Abstract
Background: Long non-coding RNAs (lncRNAs) play widespread roles in gene regulation and cellular processes. However, the functional roles of lncRNAs in hepatocellular carcinoma (HCC) are not yet well elucidated. The aim of the present study was to measure the levels of lncRNA PCAT-1 expression in HCC and evaluate its clinical significance in the development and progression of HCC. Methods: We examined the expression of PCAT-1 in 117 HCC tissues and adjacent non-tumor tissues using quantitative real-time-PCR and analyzed its correlation with the clinical parameters. Results: Our data showed that PCAT-1 expression in HCC tissues was significantly increased compared with adjacent non-tumor tissues (P<0.05). Up-regulated expression of PCAT-1 was significantly associated with TNM stage and metastasis (P<0.05), but not other clinical parameters. Moreover, Kaplan-Meier survival analysis showed that a high expression level of PCAT-1 resulted in a significantly poor overall survival of HCC patients. The multivariate Cox regression analysis demonstrated that PCAT-1 expression level was an independent prognostic factor for the overall survival rate of HCC patients. Conclusions: Our data suggested that the increased expression of PCAT-1 was associated with advanced clinical parameters and poor overall survival of HCC patients, indicating that PCAT-1 up-regulation may serve as a novel biomarker of poor prognosis in HCC patients.
Keywords: PCAT-1, lncRNA, hepatocellular carcinoma, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the most common primary hepatic malignancy and ranks the fifth most prevalent malignant tumors worldwide [1]. It is more prevalent in Central Africa and parts of Asia, and half of the worldwide instances are reported in China [2]. Nowadays surgical resection is still the main method of therapy, however, it does not have favorable effect with 5 year overall survival rate about 50% [3,4]. This tragic situation stems from a lack of early diagnosis, the deteriorated condition of the cirrhotic liver from which most HCC cases develop, and the high resistance of HCC to chemotherapy [5]. Therefore, the identification of novel molecular mechanisms involved in the development and progression of HCC may provide new strategies for the diagnosis and treatment of this life threatening disease.
Apart from about 2% protein coding genes, the vast majority of the human genome is made up of non-coding RNAs (ncRNAs), indicating that ncRNAs could play significant regulatory roles in complex organisms [6]. Among them, the long non-coding RNA (lncRNA) is an RNA molecular that is longer than 200 nucleotides and cannot be translated into a protein [7]. Recent reports suggested that lncRNAs could play an important role in cancer, such as the process of carcinogenesis, invasion, and metastasis of cancer [8]. The deregulation of lncRNAs has been found in many types of cancer. For example, Deng et al showed lncRNA 91H expression was significantly increased in colorectal cancer, the elevated expression of 91H was associated with poor prognosis and distant metastasis. Moreover, they indicated that knockdown of 91H inhibited the proliferation, migration, and invasiveness of colorectal cancer cells in vitro [9]. Chen et al lncRNA HOTAIR was increased in esophageal squamous cell carcinoma tissues and associated with poor prognosis. Furthermore, they demonstrated that knockdown of HOTAIR reduced esophageal squamous cell invasiveness and migration in vitro [10]. Shi et al revealed that GAS5 expression was down-regulated in non-small cell lung cancer tissues and correlated with tumor size and TNM stage, in addition, they indicated that GAS5 over-expression increased tumor cell growth arrest and induced apoptosis in vitro and in vivo [11]. However, the emerging functional role of lncRNAs in HCC remains poorly understood.
In the present study, we examined the expression of PCAT-1 in HCC tissues and analyzed the correlation between PCAT-1 levels, clinical features, and patient overall survival to determine whether PCAT-1 levels represent a prognostic factor for clinical outcomes in HCC patients. These data provide novel insights into the role of PCAT-1 in the progression of HCC and identify a potential therapeutic target in HCC patients.
Materials and methods
Patients and tissue samples
The study was approved by the Research Ethics Committee of The First Clinical Medical College of Fujian Medical University, Fuzhou, China. Informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to accepted ethical and legal standards. A total of 117 patients who presented with primary HCC and underwent curative hepatectomy at The First Clinical Medical College of Fujian Medical University, Fuzhou, China were included in this retrospective study. All HCC specimens and adjacent non-tumor tissues were snap-frozen in liquid nitrogen and stored at -80°C following surgery for quantitative real-time PCR (qRT-PCR). The tissue samples used in this study were retrieved from the tissue bank of the Department of Pathology. The patients had been diagnosed with HCC between 2006 and 2008. None of the patients recruited in this study had undergone preoperative chemotherapy or radiotherapy. The diagnosis was confirmed by histopathological study. Tumor stage was determined according to the 2002 International Union Against Cancer TNM classification system [12]. Tumor differentiation was graded by the Edmondson grading system. The clinicopathological features of the 117 patients are summarized in Table 1.
