Abstract
Background: Down syndrome is a condition which extra genetic material causes delays in child development, both mentally and physically. Strengthening the study of the neural defects of DS is of great significance. Methods: Ts65Dn mice were used in this study. We removed the brain and isolated their hippocampus. We customized 54 genes in one PCR arrays, included some important genes related to Alzheimer’s disease. The expression of genes were detected by RT-PCR. Results: PCR arrays contained 54 genes related to Alzheimer’s disease. After real-time PCR, three genes (Nae1, APP and Mapt) expressed differently in the hippocampus of Ts65Dn, compared with the normal mice. Nae1 was decreased significantly, while APP and Mapt were increased obviously. The levels of fold-changes of Nae1, APP and Mapt were 86.19, 4.49 and 2.89 respectively. Significantly different levels of expression were found in the Ts65Dn mice compared with the normal control group (P=0.00 for Nae1, P=0.02 for APP, P=0.01 for Mapt respectively). Conclusions: There are differential expressed genes in the hippocampus of Ts65Dn mice that may be closely related to Alzheimer’s disease. PCR array technology was used in the screening and identification of these genes.
Keywords: Down syndrome, hippocampus, PCR array, Alzheimer’s disease, Ts65Dn
Introduction
Down syndrome (DS) is one of the most common gross chromosomal abnormalities and birth defect. It is well-known that DS is a condition which extra genetic material causes delays in child development, both mentally and physically. Intellectual disabilities are the foremost and most debilitating trait, which causes the loss of cognitive abilities and the development of early onset Alzheimer’s disease (AD) [1]. The survival rate of people with DS has greatly improved in the past decades, with a current life expectancy of 60 years or longer [2]. However, the problems of mental retardation of Down syndrome patients appeared more and more prominent. Even with effective medical intervention and skills training, the IQ of DS patients could be significantly improved. However, their memory, athletic ability, and learning ability still have great defects [3,4]. It prevents the ability of language, learning and memory of DS patients, and thus restricts their life and social skills. Therefore, problems concerning cognitive mental retardation become more important to those with DS.
Alzheimer’s disease (AD) is a clinically heterogeneous neurodegenerative disease with a strong genetic component [5]. Several genes have been associated with AD risk for nearly 20 years, mainly including APP, PSEN-1, PSEN-2, ApoE, ACT, TREM2, SUMO1 [6]. Some genes were considered as a major genetic risk factor, some were used as biomarkers in AD diagnosis. They might be related to the occurrence and development of AD.
At the same time, it is well known that Down syndrome patients will also get serious problem by AD. DS patients are characterized by the early appearance of neurodegenerative diseases such as AD: 11% of DS patients have AD pathology at 40 years of age and 100% have AD lesions at >70 years of age [7]. On the other hand, it is well known that Alzheimer’s occurs in trisomy 21 since the gene encoding amyloid is in this chromosome. Thus these individuals have a higher load of amyloid that in turn results in a higher incidence of developing cognitive decline and Alzheimer’s dementia with aging [8]. So, whether Alzheimer’s diseases related genes also have to do with Down syndrome?
In present study, a target PCR array related to the Alzheimer’s diseases was constructed, and we observed the changes of gene expression in Ts65Dn mice.
Materials and methods
This study was conducted in the Changzhou Women and Children Hospital of Nanjing Medical University (Changzhou City, Jiangsu Province, China). The animals were bred in the Animal center of Jiangsu University (Zhenjiang City, Jiangsu Province, China). All efforts were made to minimize the suffering and number of animals used.
Animals
Five Ts65Dn mice carrying a partial trisomy of chromosome 16 were purchased from the Jackson Laboratories (Bar Harbor, ME, USA). Five normal C57BL/6JEi mice were used as the normal control group. Both groups were matched by age which were more than 14 weeks old. The animals’ health and comfort were monitored by the veterinary service. The animals had access to water and food according to the routine methods of animal breeding in the Ani-mal center of Jiangsu University.
Methods
Sample collection
Anesthetized animals were euthanized using 10% chloral hydrate (Changzhou First People’s Hospital, Changzhou, China). We removed the brain and isolated the hippocampus.
