Abstract
The high level of methylguanine-DNA methyltransferase (MGMT) in glioblastoma is responsible for resistance to alkylating agents, such as temozolomide (TMZ). In glioblastomas with a methylated MGMT promoter, MGMT deficiency is presumed, resulting in an enhanced effect of TMZ. The aim of the present study was to investigate whether genomic alterations work synergistically with MGMT methylation status and contribute to the response to treatment and overall prognosis in glioblastoma. The current study included a cohort of 35 glioblastoma patients, with MGMT promoter methylation present in 48% of tumors. MGMT methylation was associated with significantly longer median survival (29.0 months) compared with patients without MGMT methylated tumors (12.0 months), as well as longer median time to progression following TMZ treatment (13.2 months, compared with 5.6 months for patients with an unmethylated MGMT status). In addition, somatic variants in hot spot exonic regions of 50 key cancer genes were examined in these glioblastomas. Non-synonymous mutations in methylated MGMT glioblastomas were four times higher compared with unmethylated MGMT glioblastomas. Furthermore, significantly increased frequencies of mutations in the TP53, CDKN2A, PTEN and PIK3CA genes were detected in MGMT methylated glioblastomas. The relative significance of these mutations, and their contribution to TMZ sensitivity, adjunct to MGMT methylation, require further investigation in a larger cohort.
Keywords: methylguanine-DNA methyltransferase methylation, mutations, glioblastoma
Introduction
Glioblastoma (GBM) is the most common type of brain tumor, with high mortality rates. The aggressive biology of GBM and resistance to therapy are major clinical problems for patients diagnosed with the disease (1). At present, standard treatment includes maximal safe surgery, concomitant radiotherapy and temozolomide (TMZ) chemotherapy, where a median survival time of <15 months is observed in patients fit for this current first-line therapy (1). In terms of years of life lost, the population burden from GBM is the highest of all malignancies (2). GBM frequently affects young people (30–40 years of age), as well as the elderly (3). Physically or cognitively disabled patients require care from the early stages of the disease, which results in increased expenses. Despite advances in therapeutic strategies, no significant reductions in the age-standardised incidence and mortality rates of this disease have been observed within the last decade (3).
Patients with a good Eastern Cooperative Oncology Group performance status or Karnofsky score at the time of diagnosis undergo surgical resection (4,5). Complete removal of the tumor is challenging due to the infiltrative nature of GBM; however, 80–95% debulking is typically achieved unless the disease is in a neurologically eloquent location (6). The standard treatment, known as ‘The Stupp Protocol’, consists of concurrent temozolomide (TMZ) therapy and radiotherapy (60 Gy/30 fractions), followed by subsequent adjuvant TMZ therapy for 6–12 cycles (1). TMZ functions by alkylating the guanine residues in DNA, resulting in mispairing with thymine during DNA replication and cellular arrest in rapidly dividing cells (7,8).
Methylguanine-DNA methyltransferase (MGMT) removes mutagenic alkyl adducts and, thus, protects DNA from the damage induced by TMZ and other alkylating agents. Loss of MGMT expression commonly occurs in human neoplasia (9). The expression of MGMT is regulated by hypermethylation of the CpG islands within the promoter and enhancer regions of the gene (10–12). MGMT deficiency is presumed in GBMs with a methylated MGMT promoter, resulting in the enhanced cytotoxicity of TMZ (13). Studies have demonstrated that presence of MGMT promoter methylation is correlated with the response of patients to treatment with TMZ; thus, it is considered to be a predictive biomarker in TMZ-treated GBMs, even in elderly patients (14,15). Between 25 and 50% of GBMs are methylated at the MGMT promoter; therefore, using MGMT methylation status clinically as a predictive biomarker for GBM patients has received increased attention, with the long-term aim to use it as a biomarker to assign alkylating therapy to individual patients. However, a number of studies have reported discordance between MGMT methylation status and treatment outcome, and no clear alternative treatment is available for patients with unmethylated MGMT tumors, who derive only limited benefit from TMZ (13).