Table 1.
Correlation between PCAT-1 expression and clinicopathological features of HCC
| N=117 | PCAT-1 expression | ||||
|---|---|---|---|---|---|
|
|
|||||
| Variable | High (n=59) | Low (n=58) | χ2 | P value | |
| Gender | 1.521 | 0.217 | |||
| Male | 68 | 31 | 37 | ||
| Female | 49 | 28 | 21 | ||
| Age | 0.684 | 0.408 | |||
| ≤50 | 52 | 24 | 28 | ||
| >50 | 65 | 35 | 30 | ||
| Tumor size (cm) | 1.152 | 0.283 | |||
| ≤5 | 44 | 25 | 19 | ||
| >5 | 73 | 34 | 39 | ||
| Tumor number | 1.991 | 0.158 | |||
| Solitary | 86 | 40 | 46 | ||
| Multiple | 31 | 19 | 12 | ||
| Serum AFP (ng/ml) | 1.094 | 0.296 | |||
| <20 | 39 | 17 | 22 | ||
| ≥20 | 78 | 42 | 36 | ||
| Liver cirrhosis | 0.417 | 0.518 | |||
| Yes | 74 | 39 | 35 | ||
| No | 43 | 20 | 23 | ||
| Tumor differentiation | 1.463 | 0.226 | |||
| Well/Moderate | 62 | 28 | 34 | ||
| Poor | 55 | 31 | 24 | ||
| TNM stage | |||||
| I-II | 42 | 17 | 35 | 11.777 | 0.001 |
| III-IV | 75 | 42 | 23 | ||
| Metastasis | 5.029 | 0.025 | |||
| Yes | 24 | 17 | 7 | ||
| No | 93 | 42 | 51 | ||
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from tissues using TRIzol reagent (Invitrogen). For qRT-PCR, RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara). Real-time PCR analyses were performed with Power SYBR Green (Takara). Results were normalized to the expression of GAPDH. The PCR primers for PCAT-1 or GAPDH were as follows: PCAT-1 sense, 5’-TGAGAAGAGAAATCTA TTGGAACC-3’ and reverse, 5’-GGTTTGTC-TCCGCTGCTTTA-3’; GAPDH sense, 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse, 5’-AGTCTTCTGGGTGGCAGTGAT-3’. qRT-PCR and data collection were performed on ABI 7500. The relative expression of PCAT-1 was calculated and normalized using the 2-ΔΔCt method relative to GAPDH.
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 software. The gene expression levels in HCC were compared with those in adjacent non-tumor tissues with the use of Wilcoxon test. Measurement data were analyzed using Student’s t-test, while categorical data were studied using chi-square test. The postoperative survival rate was analyzed with Kaplan-Meier method, and differences in survival rates were assessed with log-rank test. To determine independent prognostic factors, the Cox multivariate regression analysis was used. P values lower than 0.05 were considered statistically significant.
Results
PCAT-1 expression is up-regulated in human HCC tissue
We examined PCAT-1 expression level in 117 paired HCC tissues and adjacent non-tumor tissues by qRT-PCR, and normalized to GAPDH. Figure 1A showed that the PCAT-1 expression was significantly increased in HCC tissues compared with adjacent non-tumor tissues (P<0.05). In tumor tissues, PCAT-1 expression was at a level higher than that of non-tumor tissues, with the median ratio of 2.79 compared with normal counterparts. These data indicated that abnormal PCAT-1 expression may be related to HCC pathogenesis.
Figure 1.