Total RNA extraction
Total RNA was isolated by TRIZOL (Invitrogen) extraction. After homogenization of tissue samples, insoluble material was removed from the homogenate by centrifugation at 12000× g for 10 minutes at 2 to 8°C The homogenized samples were incubated for 5 minutes at room temperature, and 0.2 ml chloroform per 1 ml of trizol reagent was added. The tubes were shaken vigorously by hand for 15 seconds and incubated at 15 to 30°C for 2 to 3 minutes. Then, the samples were centrifuged at no more than 12000× g for 15 minutes at 2 to 8°C, and the colorless upper aqueous phase was transferred to a fresh tube. The RNA was precipitated from the aqueous phase by mixing with isopropyl alcohol (0.5 ml isopropyl alcohol per 1 ml trizol reagent). Samples were incubated at 15 to 30°C for 10 minutes and centrifuged at no more than 12000× g for 10 minutes at 2 to 8°C. The supernatant was removed, and the RNA pellet was washed once with 75% ethanol, adding at least 1 ml 75% ethanol per 1 ml trizol Reagent. The samples were mixed by vortex and centrifuged at no more than 7500× g for 5 minutes at 2 to 8°C. Finally, the RNA pellets were dried by air or vacuum for 5-10 minutes, and RNA was dissolved in RNase-free water.
PCR arrays customization
PCR arrays contained 54 genes, 5 housekeeping genes, PPC and GDC. 54 genes were related to Alzheimer’s disease. The housekeeping genes were B2M, ACTB, GAPDH, RPL27, HPRT1 and OAZ1. PPC contains synthetic DNA fragments which have no homology with detected species and amplification primers, which was used as quality control. GDC was used to detect the residual genomic DNA.
PCR array experiment
Total RNA from each sample was used for reverse transcription with an RT-PCR Kit (catalog#CTB101; CT biosciences, China) on an ABI 9700 thermo cycler (ABI, Foster City, CA). For reverse transcription, 4 µl total RNA was mixed with 10 µl OligodT Primer (10 µM), and the solution was incubated at 70°C for 10 minutes and then quickly cooled on ice for 2 minutes. The cooled solution was mixed with 4 µl 5× reverse transcription buffer, 1 µl dNTP (10 mM), 0.5 µl RNasin (40 U/µl), and 0.5 µl reverse transcriptase (200 U/µl). Reverse transcription was performed at 42°C for 1 hour, followed by an inactivation reaction at 70°C for 15 minutes. The resulting cDNA was stored at -20°C until it had been used. PCR arrays were performed with customized PCR containing pre-dispensed primers (CT biosciences, China) on a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using SYBR Master Mix ((catalog#CTB101; CT biosciences, China). The thermo cycler parameters consisted of an initial denaturation at 95°C for 2 min followed by 45 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 20 s. Relative changes in gene expression were calculated using the ΔΔCt (threshold cycle) method. The housekeeping genes (B2M, ACTB, GAPDH, RPL27, HPRT1 and OAZ1) were used to normalize the amount of RNA. Fold change values were calculated using the formula: 2-ΔΔCt.
Statistical analysis
All data were collected and statistically analyzed using SPSS 13.0 software. Results of parameters were expressed as median (M), 2.5th percentile (P2.5) and 97.5th percentile (P97.5). Non-parametric tests (Mann-Whitney U test) was employed to compare differences for CP (median level) between two groups. A P-value of less than 0.05 was considered to be statistically significant.
Results
In present study, PCR arrays contained 54 genes which were related to Alzheimer’s disease. After real-time PCR, three genes (Nae1, APP and Mapt) expressed differently in the hippocampus of Ts65Dn, compared with the normal mice (Table 1). Nae1 was decreased significantly, while APP and Mapt were increased obviously. The old-changes levels of Nae1, APP and Mapt were 86.19, 4.49 and 2.89 respectively (Figure 1). The gene levels were compared by the value of 2-ΔΔCt for target genes to housekeeping genes expression in two groups. We found significantly different levels of expression in the Ts65Dn mice compared with the normal control group (P=0.00 for Nae1, P=0.02 for APP, P=0.01 for Mapt respectively). We compared the significant differences in the expression of Nae1, APP and Mapt using Box plots, as shown in (Figure 2).
Table 1.
