Abstract
The aim of this paper is to develop a supervised dimension reduction framework, called Spatially Weighted Principal Component Analysis (SWPCA), for high dimensional imaging classification. Two main challenges in imaging classification are the high dimensionality of the feature space and the complex spatial structure of imaging data. In SWPCA, we introduce two sets of novel weights including global and local spatial weights, which enable a selective treatment of individual features and incorporation of the spatial structure of imaging data and class label information. We develop an e cient two-stage iterative SWPCA algorithm and its penalized version along with the associated weight determination. We use both simulation studies and real data analysis to evaluate the finite-sample performance of our SWPCA. The results show that SWPCA outperforms several competing principal component analysis (PCA) methods, such as supervised PCA (SPCA), and other competing methods, such as sparse discriminant analysis (SDA).
Keywords: Classification, Imaging, Principal Component Analysis, Spatial Weight
1 Introduction
In various neuroimaging studies, imaging classification is to predict a set of response variables (class labels) Y by using a set of imaging data measured on each of N subjects, where is a 3-dimensional (3D) volume (or 2D surface) and d is a voxel (or pixel) of . For instance, Y may include cognitive outcome, disease status, and the early onset of disease, among others, whereas x may include magnetic resonance imaging (MRI) and positron emission tomography (PET), among many others. Moreover, imaging data usually can be represented as data on a graph such that D is a graph with {d1,..., dp} as the set of vertexes and an edge set, denoted by .
Two major challenges associated with imaging classification include (i) ultra-high dimension, but low sample size and (ii) correlated features with complex spatial structure including spatial smoothness and spatial correlation. For instance, the size of a typical T1 MRI is 256×256×256, and thus MRI contains 2563 = 16,777,216 voxels. In contrast, the number of observations in most neuroimaging studies varies from several dozens to several hundreds. Thus, it is imperative to perform dimension reduction before classification. Moreover, imaging data has an inherent and strong spatial dimension due to the inherent biological structure of objects (Friston, 2007; Lazar, 2008; Ye et al., 2009; Wang et al., 2007; Meyer and Chinrungrueng, 2005). The aim of this paper is to develop a Spatially Weighted Principal Component Analysis (SWPCA) to address the two challenges for high dimensional imaging classification.
Despite of its e cacy and popularity in image applications, principal component analysis (PCA; Jolliffe, 2002) as a general non-supervised dimension reduction technique is known to su er from major limitations. Firstly, each principal component (PC) is a linear combination of the original p features with nonzero loadings, which not only incorporates unnecessary noises but also makes it very di cult to interpret the derived PCs, especially when p >> N. Secondly, PCA treats all the features equally, and thus it may be not well-suited for some problems, in which some regions of interest are more important than others. Thirdly, PCA ignores the inherent spatial smoothness and spatial correlation of imaging data.
Many PCA variants have been proposed to address some of these limitations discussed above (Jolliffe, 2002; Zou et al., 2006; Bair et al., 2006; Skočaj et al., 2007; Shen and Huang, 2008; Leng and Wang, 2009; Pinto da Costa et al., 2011; Allen et al., 2011, etc.). For instance, Bair et al. (2006) proposed a Supervised PCA (SPCA) by conducting standard PCA on marginally selected features. However, SPCA su ers from ignoring the inherent spatial structure of imaging data. Skočaj et al. (2007) proposed a weighted PCA (WPCA) by introducing temporal and spatial weights in order to downweight individual images and individual components of x. However, they only focused on the temporal weights but failed to discuss how to choose the spatial weights which is more interesting in image analysis. Thomaz et al. (2010) introduced a supervised spatially weighted version of PCA (SSWPCA) by using a linear discriminant analysis (LDA) to determine spatial discriminant weights for x in a two-class classification setting and then applying PCA to the sample correlation matrix weighted by those spatial weights. SSWPCA is limited to binary responses and su ers from the “p >> N” problem, which requires further regularization. Recently, Pinto da Costa et al. (2011) proposed another weighted version of PCA based on a weighted rank correlation coe cient using rankings of original data, which is not appropriate for imaging data. Allen et al. (2011) proposed a generalized least squares matrix decomposition framework for two-way regularization PCA, while explicitly accounting for their structural relationship.
The aim of this paper is to develop a supervised dimension reduction method, called Spatially Weighted Principal Component Analysis (SWPCA), for imaging classification. In SWPCA, we introduce two sets of weights including global and local spatial weights, which enables the selection of individual features and the incorporation of both the spatial pattern of imaging data and class label information. We develop an e cient two-stage iterative SWPCA algorithm and its penalized version along with the associated weight determination. We evaluate the finite-sample performance of SWPCA by using two simulation studies and real data analysis, whose results strongly indicate that SWPCA outperforms several competing PCA variants and other competing methods, such as sparse discriminant analysis (SDA; Clemmensen et al., 2011).
The rest of this paper is outlined as follows. In Section 2, we develop the general SWPCA framework and its two-stage algorithm. Section 3 discusses several strategies of determining global and local weights. In Section 4, two simulation studies and real data analysis are conducted to demonstrate the improvement of our SWPCA over other commonly used PCA methods. Concluding remarks and discussions are given in Section 5.
2 Spatially Weighted Principal Component Analysis
2.1 Principal Component Analysis
Principal Component Analysis (PCA) as a basic dimension reduction tool is to project high-dimensional data to a lower dimensional space with a few uncorrelated features, called principal components (PC). Let X = (x1,..., xN)T denote an N × p data matrix of rank q ≤ min(N, p), where N is the number of observations, p is the number of features, and xi = (xij) is a p × 1 vector of features from the i-th object. Denote X̃ = X – 1NμT as the centered data matrix, where 1N is an N × 1 vector of ones and μ = (μ1, · · · ,μp)T is a p × 1 mean vector. Let Iq be a q × q identity matrix. PCA finds a lower-dimensional representation that maximizes the variance of projections. Numerically, PCA can be easily derived by Singular Value Decomposition (SVD) method as follows:
| (1) |
where the columns of A = UD = X̃V are PCs, the columns of V = (v ,..., vp)T are principal component directions (principal axes), D is a q × q diagonal matrix with singular values, and the columns of U and V are orthonormal such that UTU = VTV = Iq. In image analysis, the data matrix X consists of N images as rows, and each row (i.e., xi) represents a vectorized image of dimension p, where p is the number of pixels/voxels of the image and generally p >> N. When applying PCA to image data, the mean vector μ is the mean image and the columns of V are called eigenimages.
Alternatively, PCA can be interpreted as approximating the original data in the high-dimensional space by using a low-rank factor model. Specifically, the rank-q factor model can be written as
| (2) |
The PCA is then taken to minimize the reconstruction error (RE) defined as the squared distance between the original data and its rank-q approximation as follows:
| (3) |
where is the L2 norm of a vector and x̃ij = xij –μj. As seen from (3), can be represented as a summation of individual squared distance for each image at each location (feature), where all the features are treated equally and “independently”. In this sense, standard PCA ignores the underlying spatial pattern of image data, since remains the same no matter where each feature is located. Although the standard PCA loses the spatial information, this is not the case for SWPCA developed below, since we can explicitly incorporate such spatial information by introducing locally spatial weights, which depend on both the edge set and the spatial smoothness of imaging data.
