCLC.
It is well known that GSTP1 is a kind of phase II detoxification enzymes, which plays an important role in the detoxification of cisplatin [26], and in vitro experimental studies reported that over expression of GSTP1 could reduce the sensitivity to platinum agents in advanced NSCLC patients [27]. The polymorphism of GSTP1 A313G could alter the expression of GSTP1 and change the thermal stability and conjugation capacity of GSTP1, and the polymorphism of this gene could influence the ability to detoxify chemotherapeutic agents and modulate drug responses. Therefore, enzyme activity with the GSTP1 A313G is more effective on cisplatin treatment than that with wild-type in vitro [28], but it is a controversy that the A or G allele would enhance clinical response of cisplatin-based chemotherapy. One previous meta-analysis regarding on the association between GSTP1 A313G polymorphism and response to platinum-based chemotherapy in NSCLC patients have pointed out that GSTP1 A313G polymorphism is significantly correlated with platinum-based chemotherapy in NSCLC patients [16], and another meta-analysis with nine studies also found that GSTP1 A313G polymorphism could predict the treatment response to platinum-based chemotherapy in NSCLC patients [17]. Our study found GSTP1 A313G polymorphism was correlated with response to cisplatin-based chemotherapy and overall survival of advanced NSCLC, which is in line with previous studies.
XRCC1 gene is located on chromosome 19q13.2, and it codes for a 633 amino acid residue protein as well as serves as a scaffolding protein in base excision repair. XRCC1 protein plays an important role in a complex with many other components to facilitate BER and single-strand break-repair processes. Functional polymorphisms of XRCC1 have been identified. The XRCC1 Arg194Trp, Arg280His and Arg399Gln variants are the hot spot in the researches of XRCC1 polymorphisms, which especially links with the change of XRCC1 protein activity to impact DNA repair capacity [29]. DNA damage severity induced by cisplatin has relevance with the efficiency and toxicity of cisplatin-based chemotherapy. It is predicted that XRCC1 gene polymorpihsms affect the response to cisplatin-based chemotherapy. However, as the changes of DNA repair capacity caused by the SNPs of XRCC1 could take place in normal tissue as well as tumor tissue, there existed different results on the effect of XRCC1 polymorphisms. Currently, several studies reported the association between XRCC1 gene polymorphisms and clinical outcome of NSCLC [18,19,30-32]. Ke et al. conducted a follow-up study with 460 cases, and found that XRCC1 Arg194Trp and XRCC1Arg399Gln genes had a role in modifying the effect of platinum-based chemotherapy for NSCLC patients and were associated with longer survival time [30]. Another study conducted in a Chinese population and showed that AA genotype of XRCC1 Arg399Gln was associated with longer overall survival time of NSCLC [18]. However, some studies reported inconsistent results. One study with 352 NSCLC patients has indicated that no association was found between XRCC1 gene polymorphisms and chemotherapy response and survival of patients with NSCLC [19]. The divergence in results from different studies on XRCC1 polymorphism may be related to variation in ethnic origin of the studied population, sample size, carcinogenic exposure, genotyping method and also by chance.
Our results suggest that GSTP1 A313G and XRCC1 Arg399Gln polymorphisms might influence the response to cisplatin-based chemotherapy and affect the clinical outcome of advanced NSCLC. These gene polymorphisms could contribute to identification of patients, and are more likely to achieve favorable response to cisplatin-based chemotherapy.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.McCloskey P, Balduyck B, Van Schil PE, Faivre-Finn C, O’Brien M. Radical treatment of non-small cell lung cancer during the last 5 years. Eur J Cancer. 2013;49:1555–1564. doi: 10.1016/j.ejca.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J International Adjuvant Lung Cancer Trial Collaborative Group. Cisplatin based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 4.Geiger S, Schlemmer M, Heinemann V, Stemmler HJ. Adjuvant cisplatin based chemotherapy for resected NSCLC: one size fits all? Anticancer Drugs. 2010;21:799–804. doi: 10.1097/CAD.0b013e32833dbbfe. [DOI] [PubMed] [Google Scholar]
- 5.Arriagada R, Dunant A, Pignon JP, Bergman B, Chabowski M, Grunenwald D, Kozlowski M, Le Pechoux C, Pirker R, Pinel MI, Tarayre M, Le Chevalier T. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J. Clin. Oncol. 2010;28:35–42. doi: 10.1200/JCO.2009.23.2272. [DOI] [PubMed] [Google Scholar]