Differential apoptotic gene expressions in the Hippocampus of Ts65Dn
| Gene symbol | Description of genes | Chromosome | Ts65Dn/Control group | ||
|---|---|---|---|---|---|
|
| |||||
| Human | Mouse | Fold changes | P value | ||
| Nae1 | NEDD8 activating enzyme E1 subunit 1 | 16 | 8 | -86.19 | 0.000245 |
| App | amyloid beta (A4) precursor protein | 21 | 16 | 4.49 | 0.021300 |
| Mapt | microtubule-associated protein tau | 17 | 11 | 2.89 | 0.010808 |
| Snca | synuclein, alpha | 4 | 6 | 6.66 | 0.0593829 |
| Mme | membrane metallo endopeptidase | 3 | 3 | 2.87 | 0.0663999 |
| Cdk5 | cyclin-dependent kinase 5 | 7 | 5 | 2.79 | 0.0699807 |
| Ncstn | nicastrin | 1 | 1 | 3.26 | 0.0712695 |
| Psen1 | presenilin 1 | 14 | 12 | 11.62 | 0.0743482 |
| Nos1 | nitric oxide synthase 1 | 5 | 12 | 2.77 | 0.0841564 |
| Bace1 | beta-site APP cleaving enzyme 1 | 11 | 9 | 2.96 | 0.0868817 |
| Apoe | apolipoprotein E | 19 | 7 | 2.52 | 0.103259 |
| Adam10 | a disintegrin and metallopeptidase domain 10 | 15 | 9 | 2.52 | 0.103259 |
| Atp2a1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | 16 | 7 | 2.52 | 0.103259 |
| Capn1 | calpain 1 | 11 | 19 | 2.52 | 0.103259 |
| Eif2ak3 | eukaryotic translation initiation factor 2-alpha kinase 3 | 6 | 2 | 2.52 | 0.103259 |
| Gnaq | guanine nucleotide binding protein, alpha q polypeptide | 9 | 19 | 2.52 | 0.103259 |
| Bace2 | beta-site APP-cleaving enzyme 2 | 21 | 16 | 2.52 | 0.103259 |
| Ern1 | endoplasmic reticulum (ER) to nucleus signalling 1 | 17 | 11 | 2.52 | 0.103259 |
| Tnfrsf1a | tumor necrosis factor receptor superfamily, member 1a | 12 | 6 | 2.52 | 0.103259 |
| Apaf1 | apoptotic peptidase activating factor 1 | 12 | 10 | 2.52 | 0.103259 |
| Casp8 | caspase 8 | 2 | 1 | 2.52 | 0.103259 |
| Casp9 | caspase 9 | 1 | 4 | 2.52 | 0.103259 |
| Fadd | Fas (TNFRSF6)-associated via death domain | 11 | 7 | 2.52 | 0.103259 |
| Casp3 | caspase 3 | 4 | 8 | 2.52 | 0.103259 |
| Il1a | interleukin 1 alpha | 2 | 2 | 2.52 | 0.103259 |
| Casp7 | caspase 7 | 10 | 19 | 2.52 | 0.103259 |
| Cdc2a | cyclin-dependent kinase 1 | 10 | 20 | 2.52 | 0.103259 |
| Aph1a | anterior pharynx defective 1a homolog (C. elegans) | 1 | 3 | -59.69 | 0.1262045 |
| Il1b | interleukin 1 beta | 2 | 2 | 6.75 | 0.1390537 |
| Mapk3 | mitogen-activated protein kinase 3 | 16 | 7 | 3.21 | 0.1722344 |
| Ide | insulin degrading enzyme | 10 | 19 | 2.98 | 0.1858611 |
| Hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | X | X | 6.04 | 0.216086 |
| Ryr3 | ryanodine receptor 3 | 2 | 15 | 4.59 | 0.2207425 |
| Plcb1 | phospholipase C, beta 1 | 20 | 2 | 4.81 | 0.222985 |
| Cox4i1 | cytochrome c oxidase subunit IV isoform 1 | 16 | 8 | 5.86 | 0.2703607 |
| Cycs | cytochrome c, somatic | 7 | 6 | -3.16 | 0.2720677 |
| Mapk1 | mitogen-activated protein kinase 1 | 22 | 16 | 6.59 | 0.2937304 |