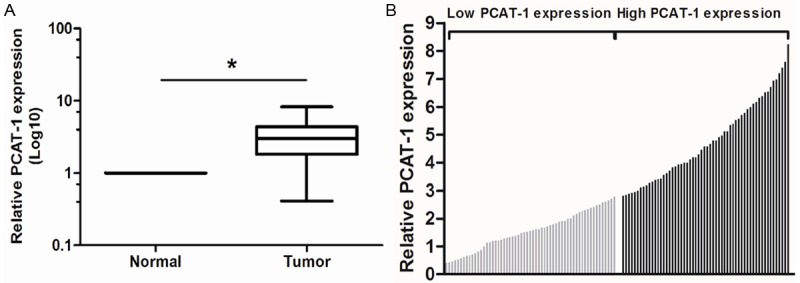
Relative PCAT-1 expression in HCC tissues and its clinical significance. A. Relative expression of PCAT-1 in HCC tissues in comparison with adjacent non-tumor tissues. PCAT-1 expression was examined by qRT-PCR and normalized to GAPDH expression. Data was presented as fold-change in tumor tissues relative to normal tissues. B. The 117 total HCC patients included in the study were divided into a low PCAT-1 group (n=58) and a high PCAT-1 group (n=59) according to the median value of relative PCAT-1 expression. *P<0.05.
Relationship between PCAT-1 expression and clinicopathological features in HCC patients
The clinical pathology findings of 117 HCC patients are shown in Table 1. According to the median ratio of relative PCAT-1 expression (2.79) in tumor tissues, the 117 HCC patients were divided into two groups: relative high PCAT-1 group (PCAT-1 expression ratio ≥ median ratio) and relative low PCAT-1 group (PCAT-1 expression ratio < median ratio) (Figure 1B). Clinicopathologic features were compared between the two groups (Table 1). The PCAT-1 expression level in HCC was associated advanced TNM stage and metastasis (P<0.05), but not with other parameters such as gender, age, tumor size, tumor number, serum AFP, liver cirrhosis and tumor differentiation (P>0.05) (Table 1).
Correlation between PCAT-1 expression and patients’ survival
We further examined whether PCAT-1 expression level correlated with outcome of HCC patients. Overall survival curves were plotted according to PCAT-1 expression level by the Kaplan-Meier analysis and log-rank test. As shown in Figure 2, the HCC patients with high PCAT-1 expression had shorter overall survival than those with low PCAT-1 expression (P<0.05). The univariate and multivariate analyses were also performed to identify factors related to patient prognosis. As shown in Table 2, the univariate analysis showed that tumor differentiation, TNM stage, metastasis and PCAT-1 expression were significantly related to postoperative survival (P<0.05). Moreover, the multivariate Cox regression analysis indicated that tumor differentiation, TNM stage, metastasis and PCAT-1 expression were independent predictors for overall survival of HCC patients (P<0.05). These findings support the hypothesis that increased PCAT-1 expression play a key role in HCC development and progression.
Figure 2.
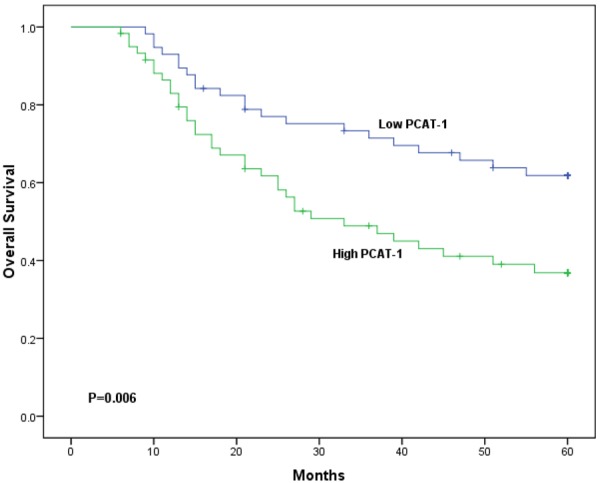
Kaplan-Meier survival curves of patients with HCC based on PCAT-1 expression status (P<0.05, log-rank test).
Table 2.