2.2 Spatially Weighted PCA (SWPCA)
In this subsection, we develop a SWPCA to find a lower dimensional representation of imaging data by explicitly accounting for their spatial feature. Since each image xi consists of p correlated features with clustered spatial structure, PCA may not be well suitable for correlated imaging data. However, SWPCA explicitly incorporates spatial information by introducing two sets of spatial weights to the reconstruction error. Such spatial weights include (i) global weights for the selective treatment of individual features and (ii) local spatial weights for the incorporation of the spatial smoothness and correlation of imaging data.
Let wj be the global spatial weight for the j-th feature of xi with . Let B(j; h) be a neighborhood of the j-th feature at scale h and ω(j, d; h) be the local spatial weight for each neighboring feature d within B(j; h) of feature j such that . The SWPCA is taken to minimize a weighted reconstruction error (WRE) given by
| (4) |
where A = (a1 ,..., aN)T and V = (v1,..., vp) are the N × q and p × q matrices, respectively. Moreover, is the d-th feature of the centered data , in which and are the weighted and simple mean images, respectively. The lower-dimensional representation of SWPCA is obtained by minimizing as follows:
| (5) |
Without making any confusion, we omit the script h in most notations.
The global and local weights play a critical role in SWPCA. Specifically, the global weights WG = {wj} play a feature selection role and enable a selective treatment of di erent features by upweighting more important features and downweighting noninformative features. Moreover, since imaging data are spatially correlated and contain spatially contiguous regions with sharp edges, the local spatial weights WL = {ω(j, d; h)} allow us to capture the spatial smoothness of imaging data and accommodate the spatial dependence among imaging features. The scale parameter h can vary in a multiscale manner, while the shape of the neighborhood sets can vary across h and di erent applications. Furthermore, SWPCA provides a supervised dimension reduction solution by incorporating outcome information via the introduced global and spatial weights. In image classification, the global weights can be assigned according to the discriminative ability of each pixel/voxel, i.e., the association between each pixel/voxel in X and the class information in Y ; the local spatial weights can be determined based on the discriminative similarity of neighboring pixel/voxel. More discussions about how class information can be incorporated in SWPCA are given in Section 3.
SWPCA can be regarded as a generalization of PCA, SPCA, and WPCA. For instance, when ω(j, d; h) = 1(j = d), where 1(·) is the indicator function of an event, SWPCA reduces to WPCA with spatial weights. If ω(j, d; h) = 1(j = d) and wj = 1 for all j and d, (4) reduces to (3) which is the standard PCA problem. If we set ω(j, d; h) = 1(j = d) for all j, d and wj = 1 only for selected “top” features and 0 for other features, SWPCA reduces to SPCA. More discussions on various choices of these weights and neighborhood scale h are given in Section 3.
We reformat as follows. Let denote an N × p locally weighted data matrix and , where Wp =diag(w1, ...,wp) and . Without loss of generality, we assume that all global spatial weights are non-zero for the sake of notation. Even for WG with zero weights, all results below are valid since only features with non-zero wj are actually included for computation. We obtain the following lemma, whose proof can be found in the Appendix.
Lemma 1 Minimizing (4) is equivalent to
| (6) |
where is the Frobenius norm.
Equation (6) is invariant to arbitrary rotations of A and V such that AVT =ÃṼT . Such invariant issue is usually solved by imposing orthonormal constraint, i.e., VTV = Iq. However, such constraint is no longer appropriate for SWPCA, when the global spatial weights are incorporated for individual treatment and selection of features that regulate V . Instead of restricting V , we impose ATA = Iq, which greatly facilitates the computation of SWPCA.
For ultra-high dimensional data, that is p >> N, PCA su ers from several major limitations. For instance, it is well-known that sample eigenvalues and eigenvectors can be inconsistent as p goes to infinity. Moreover, naive approaches to PC score prediction can be substantially biased towards 0 in the analysis of high-dimensional data. To avoid such limitations, various penalized PCA methods have been developed (Journée et al., 2010; Shen and Huang, 2008; Huang et al., 2008a; Zou et al., 2006; Johnstone and Lu, 2009; Huang et al., 2008b; Witten et al., 2009). Specifically, we consider a regularized SWPCA by including an additional penalty term on V to (6). The minimization problem becomes
| (7) |
where vck is the k-th column vector of V = (vc1,..., vcq) and is the L1-norm of the p × 1 vector vck.
2.3 Two-stage Iterative Algorithms
We develop e cient two-stage iterative SWPCA algorithms to solve the minimization problems (6) and (7). We obtain the following two lemmas pertinent to the algorithm for solving (6).
Lemma 2 Given V , A that minimizes (4) subject to AT A = Iq is A = PUT, where P and U are orthogonal matrices obtained from the SVD of XhWpV = PDUT.
Lemma 3 Given A, V that minimizes (4) is .
The above lemmas lead to our two-stage iterative SWPCA Algorithm 1 as follows.
Algorithm 1 SWPCA Algorithm
Use V derived by standard PCA as an initial value;
Given V , conduct SVD on XhWpV = PDUT and then update A = PUT ;
Given A obtained from (b), update ;
Repeat the steps (b) and (c) until convergence;
Standardize the final V to obtain Ṽ and Ã.
Our SWPCA algorithm provides a simple and efficient way for the minimization problem (4) with the constraint on A. As a special case of SWPCA, the WPCA with spatial weights can be realized by our SWPCA Algorithm 1, which overcomes the rotational ambiguity problem of the Algorithm in Skočaj et al. (2007). According to our experience, our Algorithm 1 converges much faster compared with that for WPCA. Moreover, keeping V free of scale constraint allows direct extension of our SWPCA algorithm to a sparse version for ultra-high dimensional data. We obtain the following lemma for the sparse case.
Lemma 4 Given fixed A and ATA = Iq, we have the following results:
- the minimization problem (7) is equivalent to that of the weighted Lasso given by
where vck is the k-th column vector of V and ack is the k-th column of A;(8) - the soft thresholding solution of (8) is with
where sign(·) is the sign function, (·)j denotes the j-th element of the argument, and {·}+ is the truncation function that returns the argument if it is nonnegative or 0 otherwise.(9)
Lemma 4 has several important implications. Lemma 4 (i) reformats (7) as a weighted lasso problem of Zou et al. (2006). This result yields an explicit solution in Lemma 4 (ii). Moreover, for the features with small spatial weights, the regularized SWPCA automatically increases their penalties and thus their associated 's have a higher chance to be shrunk to zero. Thus, SWPCA is very useful for eliminating many uninformative features in imaging data. Based on Lemma 4, Algorithm 1 can be extended to the penalized SWPCA algorithm as follows.
