Abstract
Objective: Overexpression of MicroRNA-196a (miR-196a) has recently been reported in different types of human cancers. However, the prognostic value of miR-196a in ovarian carcinoma remains unknown. In this study, we investigated the expression of miR-196a in ovarian carcinoma and its relationship with tumor progression and clinical prognosis. Methods: The expression level of miR-196a was examined by quantitative Real-time PCR (qRT-PCR) in surgically removed ovarian cancer tissues and ovarian cancer cell lines. The correlation between miR-196a expression and clinical features and prognosis were statistically analyzed. Results: The results showed that the miR-196a expression was significantly upregulated in tumor tissues and ovarian cancer cell lines compared with that in normal ovarian surface tissues and normal ovarian epithelial cells. Moreover, miR-196a expression was positively correlated with FIGO stage (P <0.001), tumor size (P =0.020), and lymph nodes metastasis (P =0.019). Kaplan-Meier analysis demonstrated that high levels of miR-196a expression was associated with poorer overall survival (P <0.001) and recurrent-free survival (P =0.003), especially in patients with advanced disease (P =0.002). Multivariate analysis suggested that miR-196a expression was an independent prognostic factor for overall survival of patients with ovarian carcinoma. Conclusions: In conclusion, miR-196a may play an important role in the progression of ovarian carcinoma, and could be used as an independent prognostic biomarker for patients with ovarian carcinoma.
Keywords: miR-196a, ovarian cancer, overall survival, recurrence-free survival
Introduction
Ovarian cancer is the second most common gynecologic malignancy among women and is the fourth leading cause of cancer-related deaths among women [1]. Despite significant improvements have been achieved in the diagnostics and treatment strategies including surgery, chemo-radiotherapy, targeted therapy, and multidisciplinary treatments, the clinical outcome of ovarian cancer patients is still very poor, with an overall 5 year survival rate of less than 50% from the time of diagnosis [2]. Furthermore, due to the lack of early symptoms, ovarian carcinoma is usually diagnosed at an advanced metastatic stage when the survivability of the patient is seriously compromised [3]. Therefore, it is necessary to develop novel and reliable biomarker to evaluate the prognosis and efficacy of therapeutic strategies for ovarian cancer.
MicroRNAs (miRNAs) are small noncoding RNA molecules of approximately 16-22 nucleotides that regulate expression of their target gene by inhibiting mRNA translation or inducing mRNA cleavage [4]. Although the precise functions of miRNAs are limited, it has been revealed that miRNAs play crucial roles in a variety of biological processes, including cell proliferation, differentiation, morphogenesis, embryo development, and apoptosis [5-7]. In recent years, numerous studies have implicated that certain miRNAs are aberrantly expressed in human cancers, and that dysregulation of miRNAs is associated with carcinogenesis and tumor development. In particular, miRNAs may function as oncogenes or tumor suppressors depending on the functions of their targets [8].
Increasing evidence reveals that miR-196a plays an important role in cell proliferation and tumor progression. However, the expression pattern and clinical significance of miR-196a in ovarian cancer remain unknown. In the present study, we investigated the expression levels of miR-196a in epithelial ovarian cancer (EOC) and ovarian cancer cell lines. We also assessed the relationship between miR-196a levels and clinicopathologic features of EOC. Moreover, we evaluated its relation with clinical prognosis of patients.
Materials and methods
Patients and clinical samples
All tumor and matched normal ovarian surface tissue samples were obtained from 156 EOC patients who underwent surgery at Cangzhou Central Hospital of Hebei Medical University between January 2004 and December 2007. None of these patients had received chemo-radiotherapy before surgery. For all cases, EOC specimens were dissected from the resected tumors, and non-tumor tissues were obtained from the normal-appearing ovary removed from patients. All resected 146 EOC tissues, 10 paired EOC, and corresponding normal ovarian surface tissue specimens were immediately frozen in liquid nitrogen after surgical removal and stored at -80 degree until analysis. The specimens were stained with hematoxylin and eosin and examined histopathologically. The present study was approved by the Institutional Review Board of Cangzhou Central Hospital. Written informed consent was obtained from all the members who participated in this study.