Univariate and multivariate analyses of overall survival of HCC patients
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Gender | 1.317 | 0.692-2.156 | 0.473 | |||
| Male vs Female | ||||||
| Age (years) | 0.894 | 0.447-1.538 | 0.377 | |||
| >50 vs ≤50 | ||||||
| Tumor size (cm) | 1.502 | 0.615-2.753 | 0.215 | |||
| >5 vs ≤5 | ||||||
| Tumor number | 1.208 | 0.731-1.871 | 0.269 | |||
| Multiple vs Solitary | ||||||
| AFP (ng/ml) | 1.487 | 0.642-3.175 | 0.185 | |||
| >20 vs ≤20 | ||||||
| Liver cirrhosis | 1.912 | 0.548-3.219 | 0.087 | |||
| Yes vs No | ||||||
| Tumor differentiation | 1.684 | 0.862-2.825 | 0.033 | 1.513 | 0.716-2.629 | 0.027 |
| Poor vs Well/Moderate | ||||||
| TNM stage | 3.617 | 1.218-8.391 | 0.012 | 3.275 | 1.116-7.948 | 0.009 |
| III-IV vs I-II | ||||||
| Metastasis | 4.319 | 1.478-9.247 | 0.007 | 4.168 | 1.395-8.738 | 0.003 |
| Yes vs No | ||||||
| LncRNA PCAT-1 | 3.372 | 1.268-8.548 | 0.002 | 2.977 | 1.107-7.912 | 0.001 |
| High vs Low | ||||||
Discussion
LncRNAs dysregulation may affect epigenetic information and provide a cellular growth advantage, resulting in progressive and uncontrolled tumor growth [13,14]. Effective control of both cell survival and cell proliferation is critical to the prevention of oncogenesis and to successful cancer therapy. Therefore, identification of cancer associated lncRNAs and investigation of their clinical significance and functions may provide a missing piece of the well-known oncogenic and tumor suppressor network puzzle.
PCAT-1 (prostate cancer-associated ncRNA transcripts 1), a lncRNA, has been discovered by RNA sequence recently that implicated in disease progression of patient with prostate cancer. Prensner et al showed that PCAT-1 increased in prostate cancer and promoted prostate cancer cell proliferation through association with polycomb repressive complex 2 (PRC2) and cMyc protein, indicating that PCAT-1 can be used as potential therapeutic target of prostate cancer [15,16]. Recently, they reported that PCAT-1 could produce a functional deficiency in homologous recombination through its repression of the BRCA2 tumor suppressor, which imparts a high sensitivity to small molecule inhibitors of PARP1, indicating that PCAT-1 could regulate cell response to genotoxic stress [17]. Ge et al revealed that PCAT-1 expression was increased in colorectal cancer tissues and was highly related to distance metastasis. Furthermore, they demonstrated that colorectal cancer patients with PCAT-1 higher expression have a shorter overall survival than those with lower PCAT-1 expression [18]. Those studies indicated that PCAT-1 play a critical role in tumor progression. However, the relationship between PCAT-1 expression and HCC development and/or progression is still unknown.
Our study was designed to investigate the expression and prognostic significance of PCAT-1 in HCC patients. PCAT-1 expression was retrospectively analyzed in 117 HCC patients. Results were assessed for association with clinical features and overall survival of HCC patients after surgery. Prognostic values of PCAT-1 expression and clinical outcomes were also evaluated by Cox regression analysis. Our results showed that PCAT-1 expression was elevated in HCC tissues and associated with TNM stage and metastasis. More importantly, we found that PCAT-1 expression was significantly associated with overall survival of HCC patients. In support of this, Kaplan-Meier analysis of overall survival revealed that HCC patients with high PCAT-1 expression tend to have a poorer overall survival, indicating that high PCAT-1 expression is a marker of poor prognosis for overall survival of patients with HCC. Multivariate Cox regression analysis proved that PCAT-1 was an independent prognostic factor for HCC. Taken together, our study demonstrated that over-expression of PCAT-1 in HCC is associated with more malignant phenotypes and a worse prognosis implies that it could play a oncogenic role in hepatocellular carcinogenesis.
In summary, to the best of our knowledge, the present study is the first to report that lncRNA PCAT-1 expression was up-regulated in HCC tissues and associated with biological aggressiveness and poor prognosis. Our study indicated that PCAT-1 was an independent prognostic factor of HCC patients. These findings suggested that PCAT-1 could be a potential diagnostic and therapeutic target in patients with HCC.
Acknowledgements
This work was supported by grants from Foundation of LongYan Medical Science and Technique Key Program (No. 2013LY57).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 6.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X, Chen J, Wang S. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. doi: 10.1371/journal.pone.0103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, Wang CM, Lv J, De W, Wu M, Cao XF. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 11.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2013 doi: 10.1002/mc.22120. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Greene FL. The American Joint Committee on Cancer: updating the strategies in cancer staging. Bull Am Coll Surg. 2002;87:13–15. [PubMed] [Google Scholar]
- 13.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, Lawrence TS, Chinnaiyan AM, Feng FY. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, Logothetis CJ, Araujo JC, Pisters LL, Tewari AK, Canman CE, Knudsen KE, Kitabayashi N, Rubin MA, Demichelis F, Lawrence TS, Chinnaiyan AM, Feng FY. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]