Algorithm 2 Penalized SWPCA Algorithm
Use V derived by the standard PCA as an initial value;
Given V , apply SVD on XhWpV = PDUT and then set A = PUT ;
’ Given A obtained from (b), update V = {vjk}p×q, where vjk is calculated according to (9);
’ Repeat the steps (b) and (c’) until convergence;
Standardize the final V to obtain Ṽ and Ã
The penalized SWPCA algorithm proposed above provides an efficient way to obtain a low-dimensional representation of X even for ultra-high dimensional data. Besides, our penalized SWPCA algorithm can be used to realize the sparse PCA method proposed by Shen and Huang (2008) since SWPCA is considered as a generalization of standard PCA. The sparse PCA algorithm in Shen and Huang (2008) computes PCs in a sequential way, whereas our Algorithm 2 computes all PCs simultaneously at once. Thus, Algorithm 2 may be more appealing and efficient.
One important application of SWPCA is to do prediction. We consider the data matrix with new observations and its locally weighted data matrix, which are denoted by X* and , respectively. LetÃ* be the low-dimensional projection of X* onto the principal axes Ṽ derived from SWPCA. We obtain the following lemma.
Lemma 5 Given V , the A that minimizes (4) is A = (XhWpV)(VTWpV)–1.
Based on Lemma 5,Ã* can be estimated by using . Withà , class prediction can be efficiently performed using this low-dimensional representation.
3 Spatial Weights for Imaging Classification
In this section, we discuss how to choose various spatial weights and how the class information can be incorporated for imaging classification.
3.1 Global Spatial Weights
The global weights WG = {wj} play a feature selection role in SWPCA. For instance, for classification problem, we may use each feature's discriminative importance to assign each component of WG. Specifically, let θj denote a measure of the association between the j-th feature and the class information, i.e., θj is a function of the j-th pixel/voxel of image data X and class information Y . Each wj can be defined as a function of θj as
| (10) |
Examples of θj include the Pearson correlation and test statistics, among many others. A simple example is to use the Pearson correlation between each feature and class label information. More informative features for classification (or high correlation) are assigned more weights, whereas noninformative features (e.g., correlation less than a given threshold) can be discarded by setting wj = 0. In this case, θj is the Pearson correlation and . Additionally, the importance scores used in SPCA can also be considered for WG. Another example is to fit a voxel-wise regression model with imaging data at each location as responses and the class label as covariates. Specifically, we consider an L-class classification problem and define yil = 1 if i is in class l for i = 1,..., N and l = 1,..., L and 0 otherwise. Consider a voxel-wise linear regression model by fitting xij = yTiθj + εij for i = 1,..., N and j = 1,..., p, where yi = (1, yi1,..., yi,L–1T and θj contains the discriminative information of features at location j. Then, f(θj) can be a test statistic and/or its associated p-value for testing H0 : θj = 0 at location j.
3.2 Local Spatial Weights
The local spatial weights WL = {ω(j, d; h)} play a critical role in incorporating spatial smoothness and correlation of imaging data in SWPCA. This is extremely important for imaging data, since imaging data are spatially dependent and contain contiguous regions with sharp edges in nature. Let B(j, h) be a set of neighboring locations of the j-th feature at scale h. The local weight ω(j, d; h) usually characterizes the “similarity” between feature j and features d ∈ B(j, h) and/or the “similarity” between locations d and j. Specifically, we define ω(j, d; h) at scale h as
| (11) |
where D1(d, j) denotes the spatial distance between locations d and j, D2( θd, θj) represents the discriminative similarity between θd and θj, K1(·) and K2(·) are two decreasing kernel functions, and h and CN are bandwidth parameters that may depend on N. The decreasing kernel function K1(·) gives less weight to the voxel d ∈ B(j, h), whose location is far from the voxel j. The kernel K2(·) downweights the voxels d with large D2(θd, θj), which indicates a large di erence between θd and θj. By following Polzehl and Spokoiny (2006) and Li et al. (2011), we set K1(x) = (1 – x)+ and K2(x) = exp(–x), which demonstrated excellent performance in many imaging applications. Moreover, the shape and size of B(j, h) can vary across applications and with h.
3.3 Scale Size h
The scale size h plays a critical role in the amount of features incorporated from neighboring voxels in B(j, h) for each j. A simple approach is to fix h according to some prior or empirical information. However, a small h may miss important spatial information, whereas a large h may smooth out some local details and dramatically increase the computational burden. Alternatively, we may consider a sequence of nested neighborhoods corresponding to multiple scales at each location. Specifically, let h = {h0 < h1 < ... < hS} be a sequence of scales with h0 = 0 and hS being the maximum scale. The scales can be chosen based on previous studies or empirical experiences. For example, scales can be defined as the radius of spherical neighborhood in a form of {hs = cs} with constant c > 1. In our numerical examples, we used c = 1.2 which balanced the computation intensity without losing important spatial information. One may choose an optimal h based on a specific criterion, such as WRE, and then use the PCs extracted based on the optimal h for imaging classification. Alternatively, one may integrate the PCs extracted from all scales h for imaging classification.
For imaging classification, we propose a multi-scale procedure to determine WG and WL across multiple scales. Without loss of generality, we consider the cross-sectional studies so that (xi, yi) are independent across subjects. Specifically, at a given scale h, we consider a weighted likelihood function given by
| (12) |
where X and Y , respectively, denote the imaging and class information and p(xid|yi, θj) is the likelihood function of xid given yi. Moreover, as discussed in the voxel-wise linear regression model in Section 3.1, θj may contain the discriminative information of features at j. Based on the weighted likelihood function (12), Li et al. (2011) developed a multiscale adaptive regression model (MARM) to spatially and adaptively calculate the estimate of θj, denoted by , as the scale size h ranges from h0 to hS. Instead, we borrow the spatial information learned in (12) and the estimated to spatially and adaptively construct WG and WL across h. We introduce a multi-scale algorithm as follows.
Algorithm 3 Multi-scale Algorithm
Given a series of scales h0 = 0 < h1 < ... < hS, for each feature j,
Begin with h0 = 0 with ω(j, d; h0) = 1 when d = j and 0 otherwise, calculate the initial association estimate at scale h0 denoted as and set s = 1.
At scale hs, update the association estimate by maximizing (12) based on ω(j, d; hs–1).
Calculate ω(j, d; hs) in (11) using updated and and update wj according to (10) by using . If s < S, let s=s+1.
Repeat steps (b) and (c) until the stopping criterion is met or s = S.
The stopping criterion in Algorithm 3 can be either global or local criteria. For instance, for the global criteria, we may stop the algorithm if WRE cannot be further decreased. For the local criteria at each location j, we may check the improvement of θj.
