Abstract
The aim of the present study was to determine the expression of a-disintegrin and metalloproteinase 10 (ADAM10) in human glioma tissues from surgical specimens and discuss its possible significance in glioma biology. A total of 43 glioma specimens obtained from patients between 2007 and 2010 were collected and a series of assays were performed. Of these, 22 cases were low-grade gliomas, while 21 cases were high-grade gliomas. In addition, 20 cases of meningioma were used as the control group. Reverse transcription-polymerase chain reaction (RT-PCR), western blot analysis and immunohistochemistry were used to determine the mRNA and protein expression levels of ADAM10. Besides the quantitative analysis, histological observations were also performed to localize ADAM10 expression in glioma cells. The RT-PCR and western blot analysis results demonstrated increased ADAM10 expression in the low-grade glioma samples compared with the control (P<0.05), while ADAM10 expression was further increased in the high-grade glioma samples (P<0.01 vs. control; P<0.05 vs. low-grade glioma), indicating that the mRNA and protein expression levels of ADAM10 were malignancy-dependent. The immunohistochemical analysis revealed that the ADAM10 protein was located on both the tumor cell membrane and blood vessel walls within tumor tissues. In conclusion, these results indicated that ADAM10 expression correlates with the grade of malignancy in human glioma from surgical specimens. In addition, the fact that ADAM10 protein was expressed on cell membranes and blood vessel walls within tumor tissues, indicates that its expression may be associated with invasive tumor growth and peritumoral edema formation.
Keywords: a-disintegrin and metalloprotease 10, expression, glioma, malignancy
Introduction
Gliomas comprise the majority of primary brain tumors and are the leading cause of intracranial neoplasm mortality, resulting in a poor prognosis despite advances in surgical techniques and concurrent radiochemotherapy regimens (1–3). High-grade gliomas exhibit aggressive biological features, including extensively infiltrative growth and diffuse invasion, and are frequently recurrent and intractable (3,4). Radiotherapy, chemotherapy and photodynamic therapy may increase survival rates (5,6); however, disease recurrence is virtually inevitable (7). The invasive growth of gliomas is a long-standing and formidable issue that often leads to the failure of glioma therapy (3,8). At present, the biological behavior and characteristics of gliomas, including infiltrative invasion, should be investigated, rather than methods of reducing the tumor bulk.
The invasive growth of gliomas is a complex multistage process that involves changes in the expression levels of multiple genes and includes various pathophysiological aspects, including attachment to neighboring cells, aggregate formation, adhesion to the matrix substratum, migration and invasion into the three-dimensional cellular microenvironment, which includes tumor cells, the extracellular matrix, stromal cells, cytokines, immune cells and is characterized by tissue hypoxia, acidosis and interstitial high pressure; of these components, the extracellular matrix key and may be hydrolyzed by ADAM10 during glioma invasion (9,10). As glioma cells acquire their infiltrative phenotype, they overexpress various matrix metalloproteinases (MMPs), which are a family of zinc-dependent endopeptidases that cleave extracellular matrix components (11–14). In addition to their role in extracellular matrix degradation, a number of MMPs and the associated family of ‘a-disintegrin and metalloproteases’ (ADAMs) also regulate proliferation, adhesion, migration and metastasis by initiating the cleavage of cell surface proteins (12–15). Among these genes, ADAM10 is currently attracting increased attention (12–14). As a member of the zinc-dependent protease family, the structural characteristics of ADAM10 form the basis of its biological functions (13,15,16). Previous studies revealed that ADAM10 is involved in the pathogenesis of several types of malignant solid tumors and that it may be involved in invasive growth (12,17–20). Therefore, in the present study, a series of assays were designed and conducted to determine the expression of ADAM10 and its localization in low- and high-grade gliomas. These results may provide an insight into the biological roles of ADAM10 in gliomas with different malignancies, thereby elucidating its underlying mechanisms, which may provide a promising target for gene therapy of gliomas.
Materials and methods
Specimen processing
Between May 2007 and July 2010, a total of 43 glioma specimens were collected from patients hospitalized at the Department of Neurosurgery (The First Affiliated Hospital of China Medical University, Shenyang, China) and included in this study. This study was approved by the ethics committee of The First Affiliated Hospital of China Medical University and written informed consent was obtained from all patients. These specimens included 22 cases of low-grade gliomas and 21 cases of high-grade gliomas. In addition, 20 meningioma specimens [World Health Organization (WHO) grade II (21)] were collected and used as the controls. Each specimen was stored at −70°C immediately following surgical resection, and the corresponding patient's information was obtained and registered. All the specimens were pathologically confirmed by two neuropathologists according to the 2007 WHO criteria (21). Paraffin-embedded sections were obtained from the Department of Pathology of The First Affiliated Hospital of China Medical University and reserved for the subsequent immunohistochemical assays. The frozen fresh tissue samples were subjected to reverse transcription-polymerase chain reaction (RT-PCR) and western blot assays.