Cell lines
Primary normal ovarian epithelial cells (NOEC), obtained from normal ovaries, were cultured in Ham’s F-12 (Gibco, NY) supplemented with 20% fetal calf serum (Gibco, NY) and antibiotics (120 mg/ml streptomycin and 120 mg/ml penicillin). Ovarian cancer cell lines (SKOV3, OVCAR-3, ES-2, HO8910, MCV152, A2780, CAOV-3) were grown in the DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 1% penicillin/streptomycin (Invitrogen).
Quantitative real-time reverse transcriptase-PCR
Total RNA was extracted from cultured cells and frozen ovarian cancer tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. All of the manipulations of the RNA were carried out under RNase-free conditions. The concentration and purity of RNA were measured spectrophotometrically at 260 and 280 nm. cDNA were generated from 1 µl total RNA using One Step PrimeScript miRNA cDNA Synthesis Kit (Takara, Japan) according to the manufacture’s instructions. Then, quantitative real-time PCR was performed using SYBR RT-PCR kit (Takara) on an ABI 7500 Real-Time PCR System (Applied Biosystems, USA). U6 small nuclear RNA (snRNA) was used as an internal control. The threshold cycle (Ct) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The fold change of miRNA expression was calculated using the 2-ΔΔCt method after normalization to U6 expression.
Statistical analysis
All statistical analyses were carried out using the software of SPSS version 19.0 for Windows (SPSS Inc, IL, USA). Data were expressed as mean ± SD. Student’s t test was performed to evaluate the difference of miR-196a expression between EOC and non-tumor tissues. Cases were divided into two groups, high or low, using the median expression level of miR-196a in cancer tissues as a cutoff. The Fisher’s exact test and chi-square test were performed to evaluate the associations between miR-196a expression and different clinicopathological characteristics. Survival analyses were performed using the Kaplan-Meier method, and differences between curves were compared by the log-rank test. Differences were considered statistically significant when P was less than 0.05.
Results
Upregulation of miR-196a in human ovarian cancer
In order to explore the role of miR-196a in the development of ovarian cancer, we detect the expression level of miR-196a in 10 pairs of ovarian cancer and normal human ovarian tissues by qRT-PCR assay. As shown in Figure 1A, after normalization to U6 expression levels, the expression level of miR-196a in EOC tissues was significantly lower than that in normal human ovarian tissues (median fold change of T/NT = 6.38, P <0.05). To further characterize the expression of miR-196a in ovarian cancer, we performed real-time RT-PCR analyses and found that miR-196a was markedly up-regulated to various levels in all seven ovarian cancer cell lines examined compared with NOEC (Figure 1B).
Figure 1.
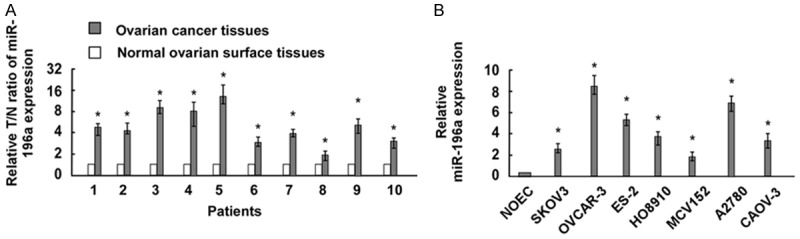
miR-196a expression is increased in ovarian cancer tissues and cell lines. A. The expression of miR-196a in 10 paired 10 pairs of ovarian cancer and normal human ovarian tissues. MiR-196a expression was normalized by U6 expression. Bars represent the means of three independent experiments. B. Real-time PCR analysis of miR-196a expression in normal ovarian epithelial cells (NOEC) and ovarian cancer cell lines (SKOV3, OVCAR-3, ES-2, HO8910, MCV152, A2780, and CAOV-3). *P <0.05.
Relationship between miR-196a expression and clinicopathological characteristics
To further investigate the role of miR-196a in the development and progression of ovarian cancer, we summarized the association between miR-196a expression and clinicopathological characteristics of EOC patients. As shown in Table 1, miR-196a expression was correlated with tumor stage, tumor size, and lymph nodes metastasis. miR-196a expression level was high in EOC patients with advanced stage tumors (P <0.001). In addition, high expression of miR-196a was more frequently detected in patients with large tumor (P =0.020). Moreover, low expression of miR-196a was more frequently detected in patients with lymph nodes metastasis (P =0.019). However, there were no significant correlations between miR-196a expression and other clinicopathologic features, such as age, histological type, or grade.