4 Numerical Examples
In this section, we conducted two simulation studies and real data analysis to examine the performance of SWPCA in L-class classification problems, where L is a positive integer. In each simulation study, we compared our SWPCA with PCA, SPCA, and WPCA. For SPCA, we used the importance scores used in Bair et al. (2006) to select “top” important features. For WPCA, we considered two WPCAs by using two different spatial weights which are based on the importance scores of SPCA and WG of SWPCA, respectively. For PCA, SPCA, and WPCA, imaging data were smoothed using an isotropic Gaussian kernel with different degrees of smoothness including no smoothing, moderate smoothing, and over-smoothing. For the sake of space, we only reported the best results under the moderate degree of smoothness. For SWPCA, we used Algorithm 3 with spherical neighborhoods and {hs = 1.2s : s = 1,..., S = 5} to determine optimal scales locally and its associated WG and WL. Specifically, we used -log10 of the FDR (False Discovery Rate) corrected p-values for testing the null hypothesis of no group di erences as our global spatial weights WG. For the local spatial weights, we chose the Euclidean norm for D1(·), the Mahalanobis norm for D2(·), and CN = log(N)χ2L–1(0.95), where χ2L–1(b) is the upper 1 – b percentile of the χ2 distribution with L – 1 degrees of freedom. For the penalized SWPCA, denoted as PSWPCA, we used the same spatial weights as those of SWPCA and varied λk across 0.5, 1.0, 2.0, 5.0, and 10.0. Extracted PCs were then used for class prediction. Since the proposed method is more efficient requiring less components, we chose number of PCs retained based on the amount of total sample variance explained by the standard PCA to assure that at least a certain amount of total variance can be accounted for. Since comparison of different classification methods is not the interest of this paper, we just used three standard classification methods including (i) linear regression (REG), (ii) k-nearest neighbor (k-NN) classification, and (iii) support vector machine (SVM) to evaluate the performance of different PCA methods.
4.1 Simulation Studies
For each simulation study, a total of 100 3D-images were simulated and randomly split into a training set with N = 60 images and a test set of 40 images. We repeated each simulation 100 times, and evaluated the classification performance of different methods by using the average misclassification rate. For simulation studies, the results with K = 2 PCs retained were presented for graphical illustration purpose to display the low-dimensional representation of the simulated data constructed by the PCs extracted by different dimension reduction methods. With 2 PCs, the standard PCA can already account for on average around 60% of the total variance in Simulation I and around 50% in Simulation II.
4.1.1 Simulation Study I
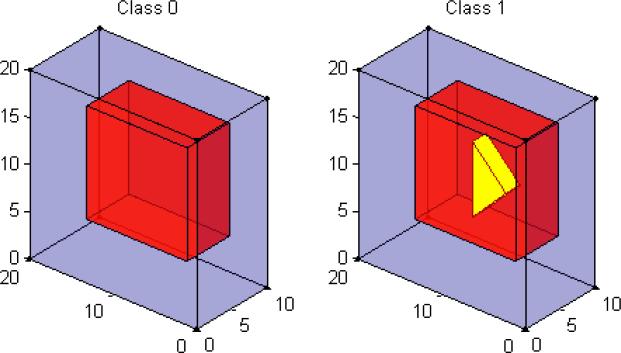
In this study, we simulated 20×20×10 3D-images from a linear regression model: xij = θj0+ θj1yi+ εij, where εij ~ N(0, 4) for i = 1,..., 100 and j = 1,...,4000. The feature dimension p = 4000 is much larger relative to the training sample size N=60. This is a two-class classification problem with L = 2. The class label yi is coded as 0 and 1. Thus, μ0 = {θj0 : j = 1,...,4000} is the true mean image of Class 0, whereas μ1 = {θj0 + θj1 : j = 1,...,4000} is the true mean image of Class 1. See Figure 1 for a graphical illustration. For μ0, we divided the 3D image into two different regions of interest (ROIs) with different shapes and then varied θj0 as 0 and 1, respectively, across these two ROIs. For μ1, we divided the 3D image into three different ROIs with different shapes and then varied θj0 + θj1 as 0, 1, and 2, respectively, across these three ROIs. Figure 1 reveals that the di erence between the two classes only lies in the yellow triangular prism region.
Figure 1.
True mean images for Simulation I. The left panel is the true mean image of Class 0: μ0, in which purple and red colors represent θj0 = 0,1, respectively; the right panel is the true mean image of Class 1: μ1, in which purple, red, and yellow colors represent θj0 + θj1 = 0, 1, 2, respectively.
We presented the classification results in Table 1 based on the results obtained from the 100 simulated datasets. We only presented the average misclassification rates and the standard deviations (SD) of misclassification errors. Table 1 reveals that the classification results from the three classification methods show similar pattern across different PCA methods. For SPCA, we varied the number of “top” voxels as 50, 100, 200, 400, and 1000, and the algorithm based on the top 50 voxels outperforms the rest. Overall, SPCAs greatly improve the class prediction over PCA by screening out many uninformative voxels. We denote two WPCAs using the importance scores of SPCA and using WG of SWPCA as their spatial weights by WPCA-1 and WPCA-2, respectively. The WPCA-1 performs better than PCA, as good as SPCA with relatively large number of top selected voxels, but worse than SPCA with less selected voxels. The WPCA-2 improves the class prediction over WPCA-1 and performs as good as SPCA based on top 50 voxels. Furthermore, SWPCA using Algorithm 1 significantly reduces the misclassification rate to only 2.6% for REG, 3.0% for 5-NN, and 3.3% for SVM with much smaller standard deviations. Finally, PSWPCA based on Algorithm 2 further reduces the misclassification rate.
Table 1.
Average Misclassification Percentage for Simulation I
| PCA | SPCA | WPCA-1 | WPCA-2 | SWPCA | PSWPCA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 50 | 100 | 200 | 400 | 1000 | ALL | ALL | ALL | ALL | |
| REG | .302 (.078) | .126 (.052) | .132 (.052) | .142 (.055) | .162 (.057) | .205 (.064) | .199 (.064) | .130 (.056) | .026 (.025) | .025 (.024) |
| k-NN | .338 (.071) | .135 (.049) | .141 (.049) | .152 (.050) | .182 (.053) | .225 (.071) | .186 (.055) | .156 (.059) | .030 (.029) | .027 (.025) |
| SVM | .327 (.078) | .140 (.054) | .147 (.055) | .159 (.055) | .183 (.059) | .226 (.072) | .215 (.067) | .152 (.055) | .033 (.029) | .028 (.026) |
Standard deviations are in parenthesis. For SPCA, the number of “top” selected voxels used in the algorithm are considered to be 50, 100, 200, 400, and 1000.