| Bid | BH3 interacting domain death agonist | 22 | 6 | -2.03 | 0.3028315 |
| Calm1 | calmodulin 1 | 14 | 12 | -5.31 | 0.3548665 |
| Psen2 | presenilin 2 | 1 | 1 | 2.01 | 0.4103427 |
| Fas | Fas (TNF receptor superfamily member 6) | 10 | 19 | 3.23 | 0.4264696 |
| Casp4 | caspase 4 | 11 | 9 | 1.7 | 0.4756739 |
| Tnf | tumor necrosis factor | 6 | 17 | 2.01 | 0.5039548 |
| Cdk5r1 | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 17 | 11 | 1.1 | 0.6050327 |
| Atf6 | activating transcription factor 6 | 1 | 1 | 1.43 | 0.6661326 |
| Lpl | lipoprotein lipase | 8 | 8 | -1.87 | 0.6721144 |
| Gsk3b | glycogen synthase kinase 3 beta | 3 | 16 | 1.78 | 0.6980795 |
| Ppp3ca | protein phosphatase 3, catalytic subunit, alpha isoform | 4 | 3 | -1.09 | 0.7517183 |
| Ppp3cc | protein phosphatase 3, catalytic subunit, gamma isoform | 8 | 14 | 1.58 | 0.8033197 |
| Itpr1 | inositol 1,4,5-trisphosphate receptor 1 | 6 | 3 | 1.19 | 0.8720633 |
| Ppp3r1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | 2 | 11 | 1.22 | 0.8949331 |
| Bad | BCL2-associated agonist of cell death | 11 | 19 | 1.15 | 0.901663 |
| Apbb1 | amyloid beta (A4) precursor protein-binding, family B, member 1 | 11 | 7 | 1.1 | 0.9243038 |
| Lrp1 | low density lipoprotein receptor-related protein 1 | 12 | 10 | 1.02 | 0.9865195 |
Figure 1.
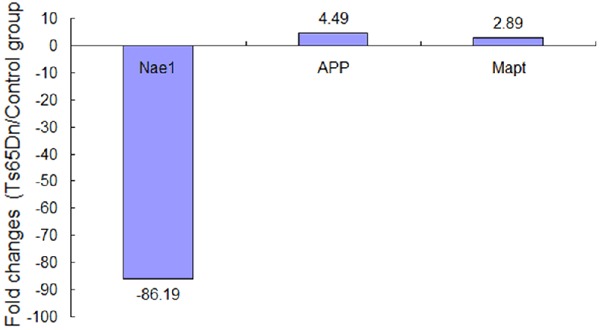
Fold change expressions of genes between Ts65Dn and control group. Compared with control group, the expressions of Nae1 were decreased and App, Mapt were increased in the hippocampus tissues of Ts65Dn.
Figure 2.
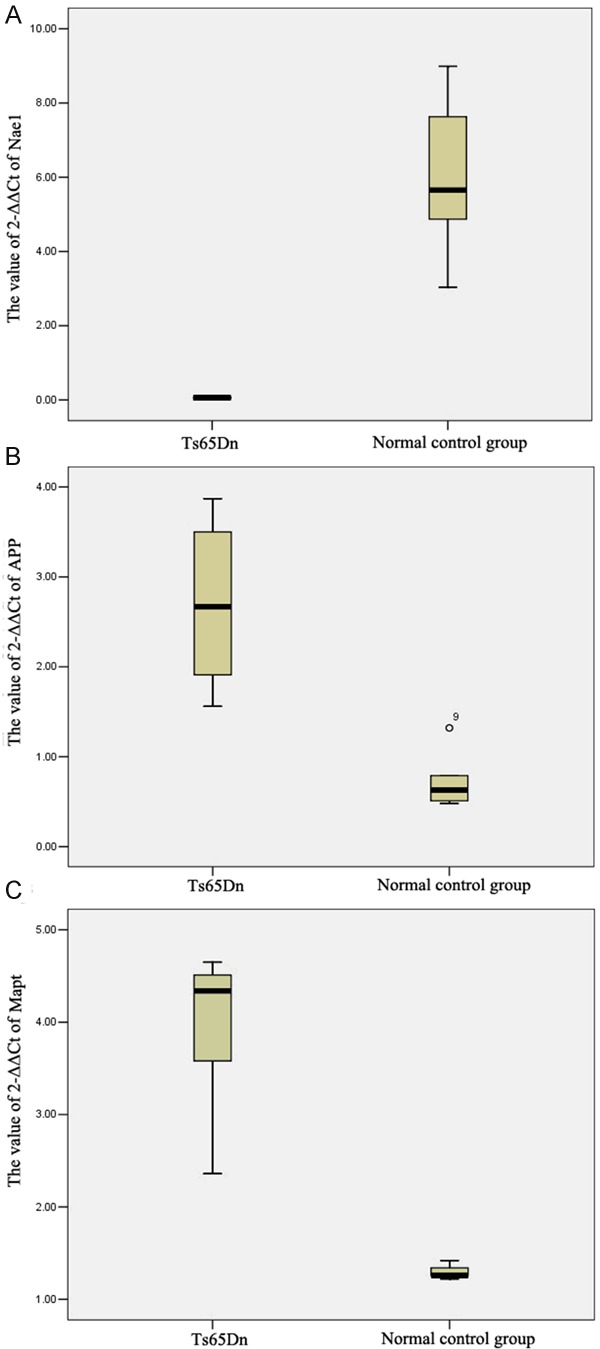
Comparison of the expressions of Nae1, APP and Mapt by Box plots. Box plots show the comparison of the value of 2-ΔΔCt of Ts65Dn and normal control group. Data were compared using the t-test. A: Nae1, B: APP, C: Mapt
In present study, we also detected the genes of ApoE, which were more than a contributing factor to neurodegeneration [9,10]. Some reports also showed that they have been linked to early onset of Alzheimer’s disease. However, it had no significant differences in expression levels. Compared with normal control group, ApoE was increased in Ts65Dn mice ant its levels of fold-changes was 2.52 (P=0.103259) (Table 1).
Discussion
In the study, we discovered three genes were differently expressed in the hippocampus of Ts65Dn by a target PCR array, which included 54 genes related to Alzheimer’s disease.