Table 1.
Correlation between miR-196a expression with clinicopathologic features of EOC
| Parameters | Category | No. | miR-196a level | P | |
|---|---|---|---|---|---|
|
| |||||
| high | low | ||||
| Age (y) | ≤50 | 57 | 30 | 27 | 0.658 |
| >50 | 89 | 43 | 45 | ||
| FIGO stage | I-II | 62 | 9 | 53 | <0.001 |
| III-IV | 84 | 64 | 20 | ||
| Differentiation | 1/2 | 97 | 45 | 52 | 0.220 |
| 3 | 49 | 28 | 21 | ||
| Tumor size | ≤5 cm | 64 | 25 | 39 | 0.020 |
| >5 cm | 82 | 48 | 34 | ||
| Histological type | Serous | 78 | 41 | 37 | 0.859 |
| Endometrioid | 48 | 22 | 26 | ||
| Mucinous | 13 | 7 | 6 | ||
| Clear cell | 7 | 3 | 4 | ||
| LN Metastasis | No | 119 | 54 | 65 | 0.019 |
| Yes | 27 | 19 | 8 | ||
Association of miR-196a expression with prognosis of EOC patients
To evaluate the prognostic value of miR-196a expression in EOC, survival curves were constructed by Kaplan-Meier method and compared by the log-rank test. As shown in Figure 2, patients with higher miR-196a expression had shorter overall (P <0.001, Figure 2A) and recurrence-free (P =0.003, Figure 2B) survival time than that with lower miR-196a expression. Among patients with stage III-IV EOC, the 5-year overall survival rate was significantly lower in patients with high miR-196a tumors than in those with low miR-196a tumors (P =0.002, Figure 2D). However, no difference was observed in the survival of patients with stage I-II EOC according to miR-196a status (P =0.389, Figure 2C). Multivariate analysis showed that miR-196a expression was an independent prognostic factor for the overall survival of EOC patients (Table 2). These results suggested that miR-196a expression could be used as a powerful independent prognostic factor in EOC patients.
Figure 2.
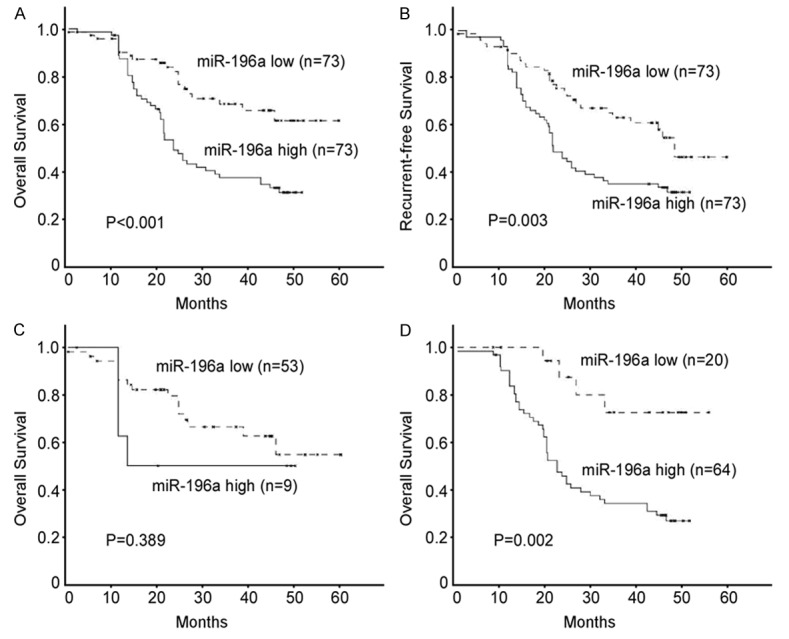
Correlation between expression levels of miR-196a and patients’ survival. Patients with higher miR-196a expression were closely correlated with poorer overall (A) and recurrence-free survival (B) than patients with tumor with lower miR-196a expression. (C, D) Overall survival in relation to the miR-196a status in stage I-II (C) and stage III-IV (D) patients.