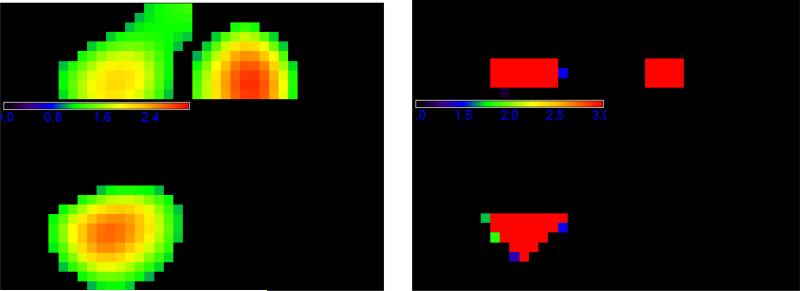
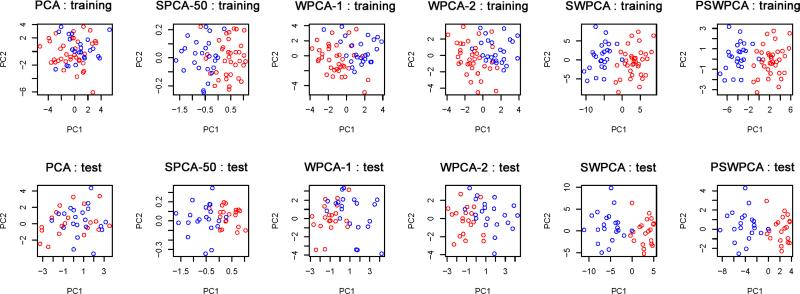
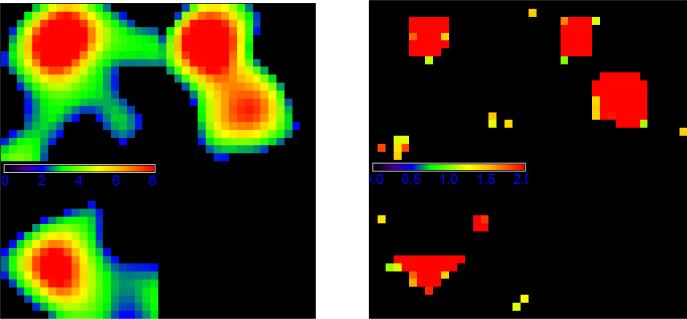
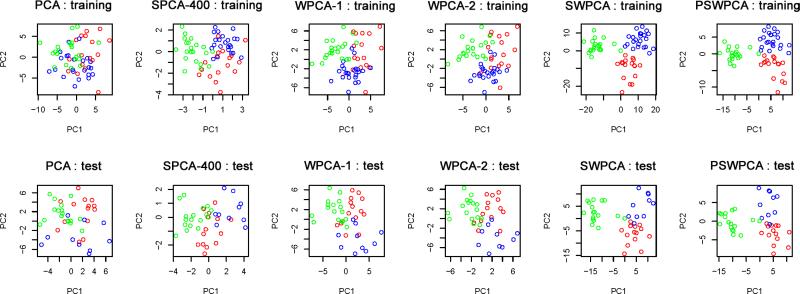
The two different sets of spatial weights used in WPCA-1 and WPCA-2 are illustrated in Figure 2. Figure 2 shows that WG of SWPCA clearly identifies those “true” voxels which are located in the triangular prism. In contrast, the importance scores of SPCA can only roughly locate the region but not the shape, along with many false positive voxels. Let nt denote the number of true informative voxels. In Simulation I, nt = 75 voxels forms the triangular prism. To further evaluate the “feature selection” ability of the weights, we calculate the true positive rate (TP) as the ratio of the true informative voxels to the top ranked nt voxels. We have TPSPCA=.63, and TPSW PCA=.90. These two numbers also explain why WPCA-2 works better than WPCA-1. Moreover, the performance of SPCA is sensitive to the threshold used to determine significant features in SPCA, whereas WPCAs and SWPCAs can include all the voxels by weighing differently without concerning the thresholding issue. To visualize the performance of dimension reduction, we plotted the first two extracted PCs obtained from different PCA methods for one simulated dataset in Figure 3. We observed that the two PCs of SWPCA and PSWPCA can easily separate the two classes, whereas those of other PCA methods cannot.
Figure 2.
Weight illustration for Simulation I: a three-view slice illustration at coordinate (13, 7, 3) of the spatial weights used for WPCA-1 (left panel) and WPCA-2 and SWPCA (right panel). The left panel contains the importance scores of SPCA, while the right panel contains the WG of SWPCA, i.e., the FDR-corrected -log 10 p-value map.
Figure 3.
Two-dimensional representation for Simulation I. The first two PCs from PCA, SPCA-50 (SPCA based on top 50 voxels), WPCA-1, WPCA-2, SWPCA, and PSWPCA are plotted. The training set (top panels) and test set (bottom panels) are used to extract the PCs. Points with blue and red colors represent true Class 0 and Class 1, respectively.
For additional comparisons, we also applied two non-PCA-type of supervised dimension reduction methods including sparse partial least squares (SPLS; Chung and Keles, 2010) and sparse discriminant analysis (SDA; Clemmensen et al., 2011) to the simulated datasets. See Table 2 for detailed results. For SPLS, since different classifiers can be used, we also applied REG (SPLS-REG), k-NN (SPLS-kNN), and SVM (SPLS-SVM) besides the default one in order to have additional comparisons with various PCA-type of methods presented in Table 1. Table 2 shows that SPLS and SDA yield similar classification performance as SPCA, but they significantly underperform our proposed SWPCA and PSWPCA.
Table 2.
Average Misclassification Percentage for Simulation I (Non-PCA Methods)
| SPLS-REG | SPLS-kNN | SPLS-SVM | SPLS | SDA |
|---|---|---|---|---|
| .130 (.052) | .139 (.056) | .156 (.066) | .128 (.050) | .120 (.050) |
Standard deviations are in parenthesis.
4.1.2 Simulation Study II
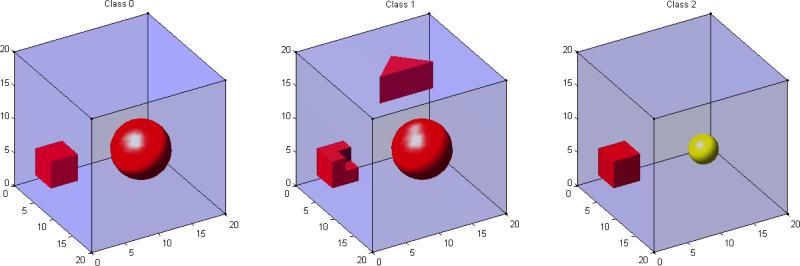
In this simulation study, we simulated 20 × 20 × 20 3D-images from L = 3 classes (coded as 0, 1, 2) according to a linear regression model: xij = θj0 + θj1yi1 + θj2yi2 + εij, where εij ~ N(0, 9) for i = 1,...,100 and j = 1,...,8000 and yi1 and yi2 are dummy variables for Class 1 and Class 2, respectively. Thus, μ0 = {θj0} is the true mean image of Class 0, μ1 = {θj0 + θj1} is the true mean image of Class 1, and μ2 = {θj0 + θj2} is the true mean image of Class 2. Figure 4 shows the true mean images for each class in 3D. For μ0, we divided the 3D image into two different ROIs with different shapes and then varied θj0 as 0 and 1, respectively, across these two ROIs. For μ1, we divided the 3D image into two different ROIs with different shapes and then varied θj0 + θj1 as 0 and 1, respectively, across these two ROIs. For μ2, we divided the 3D image into three different ROIs with different shapes and then varied θj0 + θj2 as 0, 1, and 2, respectively, across these three ROIs. Figure 4 reveals that the di erences between the three classes lie in the yellow and red regions.
Figure 4.
True mean images for Simulation II. The left panel is the true mean image of Class 0: μ0, in which two ROIs with purple and red colors represent θj0 = 0 and 1, respectively; the middle panel is the true mean image of Class 1: μ1, in which two ROIs with purple and red colors represent θj0 + θj1 = 0 and 1, respectively; the right panel is the true mean image of Class 2: μ2, in which three ROIs with purple, red, and yellow colors represent θj0 + θj2 = 0, 1, and 2, respectively.