Based on the signatory gene expression profiles using data generated from The Cancer Genome Atlas (TCGA) (16), four distinct molecular subtypes of GBM were described: Classical, mesenchymal, neural and proneural (17). These subtypes have also been associated with characteristic genetic alterations (17). The classical subtype is associated with an astrocytic expression profile, with frequent epidermal growth factor receptor (EGFR) amplification, concomitant chromosome 7 amplification and chromosome 10 loss, and focal deletions of 9p encompassing cyclin-dependent kinase inhibitor 2A (CDKN2A). Notably, the classical subtype is not associated with TP53 mutations, which are common in GBMs (17). The mesenchymal subtype is typified by the expression of mesenchymal markers, with frequent deletions or mutations of the neurofibromin 1 (NF1) and phosphatase and tensin homolog (PTEN) genes. The neural subtype exhibits expression of neuronal markers and displays various mutations and copy number alterations, including amplification of EGFR and deletion of PTEN. The proneural subtype exhibits an oligodendrocytic expression signature and is characterized by TP53 mutations. In addition, focal amplifications of the chromosome 4q12 region, which contains the oligodendrocytic development gene α-type platelet-derived growth factor receptor (PDGFRA), or mutations of the isocitrate dehydrogenase 1 gene (IDH1) are observed. The TCGA group has recently published an updated comprehensive genomic, epigenomic, transcriptomic and proteomic analysis of >500 GBMs (18). Patients were categorized into the four different groups and the overall survival was stratified based on MGMT methylation. Notably, the findings indicated that MGMT methylation may be a predictive biomarker for response to treatment, but only in the classical subtype of GBM.
In the present study, MGMT methylation and somatic mutations were assessed using semiconductor sequencing technology in the same tumor DNA. The cohort included biopsies from 35 GBM patients, who had been treated uniformly with standard concurrent radiotherapy plus TMZ. The association between MGMT methylation status/mutation profile and survival was also evaluated.
Materials and methods
Patient samples
Formalin-fixed paraffin-embedded tissues from patients with GBM, which had been obtained during the initial surgical resection, were collected from the Australian Genomics and Clinical Outcomes of Glioma biobank (www.agogbio.org.au). All the patients provided written informed consent and the study was approved by the Institutional Human Research Ethics Committee of South Eastern Sydney Local Health District (SESLD) Human Research Ethics Committee (HREC), University of New South Wales (Sydney, Australia). Specimens were obtained prior to any chemotherapy or radiotherapy, and the patient information, including age, gender, extent of tumor removal, pathology and treatment, was recorded. Only patients receiving maximal debulking of the tumor (>85%) and undergoing concurrent chemoradiotherapy (60 Gy in 30 fractions with 75 mg/m2/day TMZ) were included in this study. The number of adjuvant cycles of TMZ, determined by clinical variables including toxicity, response to treatment and progression, was noted. Time to progression (TTP; measured from the start of chemoradiotherapy to the time of clinical progression) was determined by the treating oncologist (Professor Anna Nowak).
MGMT promoter methylation
Bisulfite modification prior to CpG pyrosequencing was performed to assess the percentage level of MGMT promoter methylation for each tumor specimen, as described previously (19). Chemically methylated or unmethylated universal human genomic DNA (EMD Millipore, Billerica, MA, USA) controls were included with each batch. The CpG pyrosequencing methylation assay was performed using the PyroMarkTM MGMT kit (Qiagen, Alameda, CA, USA) on a PSQTM96 MA system (Qiagen) according to the manufacturer's instructions.
Detection of EGFR gene amplification
EGFR amplification was recorded in the patient pathology reports by PathWest Laboratory Medicine (Nedlands, Australia) using the Multiplex Ligation-dependent Probe Amplification assay (MLPA; MCR-Holland, Amsterdam, Netherlands), as described previously (20).