Table 2.
Multivariate Cox regression analysis of OS and RFS in EOC patients
| Prognostic variables | OS | RFS | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (>50 vs. ≤50) | 1.346 (0.216-4.159) | 0.647 | 1.417 (0.257-5.247) | 0.672 |
| FIGO Stage (III-IV vs. I-II) | 2.448 (0.813-6.276) | 0.036 | 2.549 (0.974-7.172) | 0.045 |
| Differentiation (Grade 3 vs. 1/2) | 1.245 (0.024-4.367) | 0.757 | 1.313 (0.025-6.211) | 0.424 |
| Tumor size (>5 cm vs. ≤5 cm) | 2.465 (1.158-5.560) | 0.074 | 2.624 (1.160-6.720) | 0.085 |
| Histological type (Serous vs. others) | 1.874 (0.365-2.172) | 0.765 | 1.679 (0.449-1.861) | 0.895 |
| LN Metastasis (+ vs. -) | 3.084 (1.155-7.831) | 0.014 | 3.376 (1.714-8.517) | 0.054 |
| miR-196a expression (high vs. low) | 2.731 (0.804-9.637) | 0.025 | 2.432 (0.638-8.537) | 0.076 |
Discussion
Growing evidence has strongly implicated miRNAs have the ability to regulate a large number of genes in cancer development, which explains the control of multiple critical functions, including cancer cell proliferation, migration, and resistance to therapeutic interventions [9]. Identification of miRNAs associated with clinicopathological features and prognosis of cancer patients may underlying biologic mechanisms involved in the development of human cancer. In the present study, we investigated the relationships between miR-196a expression and clinical features, and the prognosis of patients with EOC. Our data suggest a critical role of miR-196a in the progression of human ovarian carcinoma.
Numerous studies have shown that miRNAs may function as oncogenes or tumor suppressors to regulate the expression of their downstream target genes at the post-transcriptional level [10-12]. Recent, several studies have demonstrated that miRNAs display diverse functions in ovarian cancer, depends on individual miRNA-mRNA pairs. Li et al. found that miR-17-5p modulates the cell cycle progression and apoptosis in human ovarian cancer by up-regulating YES1 expression [13]. Feng et al. suggested that miR-25 remarkably promotes proliferation, migration, and invasion of ovarian cancer cells by direct targeting large tumor suppressor 2 [14]. In contrast, Ge et al. proposed that miR-302b functions as a tumor suppressor by targeting RUNX1 and modulating the activity of the STAT3 signaling pathway in EOC [15]. Li et al. reported that overexpression of miR-128 resensitized ovarian cancer cells to cisplatin and reduced the expression of cisplatin-resistant-related proteins ABCC5 and Bmi-1, whereas miR-128 inhibitors increased cisplatin resistance in tumor cells [16]. These findings underline the complex effects of miRNAs in EOC development and progression. In our present study, we showed that miR-196a was markedly up-regulated in both ovarian cancer cell lines and ovarian cancer specimens as compared with normal ovarian cells and paired normal ovarian surface tissues, respectively. These data suggest that miR-196a play critical roles in the pathogenesis in cervical cancer.
MiR-196a is located in the intergenic regions in HOX gene clusters, which are major regulators of embryogenesis and oncogenesis [17]. It has been reported to be involved in the development and progression of a several malignant tumors, including breast cancer, leukemia, colorectal cancer, and cervical cancer [18-21]. Here, we associated clinicopathological characteristics of the patients with miR-196a expression and observed that miR-196a expression correlated with tumor stage, tumor size, and lymph nodes metastasis, which strongly suggested that miR-196a is involved in the development and progression of EOC. Our data is supported by Huang et al., who demonstrated that miR-196a may play a crucial role in pancreatic cancer proliferation and migration through its downstream target, nuclear factor-kappa-B-inhibitor alpha [22]. Zhang et al. also shown that miR-196a could regulates cervical cancer cell proliferation and migration by targeting netrin 4 [23]. Collectively, these studies suggested an oncogenic role of miR-196a in EOC.