We presented the classification results in Table 3 based on the results obtained from the 100 simulated datasets. In Table 3, three classification methods, REG, k-NN, and SVM, show similar classification results across different PCA methods. For SPCA, the algorithm based on the 400 or 1,000 top voxels outperforms the rest. Overall, SPCA moderately improves the class prediction over the standard PCA. The WPCA-1 algorithm outperforms PCA and is comparable to SPCAs in cases of 200 or more top voxels. The WPCA-2 algorithm shows better performance than SPCA and WPCA-1. Furthermore, SWPCA and PSWPCA outperform other competing methods and greatly reduce the misclassification rate. The results for the non-PCA methods are given in Table 4. Similarly as Simulation I, SPLS and SDA yield similar but significantly poorer classification performance comparing with the best SWPCA results.
Table 3.
Average Misclassification Percentage for Simulation II
| PCA | SPCA | WPCA-1 | WPCA-2 | SWPCA | PSWPCA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | (50) | (100) | (200) | (400) | (1000) | ALL | ALL | ALL | ALL | |
| REG | .461 (.078) | .372 (.126) | .343 (.118) | .302 (.102) | .275 (.084) | .274 (.069) | .305 (.075) | .246 (.069) | .096 (.076) | .092 (.080) |
| k-NN | .514 (.093) | .381 (.118) | .364 (.112) | .315 (.105) | .295 (.086) | .299 (.075) | .332 (.077) | .268 (.069) | .099 (.079) | .085 (.078) |
| SVM | .477 (.097) | .372 (.117) | .346 (.112) | .306 (.105) | .278 (.078) | .288 (.067) | .317 (.074) | .258 (.073) | .099 (.076) | .083 (.078) |
Standard deviations are in parenthesis. For SPCA, the number of “top” selected voxels used in the algorithm are considered to be 50, 100, 200, 400, and 1000.
Table 4.
Average Misclassification Percentage for Simulation II (Non-PCA Methods)
| SPLS-REG | SPLS-kNN | SPLS-SVM | SPLS | SDA |
|---|---|---|---|---|
| .341 (.119) | .356 (.125) | .337 (.120) | .339 (.112) | .277 (.076) |
Standard deviations are in parenthesis.
Figure 5 illustrates two different sets of spatial weights used in WPCA-1 and WPCA-2. It shows that WG of SWPCA identifies the “true” voxels located in the triangular and cubic shapes. However, the importance scores of SPCA roughly identifies the location of true voxels, but not the shapes. In this simulation study, there are 409 truly informative voxels, and we have TPSPCA=.59 and TPSWP CA=.81. In Figure 6, we draw the plots of the first two extracted PCs obtained from all PCA methods for one simulated dataset. From Figure 6, we observe that SWPCA and PSWPCA perform well in separating three classes, but other PCA method do not.
Figure 5.
Weight illustration for Simulation II: a three-view slice illustration at coordinate (7, 6, 16) of the spatial weights used for WPCA-1 (left panel) and WPCA-2 and SWPCA (right panel). The left panel contains the importance scores of SPCA, while the right panel contains the WG of SWPCA, i.e., the FDR-corrected -log 10 p-value map.
Figure 6.
Two-dimensional representation for Simulation I. The first two PCs for PCA, SPCA-400 (SPCA based on top 400 voxels), WPCA-1, WPCA-2, SWPCA and PSWPCA are plotted. The training set (top panels) and test set (bottom panels) are used to extract the PCs. Points with blue, red, and green colors represent Class 0, Class 1, and Class 2, respectively.
4.2 Real Data Analysis
We applied SWPCA to the Alzheimer's Disease Neuroimaging Initiative (ADNI) data. Alzheimer's Disease (AD) is the most common form of dementia, which progressively causes problems in memory, thinking, behavior, and eventually leads to death. The ADNI study is a large scale multi-site study collecting clinical, imaging, and laboratory data at multiple time points from cognitively normal controls (CN), individuals with amnestic mild cognitive impairment (MCI), and subjects with AD. One of the goals of ADNI is to develop improved methods to track the longitudinal course of AD based on imaging and biomarker data. More information about data acquisition can be found at the ADNI website (www.loni.ucla.edu/ADNI).
“Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year publicprivate partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their e ectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. ADNI is the result of e orts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org. ”
A subset of the ADNI data including AD patients and CN controls was used here to illustrate the empirical utility of our proposed methods in imaging classification. After removing subjects with missing or low quality imaging data, the data set consists of 390 subjects (218 CN controls and 172 AD patients). Among them, there are 206 males whose mean age is 75.46 years with standard deviation 6.34 years and 184 females whose mean age is 75.50 years with standard deviation 6.40 years. T1-weighted images at the baseline were used for all subjects. The T1-weighted images were preprocessed by standard steps including AC (anterior commissure) and PC (posterior commissure) correction, N2 bias field correction, skull-stripping, intensity inhomogeneity correction, cerebellum removal, segmentation, and registration. After segmentation, the brain were segmented into four different tissues: grey matter (GM), white matter (WM), ventricle (VN), and cerebrospinal fluid (CSF). The imaging pipeline was described in detail in Wang et al. (2011).
We quantified the local volumetric group di erences by generating RAVENS-maps (Davatzikos et al., 2001) for the whole brain and four different types of segmented tissue (GM, WM, VN, and CSF), respectively, using the deformation field obtained during registration. RAVENS methodology is based on a volume-preserving spatial transformation, which ensures that no volumetric information is lost during processing spatial normalization, since this process changes an individual's brain morphology to conform it to the morphology of a template. We obtained the 256 × 256 × 256 RAVENS-maps and then down-sampled them to 128 × 128 × 128 for analysis. A sample RAVENS-map is displayed in the left panel of Figure 7.
Figure 7.
Data and weight illustration for ADNI study. The left panel is a three-view slice illus tration at coordinate (49, 57, 32) of a sample RAVENS-map; the middle panel shows the important scores of SPCA; the right panel illustrates the FDR-corrected – log 10 p-value map used as WG for WPCA-2, SWPCA and PSWPCA.