Down syndrome is characterized by the early appearance of neurodegenerative diseases such as Alzheimer’s disease. Cognitive disabilities in DS from appearance to result mainly two pathological processes neurogenesis impairment and Alzheimer-like degeneration [8]. On the other hand, it is well known that Alzheimer’s occurs in trisomy 21 since the gene encoding amyloid is in this chromosome. Therefore, they might have similar pathological processes between the two kinds of disease. Recently, some genes have been associated with AD risk were paid more and more attention, such as APP, ApoE, PSEN-1, PSEN-2, ACT, TREM2, SUMO1[6]. So, whether they also have to do with Down syndrome? In this study, we costumed them in one chip and detected their expression by PCR.
There are so many molecules involved in the process of Alzheimer’s disease that real-time quantitative PCR may not be suitable for observing them. A high-throughput platform with efficient detection capacity is needed to address this issue. Recently, PCR arrays have been considered the most reliable tools for analyzing the expression for a focused panel of genes, especially in signal transduction pathways, biological process or disease related gene networks [11]. PCR arrays have been used for cancer, immunology, stem cells, toxicology, and many other areas of biological and medical research [12,13].
In PCR arrays, we found that the Nae1, APP and Mapt genes showed significantly altered expression in Ts65Dn mice. They might be play important roles in the early appearance of Alzheimer’s disease in Down syndrome patients. Nae1 encodes the protein NEDD8-activating enzyme E1 regulatory subunit, which is bound to the beta-amyloid precursor protein. Beta-amyloid precursor protein is a cell surface protein with signal-transducing properties, and it is thought to play a role in the pathogenesis of Alzheimer’s disease [14]. However, there has been no reports about Nae1 and Down syndrome until now. APP is a familiar gene. It encodes a cell surface receptor and transmembrane precursor protein, which is the major component of amyloid plaques found in the brains of Alzheimer’s patients [15]. Mutations in APP have been linked is ralative to early onset Alzheimer’s disease [16]. As a cell surface receptor, it performs physiological functions on the surface of neurons relevant to neurite growth, neuronal adhesion and axonogenesis. Furthermore, it has been associated with other diseases including Lewy body dementia, inclusion body myositis and cerebral amyloid angiopathy. Mapt gene encodes the microtubule-associated protein tau which is differentially expressed in the nervous system, depending on is relative to the stage of neuronal maturation and neuron type. Its main functions are promoting microtubule assembly and stability, and may be involved in the establishment and maintenance of neuronal polarity. According to the current reports, Mapt gene mutations have been associated with several neurodegenerative disorders [17] such as Alzheimer’s disease, Parkinson’s disease [18], frontotemporal dementia, cortico-basal degeneration and progressive supranuclear palsy.