Importantly, our study demonstrated that miR-196a expression was significantly associated with overall and recurrent-free survival of patients with EOC. Kaplan-Meier analysis of clinical survival showed that miR-196a expression was associated with poor clinical survival of patients with EOC, especially in Stage III-IV patients, indicating that high miR-196a expression cells might be more aggressive and progress more quickly and less sensitive to therapy [24]. Moreover, multivariate Cox analysis proved that miR-196a was an independent prognostic indicator for EOC patients. Thus, miR-196a could be used as a marker of poor prognosis for patients with EOC.
In summary, this is the first report demonstrating that the upregulation of miR-196a was associated with advanced clinicopathological features and poor prognosis of EOC, implying that it may serve as a promising prognostic biomarker and potential therapeutic target for EOC. Further studies are needed to elucidate the function and underlying mechanism of miR-196a in EOC.
Disclosure of conflict of interest
None.
References
- 1.Knutson KL, Karyampudi L, Lamichhane P, Preston C. Targeted immune therapy of ovarian cancer. Cancer Metastasis Rev. 2015;34:53–74. doi: 10.1007/s10555-014-9540-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan S, Selvan ST, Archunan G, Gulyas B, Padmanabhan P. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics. 2013;3:930–942. doi: 10.7150/thno.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karunakaran D, Rayner KJ. MicroRNAs in cardiovascular health: from order to disorder. Endocrinology. 2013;154:4000–4009. doi: 10.1210/en.2013-1299. [DOI] [PubMed] [Google Scholar]
- 6.Kane NM, Thrasher AJ, Angelini GD, Emanueli C. Concise review: MicroRNAs as modulators of stem cells and angiogenesis. Stem Cells. 2014;32:1059–1066. doi: 10.1002/stem.1629. [DOI] [PubMed] [Google Scholar]
- 7.Liu H. MicroRNAs in breast cancer initiation and progression. Cell Mol Life Sci. 2012;69:3587–3599. doi: 10.1007/s00018-012-1128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Cabay RJ, Jin Y, Wang A, Lu Y, Shah-Khan M, Zhou X. MicroRNA Deregulations in Head and Neck Squamous Cell Carcinomas. J Oral Maxillofac Res. 2013;4:e2. doi: 10.5037/jomr.2013.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C, Wu J, Hu B, Cheng SY, Li M, Li J. TGF-beta induces miR-182 to sustain NF-kappaB activation in glioma subsets. J Clin Invest. 2012;122:3563–3578. doi: 10.1172/JCI62339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 13.Li L, He L, Zhao J, Liu M, Li X, Tang H. miR-17-5p up-regulates YES1 to modulate the cell cycle progression and apoptosis in ovarian cancer cell lines. J Cell Biochem. 2015;116:1050–9. doi: 10.1002/jcb.25060. [DOI] [PubMed] [Google Scholar]
- 14.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 15.Ge T, Yin M, Yang M, Liu T, Lou G. MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cell Physiol Biochem. 2014;34:2209–2220. doi: 10.1159/000369664. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Chen H, Wu N, Zhang WJ, Shang LX. Deregulation of miR-128 in ovarian cancer promotes cisplatin resistance. Int J Gynecol Cancer. 2014;24:1381–1388. doi: 10.1097/IGC.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 17.Alonso CR. Hox proteins: sculpting body parts by activating localized cell death. Curr Biol. 2002;12:R776–778. doi: 10.1016/s0960-9822(02)01291-5. [DOI] [PubMed] [Google Scholar]
- 18.Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;110:1260–1268. doi: 10.1038/bjc.2013.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 20.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, Jurisica I, Penn LZ, Liu FF. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 21.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang F, Tang J, Zhuang X, Zhuang Y, Cheng W, Chen W, Yao H, Zhang S. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS One. 2014;9:e87897. doi: 10.1371/journal.pone.0087897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Zheng F, Yu G, Yin Y, Lu Q. miR-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells. Biochem Biophys Res Commun. 2013;440:582–588. doi: 10.1016/j.bbrc.2013.09.142. [DOI] [PubMed] [Google Scholar]
- 24.Suh Y, Raulf N, Gaken J, Lawler K, Guerrero Urbano T, Bullenkamp J, Gobeil S, Huot J, Odell E, Tavassoli M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int J Cancer. 2014 doi: 10.1002/ijc.29397. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]