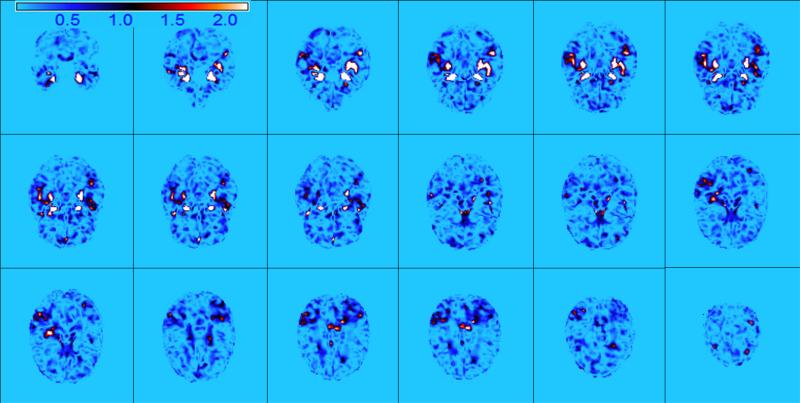
Our goal is to study the empirical performance of SWPCA in classifying subjects from ADNI to AD or CN group based on the whole RAVENS images. We randomly split the imaging data into a training set of 195 images and a test set with the remaining 195 images and repeated this 100 times to calculate the average misclassification rate. For this real data analysis, five PCs were included for classification, which can account for around 90% of the total variance on average in the standard PCA. For PCA, WPCA, and SPCA, the RAVEN images were smoothed by using an isotropic Gaussian kernel with different degrees of smoothness as in the simulation studies. For ultra-high dimensional data like our example, we may pre-filter and assign zero weight to the less “important” voxels instead of assigning non-zero weights to all the voxels in order to improve computational e ciency. For example, for WPCA-2 and SWPCA based on the p-value map, we thresholded all FDR-corrected – log10 p-values at the significance level of 0.01 for WG. For SPCA, we selected the same number of voxels as the number of non-zero WG used in SWPCA according to the importance scores. Figure 7 shows some selected views of the importance score image of SPCA and the FDR-corrected – log10 p-value map used in WPCA-2 and SWPCA. Figure 8 presents more slice views of the global spatial weights used in SWPCA and illustrates that some regions of interest in AD studies, such as hippocampus and amygdala, were highly weighted.
Figure 8.
Weight illustration for ADNI study. Selected axial slices show the FDR-corrected -log 10 p-value map used in SWPCA which correctly identifies some important regions reported in the literature for AD, such as hippocampus and amygdala.
In Table 5, we present the classification results that show the similar performance of PCA methods as in the simulation studies. SPCA slightly improves the classification rate over the standard PCA. Both SPCA and WPCA-1 based on the same importance scores show quite similar performance. WPCA-2 performs slightly better than WPCA-1. In addition, SWPCA and PSWPCA outperform all other PCA methods with much lower misclassification rates. Notice that even using simple classification procedures, SWPCA/PSWPCA directly applied to the whole image can already lead to a misclassification percentage around 20% for ADNI data. To illustrate our proposed dimension reduction tool, we applied simple classification procedures. It suggests that the classification performance can be more improved by incorporating more sophisticated procedures.
Table 5.
Average Misclassification Percentage for ADNI Data
| PCA | SPCA | WPCA-1 | WPCA-2 | SWPCA | PSWPCA | |
|---|---|---|---|---|---|---|
| REG | .329 (.029) | .312 (.043) | .307 (.052) | .274 (.029) | .213 (.034) | .198 (.033) |
| k-NN | .382 (.028) | .343 (.045) | .344 (.052) | .313 (.030) | .254 (.035) | .227 (.041) |
| SVM | .329 (.029) | .313 (.042) | .310 (.042) | .274 (.030) | .216 (.033) | .215 (.032) |
Standard deviations are in parenthesis.
Numerical examples illustrated that SWPCA and its penalized version PSWPCA provided substantial improvement over other commonly used dimension reduction methods in image classification by incorporating spatial and class information via introduced global and local spatial weights. Computationally, SWPCA algorithm provides an efficient implementation, which produces all PCs simultaneously unlike other methods like Shen and Huang (2008) that compute PC one-by-one in a sequential way and converges much faster than the WPCA algorithm by Skočaj et al. (2007) since it does not have arbitrary rotation problem. In practice, the computing time of SWPCA varies depending on the analysis and method used for weight determination.
5 Discussion
This article is to develop a general SWPCA framework to generate low-dimensional representations for high-dimensional imaging classification. By incorporating the global and local spatial weights, SWPCA enables a selective treatment and selection of individual features, accommodates the complex dependence among features of imaging data, and has the ability of utilizing the underlying spatial pattern possessed by imaging data and class label information. SWPCA integrates feature selection, smoothing, and feature extraction in a single framework. In the simulation studies and real data analysis, SWPCA shows substantial improvement over PCA, SPCA, and WPCA.
The contributions of this article are two-fold. Firstly, from an image analysis point of view, our proposal timely responds to a number of growing needs of neuroimaging classification. It may also provide a systematic solution to the integrative analysis of multi-modality imaging data and imaging genetics data (Friston, 2009; Casey et al., 2010). Secondly, from a statistical methodology point of view, our proposal provides a novel and broad framework for the use of covariates with graphic structure to predict clinical outcomes. A large number of models and extensions are potential outcomes within this framework. Although there has been imaging studies utilizing tensor/matrix structure (Li et al., 2005; Park and Savvides, 2007; Li et al., 2010; Zhou et al., 2013), our proposal, to the best of our knowledge, is the first work that integrates the spatial and graphic structure of imaging data into a statistical supervised learning paradigm. Our work can be viewed as a logic extension from the classical classification methods to a functional classification model.
Several important issues need to be addressed in future research. First, we will systematically investigate the theoretical properties of SWPCA and its variations by extending the existing results in the literature (Johnstone, 2001; Baik and Silverstein, 2006; Paul and Johnstone, 2007; Jung and Marron, 2009; Benaych-Georges and Nadakuditi, 2011). Second, we will extend our SWPCA from a classification framework to a regression framework in order to predict more complex univariate and multivariate clinical outcomes. Third, we will develop new global weighting methods based on some joint important feature selection methods and more complex screening methods, such as a robust rank correlation screening in (Li et al., 2012). Many more complexities and new statistical tools will definitely come out these new developments.
Acknowledgments
We thank the Editor, the Associate Editor, and three anonymous referees for valuable suggestions, which greatly helped to improve our presentation. Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp–content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
The research of Drs. Zhu and Ahn was partially supported by NIH grants RR025747-01, CA74015, P01CA142538-01, and MH086633. The research of Dr. Guo was supported by NIH grants NIH-NCATS,UL1TR001082, TL1TR001081, and KL2TR001080. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimers Association; Alzheimers Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Ho mann-La Roche Ltd and its a liated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
A Appendix: Proofs
A.1 Proof of Lemma 1
The WRE can be rewritten as follows:
where C is a scalar independent of A and V . Thus, minimizing is equivalent to minimizing (6) as stated in Lemma 1.
A.2 Proof of Lemma 2
Lemma 2 can be proved by using Theorem 4 of Zou et al. (2006) as follows. We kept their original notations here.
Theorem 4 (Reduced Rank Procrustes Rotation; Zou et al., 2006): Let Mnχp and Nnχk be two matrices. Consider the constrained minimization problem:
Suppose the SVD of MTN is UDVT , then  = UAT.
Based on Lemma 1 and the discussion in Section 2.2, we have a similar minimization problem of A given V :
| (13) |
If we set and in Theorem 4, then  that minimizes (13) given V subject to ATA = Iq is  = PUT, where P and U are orthogonal matrices from SVD of .
A.3 Proof of Lemma 3
Assuming A is given, we take the derivative of in (6) with respect to V . By setting it to zero, we obtain the solution
In Algorithm 1, since A derived from Step (b) is subject to ATA = Iq, V becomes XTh A in Step (c).