Targeted resequencing of DNA
In order to amplify the target regions of 50 oncogenes and tumor suppressor genes that are implicated in solid tumors, amplicon libraries for individual patient DNA samples were prepared using the Ion AmpliSeq Cancer Hot Spot Panel v2 (Life Technologies, Grand Island, NY, USA), as described previously (21). Subsequently, semi-conductor sequencing was performed using the Ion Personal Genome Machine (PGM) 200 Sequencing kit (Life Technologies) and the Ion PGM Sequencer (21). Torrent Suite™ v3.6.2 software (Life Technologies) was used to parse barcoded reads, align reads to the reference genome (human genome build 19), base call and generate run metrics, including the chip loading efficiency, total read counts, quality and total coverage. In addition, the ANNOVAR™ (Biobase Biological Databases; http://www.biobase-international.com) and Oncomine Gene Browser (Life Technologies) software were used to identify variants, as well as predict amino acid changes and clinical significance. The Integrative Genomics Viewer (Broad Institute, Cambridge, MA, USA; http://www.broadinstitute.org/igv/) was used to visualize the alignment of the reads, detect the presence of variants against the reference genome and verify the integrity of variant calls by detecting possible strand bias and sequencing errors (21).
Statistical analysis
The primary end point was the overall survival. A Kaplan-Meier survival analysis was used to generate survival curves and estimates of median survival periods. All statistical analyses were performed using SPSS software (version 22.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristic
A total of 35 patients with a diagnosis of GBM were included in the current study (Table I). The median age was 58 years (range, 31–80 years) and the study included 25 males and 10 females. The median overall survival of the cohort was 13.0 months [95% confidence interval (CI), 9.7–16.3]. The median TTP of the cohort was 6.0 months (95% CI, 4.7–7.3).
Table I.
Summary of patient characteristics and treatment results.
| Characteristic | Value |
|---|---|
| Patients, n | 35 |
| Age, years | |
| Median | 58 |
| Range | 30.5–79.6 |
| Gender, n | |
| Male | 25 |
| Female | 10 |
| Overall survival, months | |
| Median | 13 |
| 95%CI | 9.7–16.3 |
| Time to progression, months | |
| Median | 6 |
| 95% CI | 4.7–7.3 |
CI, confidence interval.
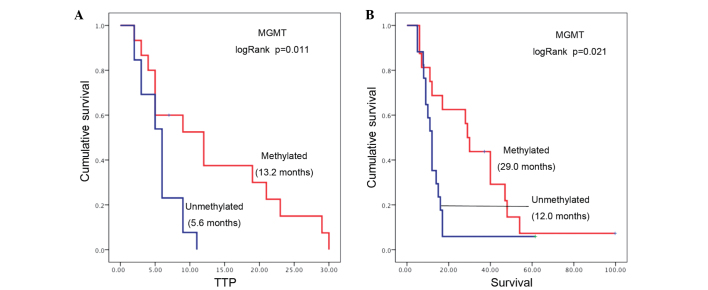
The MGMT promoter methylation status was detected in 33 out of the 35 patients, with 16 tumors (48%) found to be methylated at the MGMT promoter region. Tumor MGMT methylation was associated with significantly longer survival rates (median survival, 29.0 months; 95% CI, 20.1–32.9) compared with patients without MGMT methylated tumors (median survival, 12.0 months; 95% CI, 10.1–13.9; log-rank P=0.025; Fig. 1A). A strong association was also observed between MGMT methylation and longer TTP, since the median TTP for patients with methylated MGMT tumors was 13.2 months (95% CI, 4.1–19.9) compared with 5.6 months (95% CI, 4.5–7.5) for patients with an unmethylated tumor MGMT status (log-rank P=0.011; Fig. 1B).
Figure 1.
Kaplan-Meier survival curves based on (A) TTP and (B) overall survival for glioblastoma patients stratified by MGMT methylation. Results of log-rank tests and the median survival for each group is provided. MGMT, methylguanine-DNA methyltransferase; TTP, time-to-progression.
Overall gene alterations
Of the 35 specimens sequenced, multiple (>2) non-synonymous mutations were detected in 10 specimens. In the present study, mutations were identified in the TP53 (20%), CDKN2A (11%) and PTEN (9%) genes, as well as mutations and/or amplifications in the EGFR gene (29%). In addition, two tumors exhibited mutations in IDH1. Mutations in MPL (6%), SMARCB1 (9%), NOTCH1 (9%), PIK3CA (9%), MET (6%) and RB1 (6%) were also detected (Fig. 2).
Figure 2.