- 6.Filipits M, Pirker R. Predictive markers in the adjuvant therapy of non-small cell lung cancer. Lung Cancer. 2011;74:355–363. doi: 10.1016/j.lungcan.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Maus MK, Mack PC, Astrow SH, Stephens CL, Zeger GD, Grimminger PP, Hsiang JH, Huang E, Li T, Lara PN, Danenberg KD, Gandara DR. Histology related associations of ERCC1, RRM1, and TS biomarkers in patients with non small-cell lung cancer: implications for therapy. J Thorac Oncol. 2013;8:582–586. doi: 10.1097/JTO.0b013e318287c3c5. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A, Spanish Lung Cancer G. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 9.Rouquette I, Mazieres J. A brief overview of a lung cancer biomarker: thymidylate synthase. Rev Mal Respir. 2011;28:773–777. doi: 10.1016/j.rmr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Yokomise H, Liu D, Chang S, Go T, Ishikawa S, Misaki N, Nakashima N. Biomarkers as prognostic factors for cN2 or 3 non-small cell lung cancer treated by induction chemoradiotherapy and surgery. Anticancer Res. 2013;33:1107–1115. [PubMed] [Google Scholar]
- 11.Boyer TD. The glutathione S-transferases: an update. Hepatology. 1989;9:486–96. doi: 10.1002/hep.1840090324. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 13.Peklak-Scott C, Smitherman PK, Townsend AJ, Morrow CS. Role of glutathione S-transferase P1-1 in the cellular detoxification of cisplatin. Mol Cancer Ther. 2008;7:3247–3255. doi: 10.1158/1535-7163.MCT-08-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Lippard SJ. Photoaffinity labeling reveals nuclear proteins that uniquely recognize cisplatin-DNA interstrand cross-links. Biochemistry. 2009;48:4916–4925. doi: 10.1021/bi900389b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin JY, Huang Q, Zhao YC, Zhou HH, Liu ZQ. Meta-analysis on pharmacogenetics of platinum-based chemotherapy in non small cell lung cancer (NSCLC) patients. PLoS One. 2012;7:e38150. doi: 10.1371/journal.pone.0038150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Xian L. The association between the GSTP1 A313G and GSTM1 null/present polymorphisms and the treatment response of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients: a meta-analysis. Tumour Biol. 2014;35:6791–6799. doi: 10.1007/s13277-014-1866-4. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Wu J, Shi GY, Zhou HF, Yu Y. Role of XRCC1 and ERCC5 polymorphisms on clinical outcomes in advanced non-small cell lung cancer. Genet Mol Res. 2014;13:3100–3107. doi: 10.4238/2014.April.17.6. [DOI] [PubMed] [Google Scholar]
- 19.Du Y, Su T, Zhao L, Tan X, Chang W, Zhang H, Cao G. Associations of polymorphisms in DNA repair genes and MDR1 gene with chemotherapy response and survival of non-small cell lung cancer. PLoS One. 2014;9:e99843. doi: 10.1371/journal.pone.0099843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffaud F, Therasse P. New guidelines to evaluate the response to treatment in solid tumors. Bull Cancer. 2000;87:881–886. [PubMed] [Google Scholar]
- 21.Wheeler HE, Gamazon ER, Stark AL, O’Donnell PH, Gorsic LK, Huang RS. Genome-wide meta-analysis identifies variants associated with platinating agent susceptibility across populations. Pharmacogenomics J. 2013;13:35–43. doi: 10.1038/tpj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108:1695–1703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booton R, Ward T, Heighway J, Ashcroft L, Morris J, Thatcher N. Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol. 2006;1:679–683. [PubMed] [Google Scholar]
- 24.Bradbury PA, Kulke MH, Heist RS, Zhou W, Ma C, Xu W, Marshall AL, Zhai R, Hooshmand SM, Asomaning K, Su L, Shepherd FA, Lynch TJ, Wain JC, Christiani DC, Liu G. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics. 2009;19:613–625. doi: 10.1097/FPC.0b013e32832f3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Huo X, Lin Y, Ban H, Lin Y, Li W, Zhang B, Au WW, Xu X. Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int J Hyg Environ Health. 2010;213:140–145. doi: 10.1016/j.ijheh.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oguri T, Fujiwara Y, Katoh O, Daga H, Ishikawa N, Fujitaka K, Yamasaki M, Yokozaki M, Isobe T, Ishioka S, Yamakido M. Glutathione S-transferase-pi gene expression and platinum drug exposure in human lung cancer. Cancer Lett. 2000;156:93–99. doi: 10.1016/s0304-3835(00)00447-x. [DOI] [PubMed] [Google Scholar]
- 28.McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006;25:1639–1648. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen MR, Jones IM, Mohrenweiser H. Non conservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 30.Ke HG, Li J, Shen Y, You QS, Yan Y, Dong HX, Liu JH, Shen ZY. Prognostic significance of GSTP1, XRCC1 and XRCC3 polymorphisms in non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2012;13:4413–4416. doi: 10.7314/apjcp.2012.13.9.4413. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Ma W, Li Y, Wu J, Shi GY. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genet Mol Res. 2014;13:228–236. doi: 10.4238/2014.January.14.2. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Hu L, Xu J, Shen H, Hu Z, Ma H, Shu Y, Shao Y, Yin Y. Polymorphisms in the base excision repair pathway modulate prognosis of platinum-based chemotherapy in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2013;71:1287–1295. doi: 10.1007/s00280-013-2127-8. [DOI] [PubMed] [Google Scholar]