A.4 Proof of Lemma 4
Since we can write:
minimizing is equivalent to the following minimization problem. For each k = 1,..., q, we have
where xhj is the j-th column of Xh and vjk is the j-th element of vck. This completes the proof of part (i).
Part (ii) can be easily obtained by applying the lemma below to each element of vck.
Lemma The minimizer of (β–y)2 + λ |β| is
The proof of this lemma is straightforward, so we omitted here.
A.5 Proof of Lemma 5
Assuming V is given, we can obtain the solution of A by taking the derivative of (6) with respect to A.
References
- Allen GI, Grosenick L, Taylor J. Tech. rep. Rice University; 2011. A generalized least squares matrix decomposition. [Google Scholar]
- Baik J, Silverstein J. Eigenvalues of large sample covariance matrices of spiked population models. Journal of Multivariate Analysis. 2006;97:1382–1408. [Google Scholar]
- Bair E, Hastie T, Paul D, Tibshirani R. Prediction by supervised principal components. Journal of the American Statistical Association. 2006;101:119–137. [Google Scholar]
- Benaych-Georges F, Nadakuditi R. The eigenvalues and eigenvectors of finite, low rank perturbations of large random matrices. Advances in Mathematics. 2011;227:494–521. [Google Scholar]
- Casey B, Soliman F, Bath KG, Glatt CE. Imaging genetics and development: Challenges and promises. Human Brain Mapping. 2010;31:838–851. doi: 10.1002/hbm.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Keles S. Sparse Partial Least Squares Classification for High Dimensional Data. Statistical Applications in Genetics and Molecular Biology. 2010;9:1–32. doi: 10.2202/1544-6115.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmensen L, Hastie T, Witten D, Ersbøll B. Sparse discriminant analysis. Technometrics. 2011;53:406–413. [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick S. Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Friston KJ. Statistical Parametric Mapping: the Analysis of Functional Brain Images. Academic Press; London: 2007. [Google Scholar]
- Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- Huang JZ, Shen H, Buja A. Functional principal component analysis via regularized best basis approximation. Electronic Journal of Statistics. 2008a;2:678–695. [Google Scholar]
- Huang JZ, Shen H, Buja A. Functional principal components analysis via penalized rank one approximation. Electronic Journal of Statistics. 2008b;2:678–695. [Google Scholar]
- Johnstone I. On the distribution of the largest eigenvalue in principal components analysis. The Annals of Statistics. 2001;29:295–327. [Google Scholar]
- Johnstone IM, Lu AY. On consistency and sparsity for principal components analysis in high dimensions. Journal of the American Statistical Association. 2009;104:682–693. doi: 10.1198/jasa.2009.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe I. Principal component analysis. 2nd ed Springer-Verlag; 2002. [Google Scholar]
- Journée M, Nesterov Y, Richtárik P, Sepulchre R. Generalized power method for sparse principal component analysis. Journal of Machine Learning Research. 2010;11:517–553. [Google Scholar]
- Jung S, Marron J. PCA consistency in high dimension, low sample size context. The Annals of Statistics. 2009;37:4104–4130. [Google Scholar]
- Lazar NA. The Statistical Analysis of Functional MRI Data. Springer; New York: 2008. [Google Scholar]
- Leng C, Wang H. On general adaptive sparse principal component analysis. Journal of Computational and Graphical Statistics. 2009;18:201–215. [Google Scholar]
- Li B, Kim MK, Altman N. On dimension folding of matrix or array valued statistical objects. The Annals of Statistics. 2010;38:1097–1121. [Google Scholar]
- Li G, Peng H, Zhang J, Zhu L. Robust rank correlation based screening. Ann. Statist. 2012;40:1846–1877. [Google Scholar]
- Li Y, Du Y, Lin X. Kernel-based multifactor analysis for image synthesis and recognition. Tenth IEEE International Conference on Computer Vision. 2005:114–119. [Google Scholar]
- Li Y, Zhu H, Shen D, Lin W, Gilmore JH, Ibrahim JG. Multiscale adaptive regression models for neuroimaging data. Journal of the Royal Statistical Society: Series B. 2011;73:559–578. doi: 10.1111/j.1467-9868.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer FG, Chinrungrueng J. Spatiotemporal clustering of fMRI time series in the spectral domain. Medical Image Analysis. 2005;9:51–68. doi: 10.1016/j.media.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Park SW, Savvides M. Individual kernel tensor-subspaces for robust face recognition: a computationally efficient tensor framework without requiring mode factorization. IEEE Transactions on Systems, Man, and Cybernetics. Part B. 2007;37:1156–1166. doi: 10.1109/tsmcb.2007.904575. [DOI] [PubMed] [Google Scholar]
- Paul D, Johnstone I. Technical Report. UC Davis; 2007. Asymptotics of sample eigenstructure for a large dimensional spiked covariance model. [Google Scholar]
- Pinto da Costa JF, Alonso H, Roque L. A weighted principal component analysis and its application to gene expression data. IEEE Transactions on Computational Biology and Bioinformatics. 2011;8:246–252. doi: 10.1109/TCBB.2009.61. [DOI] [PubMed] [Google Scholar]
- Polzehl J, Spokoiny VG. Propagation-separation approach for local likelihood estimation. Probability Theory and Related Fields. 2006;135:335–362. [Google Scholar]
- Shen H, Huang J. Sparse principal component analysis via regularized low rank matrix approximation. Journal of Multivariate Analysis. 2008;99:1015–1034. [Google Scholar]
- Skočaj D, Leonardis A, Bischof H. Weighted and robust learning of subspace representations. Pattern Recognition. 2007;40:1556–1569. [Google Scholar]
- Thomaz CE, Giraldi GA, da Costa JFP, Gillies DF. Tech. rep. Department of Computing Imperial College London; 2010. A simple and efficient supervised method for spatially weighted PCA in face image analysis. [Google Scholar]
- Wang D, Shib L, Yeunga DS, Tsanga EC, Hengb PA. Ellipsoidal support vector clustering for functional MRI analysis. Pattern Recognition. 2007;40:2685–2695. [Google Scholar]
- Wang Y, Nie J, Yap P, Shi F, Guo L, Shen D. Robust deformable-surface-based skull-stripping for large-scale studies. In: Fichtinger G, Martel A, Peters T, editors. Medical Image Computing and Computer-Assisted Intervention. Vol. 6893. Springer; Berlin / Heidelberg: 2011. pp. 635–642. [DOI] [PubMed] [Google Scholar]
- Witten D, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10:515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lazar N, Li Y. Geostatistical analysis in clustering fMRI time series. Statistics in Medicine. 2009;28:2490–2508. doi: 10.1002/sim.3626. [DOI] [PubMed] [Google Scholar]
- Zhou H, Li L, Zhu H. Tensor regression with applications in neuroimaging data analysis. Journal of American Statistical Association. 2013 doi: 10.1080/01621459.2013.776499. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T, Tibshirani R. Sparse principal component analysis. Journal of Computational and Graphical Statistics. 2006;15:265–286. [Google Scholar]