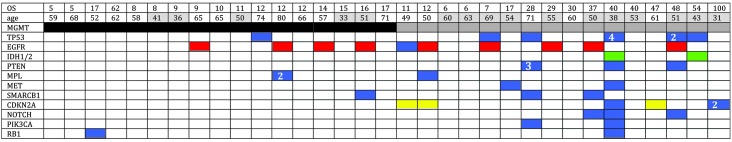
Overall gene alteration patterns for 11 target genes in 33 samples. Blue bars represent single nucleotide polymorphisms, red bars represent amplification, yellow bars represent deletions, grey bars represent MGMT methylation and black bars represent MGMT unmethylation. Numbers inside the bars show the alteration number. Sample number (no.), age (years) and OS (months) are also provided. MGMT, methylguanine-DNA methyltransferase; OS, overall survival.
The mutation profiles were mapped according to MGMT promoter methylation (Fig. 2), which revealed that non-synonymous mutations were more frequent in the MGMT methylated GBMs compared with the MGMT unmethylated GBMs. Furthermore, mutations in the CDKN2A, PTEN and PIKC3A genes were only identified in MGMT methylated GBMs. Amplification and/or mutation in the EGFR gene and deletion of CDKN2A are hallmarks of the classical tumor subtype. A recent study has reported that the predictive value of MGMT methylation was only applicable in the classical subtype (15). In the current study, the frequency of EGFR/CDKN2A gene aberrations was found to be higher in the MGMT methylated group of tumors (56%) when compared with the MGMT unmethylated tumors (24%).
Discussion
The presence of MGMT promoter methylation in GBM is a predictor of response to TMZ treatment and of overall survival. In the present study, the mutations detected in MGMT methylated GBMs were four times higher compared with MGMT unmethylated GBMs. This observation may be attributed to a lack of DNA repair function upon presence of lower levels of the MGMT protein.
In 2005, MGMT promoter methylation was reported to be associated with a survival benefit from alkylating agent chemotherapy in GBM patients, in a companion study based on a phase III concurrent radiation therapy and TMZ trial (13). A number of studies have demonstrated an even higher survival benefit in patients treated with alkylating agents, who presented tumors with MGMT promoter methylation, compared with patients having tumors with an unmethylated MGMT promoter (13,22,23). Therefore, MGMT promoter methylation is a favorable prognostic marker, in addition to a favorable predictive marker for response to TMZ treatment in patients with GBM. The molecular basis for the differential response to TMZ treatment in patients is well established. Alkylating drugs, including TMZ, induce DNA damage by introducing alkyl adducts into the DNA molecules. This causes genetic mutations and cross-links between DNA strands that inhibit DNA replication and thereby trigger cell death. However, MGMT removes the alkyl adducts from the DNA as they are introduced, preventing subsequent mutational damage. Tumor cells expressing MGMT are thus resistant to alkylating drugs (24).
In the current study, the TTP following chemoradiotherapy and subsequent overall survival were significantly increased in patients with MGMT methylation compared with patients without methylated tumors. Thus, the present study confirmed the role of MGMT methylation and its associated sensitivity to TMZ (25,26).
The current study investigated the prevalence and types of genomic alterations in GBM and whether these correlate with MGMT methylation. The results demonstrated that 75% of MGMT methylated tumors harbored one or more somatic mutations, compared with 35% of unmethylated cases. Overall, the mutations identified in MGMT methylated were four times higher compared with MGMT unmethylated tumors. Notably, mutations were identified in the TP53, CDKN2A, PTEN and PIK3CA genes in MGMT methylated cases. Since these cases also exhibited improved overall survival, further investigations should determine whether these genomic aberrations work synergistically with MGMT methylation status and contribute to the response to treatment and overall prognosis. MGMT methylation may play a dual role, increasing sensitivity to TMZ and increasing genetic instability, particularly by facilitating the appearance of G to A transition mutations. The TCGA dataset of GBMs revealed increased frequencies of TP53 and PTEN point mutations in MGMT methylated GBMs compared with MGMT unmethylated GBMs (16). In the current study, mutations in PTEN and TP53 were also found to be associated with MGMT promoter methylation, confirming the findings of the TCGA. MGMT methylation is commonly associated with G:C to A:T mutations in KRAS and TP53 genes in colorectal cancer (27,28). Therefore, MGMT promoter methylation may be an early event in the development of methylated GBM, preceding other characteristic mutations in this subgroup.