In present study, we did a interesting but preliminary research. We found three gene which were related to Alzheimer’s disease also differently expressed in the brain of Down syndrome mice. The functions of three genes played a important role in several neurodegenerative disorders. We need further research to prove whether they also have to do with Down syndrome.
Acknowledgements
We thank all the project participants for their contributions. This study was supported by grants from the major projects of Jiangsu Maternal and Child Health (F201217), the project of the health department of Jiangsu Province (H201352), the Changzhou Research Program of Applied Basic (CJ20140055) and the youth project of Health Bureau of Changzhou City (QN201405).
Disclosure of conflict of interest
None.
References
- 1.Nieuwenhuis-Mark RE. Diagnosing Alzheimer’s dementia in Down syndrome: problems and possible solutions. Res Dev Disabil. 2009;30:827–838. doi: 10.1016/j.ridd.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Bittles AH, Bower C, Hussain R, Glasson EJ. The four ages of Down syndrome. Eur J Public Health. 2007;17:221–225. doi: 10.1093/eurpub/ckl103. [DOI] [PubMed] [Google Scholar]
- 3.Couzens D, Cuskelly M, Haynes M. Cognitive development and Down syndrome: age-related change on the Stanford-Binet test (fourth edition) Am J Intellect Dev Disabil. 2011;116:181–204. doi: 10.1352/1944-7558-116.3.181. [DOI] [PubMed] [Google Scholar]
- 4.Rihtman T, Tekuzener E, Parush S, Tenenbaum A, Bachrach SJ, Ornoy A. Are the cognitive functions of children with Down syndrome related to their participation? Dev Med Child Neurol. 2010;52:72–78. doi: 10.1111/j.1469-8749.2009.03356.x. [DOI] [PubMed] [Google Scholar]
- 5.Karch CM, Cruchaga C, Goate AM. Alzheimer’s Disease Genetics: From the Bench to the Clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DH, Yeo SH, Park JM, Choi JY, Lee TH, Park SY, Ock MS, Eo J, Kim HS, Cha HJ. Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene. 2014;545:185–93. doi: 10.1016/j.gene.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Coppus A, Evenhuis H, Verberne GJ, Visser F, van Gool P, Eikelenboom P, van Duijin C. Dementia and mortality in persons with Down’s syndrome. J Intellect Disabil Res. 2006;50:768–777. doi: 10.1111/j.1365-2788.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 8.Contestabile A, Benfenati F, Gasparini L. Communication breaks-Down: From neurodevelopment defects to cognitive disabilities in Down syndrome. Prog Neurobiol. 2010;91:1–22. doi: 10.1016/j.pneurobio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Mahley RW, Huang Y. Apolipoprotein (apo) E4 and Alzheimer’s disease: unique conformational and biophysical properties of apoE4 can modulate neuropathology. Acta Neurol Scand Suppl. 2006;185:8–14. doi: 10.1111/j.1600-0404.2006.00679.x. [DOI] [PubMed] [Google Scholar]
- 10.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svensson K, Granberg M, Karlsson L, Neubauerova V, Forsman M, Johansson A. A real-time PCR array for hierarchical identification of Francisella isolates. PLoS One. 2009;4:e8360. doi: 10.1371/journal.pone.0008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi W, Li X, Hou X, Peng H, Jiang Q, Shi M, Ji Y, Liu X, Liu J. Differential apoptosis gene expressions of rhabdomyosarcoma cells in response to enterovirus 71 infection. BMC Infect Dis. 2012;12:327. doi: 10.1186/1471-2334-12-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Xu H, Lam SH, Gong Z. Selection of reliable biomarkers from PCR array analyses using relative distance computational model: methodology and proof-of-concept study. PLoS One. 2013;8:e83954. doi: 10.1371/journal.pone.0083954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 2000;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jönsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 16.Nicolas M, Hassan BA. Amyloid precursor protein and neural development. Development. 2014;141:2543–2548. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- 17.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, Luk C, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, Singleton AB, Cookson MR, Pittman AM, de Silva R, Weale ME, Hardy J, Ryten M. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin MK, Farrer MJ. Genetics and genomics of Parkinson’s disease. Genome Med. 2014;6:48. doi: 10.1186/gm566. [DOI] [PMC free article] [PubMed] [Google Scholar]