In the present study, higher frequencies of EGFR and CDKN2A alterations were observed in the MGMT methylation group. Although the tumors were not categorized into proneural, neural, mesenchymal or classical subtypes (17), the higher frequencies of EGFR/CDKN2A indicate that the MGMT methylation group may be enriched for the classical subtype. Furhtermore, the current study is in accordance with the findings of Brennan et al (18), demonstrating that the classical subtype is more responsive to TMZ, which is reflected by the longer TTP and overall survival observed.
In conclusion, MGMT methylation was strongly associated with the response to TMZ treatment and overall survival in GBMs, as previously reported. Methylation was also found to be associated with a significantly higher mutation rate. Therefore, it is hypothesized that methylation may be one of the earliest preneoplastic events in this subgroup. A larger cohort of patients is required to further investigate the association of MGMT methylation and specific mutations (including those in EGFR, CDKN2A and TP53) with overall survival in TMZ-treated GBMs.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Burnet NG, Jefferies SJ, Benson RJ, et al. Years of life lost (YLL) from cancer is an important measure of population burden – and should be considered when allocating research funds. Br J Cancer. 2005;92:241–245. doi: 10.1038/sj.bjc.6602321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leibetseder A, Ackerl M, Flechl B, et al. Outcome and molecular characteristics of adolescent and young adult patients with newly diagnosed primary glioblastoma: a study of the Society of Austrian Neurooncology (SANO) Neuro Oncol. 2013;15:112–121. doi: 10.1093/neuonc/nos283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinsella TJ, Collins J, Rowland J, et al. Pharmacology and phase I/II study of continuous intravenous infusions of iododeoxyuridine and hyperfractionated radiotherapy in patients with glioblastoma multiforme. J Clin Oncol. 1988;6:871–879. doi: 10.1200/JCO.1988.6.5.871. [DOI] [PubMed] [Google Scholar]
- 5.Hammoud MA, Sawaya R, Shi W, Thall PF, Leeds NE. Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J Neurooncol. 1996;27:65–73. doi: 10.1007/BF00146086. [DOI] [PubMed] [Google Scholar]
- 6.Gulati S, Jakola AS, Nerland US, Weber C, Solheim O. The risk of getting worse: surgically acquired deficits, perioperative complications, and functional outcomes after primary resection of glioblastoma. World Neurosurg. 2011;76:572–579. doi: 10.1016/j.wneu.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6:2585–2597. [PubMed] [Google Scholar]
- 8.Roos WP, Kaina B. DNA damage-induced apoptosis: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 10.Felsberg J, Thon N, Eigenbrod S, et al. German Glioma Network: Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129:659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur J Cancer. 2000;36:2294–2300. doi: 10.1016/S0959-8049(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 13.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 14.Reifenberger G, Hentschel B, Felsberg J, et al. German Glioma Network: Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131:1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 15.Stummer W, Nestler U, Stockhammer F, et al. Favorable outcome in the elderly cohort treated by concomitant temozolomide radiochemotherapy in a multicentric phase II safety study of 5-ALA. J Neurooncol. 2011;103:361–370. doi: 10.1007/s11060-010-0400-9. [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhaak RG, Hoadley KA, Purdom E, et al. Cancer Genome Atlas Research Network: Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan CW, Verhaak RG, McKenna A, et al. TCGA Research Network: The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald KL, Rapkins RW, Olivier J, et al. The T genotype of the MGMT C>T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. Eur J Cancer. 2013;49:360–368. doi: 10.1016/j.ejca.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Jeuken J, Sijben A, Alenda C, et al. Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol. 2009;19:661–671. doi: 10.1111/j.1750-3639.2009.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabone T, Abuhusain HJ, Nowak AK, et al. Multigene profiling to identify alternative treatment options for glioblastoma: a pilot study. J Clin Pathol. 2014;67:550–555. doi: 10.1136/jclinpath-2014-202173. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 23.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 24.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 25.Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PloS one. 2012;7:e33449. doi: 10.1371/journal.pone.0033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 27.Rosty C, Young JP, Walsh MD, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8:e65479. doi: 10.1371/journal.pone.0065479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]