RNA extraction and semi-quantitative RT-PCR
A tumor sample (100 mg) from each patient, previously stored at −70°C, was used for semi-quantitative RT-PCR detection according to the following method: The tissue sample was initially macerated in 1 ml of RNAiso Plus (Takara Bio, Inc., Otsu, Japan) in a 1.5 ml RNase-free centrifuge tube. Each centrifuge tube was vortexed for 15 sec to mix the contents and kept still for 5 min. Subsequently, the tubes were centrifuged at 12,000 × g at 4°C for 5 min. The supernatant from each tube was collected and transferred to a new 1.5-ml tube and 1/5 of the chloroform volume was added. Next, the contents were mixed with 0.5 ml isopropanol, and a white pellet was obtained and washed with 75% ethanol, following centrifugation at 12,000 × g at 4°C for 15 min, according to the manufacturer's instructions. The supernatant was removed and the resulting pellet was then air-dried and dissolved in 40 µl of RNase-free water. A UV spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, Inc., Wilmington, DE, USA) was used to measure RNA purity and content.
RT-PCR was performed on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems Life Technologies, Foster City, CA, USA) using a two-step PrimeScript RT-PCR kit (Takara Bio, Inc.) according to the manufacturer's instructions. Briefly, 1 µg of RNA from each sample was converted into cDNA in a reverse transcription reaction. PCR for each gene was performed in a 20 µl reaction mixture, containing 2 µl cDNA template, 1.6 µl dNTP, 0.4 µl of each primer, 0.1µl TaKaRa Ex Taq, 2 µl 10X Ex Taq buffer (all reagents obtained from Takara) and 13.5 µl sterilized distilled water, and the conditions were as follows: 94°C for 2 min, with cycling conditions of 94°C for 30 sec, 55°C for 30 sec and 72°C for 2 min, for a total of 30 cycles, and a final extension step of 10 min. The amplified DNA product from each specimen was subjected to 1.5% agarose gel electrophoresis. Subsequently, the samples were visualized by ethidium bromide staining (Sigma-Aldrich, St. Louis, MO, USA) and images were captured using Gel Doc 2000 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) under ultraviolet light and digitized with a Hewlett-Packard Scanjet scanner (Palo Alto, CA, USA) as a 256-level grayscale image in the gel analysis software, SigmaGel versino 1.0 (Jandel Scientific, Erkrath, Germany). Gene-specific primers were designed and synthesized by Takara Bio, Inc., as listed in Table I. All the results were normalized against the expression levels of the housekeeping gene, β-actin. The experiments were performed in triplicate for each sample.
Table I.
Primer sequences.
| Gene | Primer sequences | Length (bp) |
|---|---|---|
| ADAM10 (F) | 5′-TGGGTCAAAAAGAAAATGGC-3′ | 281 |
| ADAM10 (R) | 5′-CCCAGGTTTCAGTTTGCATT-3′ | |
| β-actin (F) | 5′-AAATCGTGCGTGACATTAA-3′ | 474 |
| β-actin (R) | 5′-CTCGTCATACTCCTGCCTG-3′ |
ADAM10, a-disintegrin and metalloproteinase 10; F, forward; R, reverse.
Western blot analysis
To determine the level of ADAM10 protein expression, the samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis, as described previously (22,23). Equal quantities of tissue lysates were resolved by SDS-PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes (Roche Diagnostics, Indianapolis, IN, USA) using electroblotting, probed with specific primary antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies, and then analyzed. The primary antibodies used were monoclonal rabbit anti-human ADAM10 (1:1,000; Abcam, Cambridge, UK) and monoclonal rabbit anti-human β-actin antibody (1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and were detected using HRP-conjugated mouse anti-rabbit immunoglobulin G antibody (1:500; Sigma-Aldrich) in blocking solution. The 75 kDa band was used to analyze changes in ADAM10 expression in all the experiments. Immunoreactive protein bands were visualized using an enhanced chemiluminescence reagent (Amersham ECL-PLUS; GE Healthcare Life Sciences, Little Chalfont, UK), scanned with a FujiFilm LAS300 Intelligent Dark Box scanner (Fujifilm, Tokyo, Japan) and quantified for pixel density by the optical density (OD) method, using the Chemi Genius gel imaging system (Chemi ImagerTM 5500; ProteinSimple, San Jose, CA, USA).
Immunohistochemical assay
ADAM10 protein localization and expression were determined using an immunohistochemical method. Immunohistochemical analysis was performed using the Histostain-SP kit (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. The primary antibody (rabbit anti-human ADAM10; Abcam) was diluted at 1:500 and ADAM10 binding was visualized using the standard avidin/biotinylated enzyme complex-HRP staining procedure, with 3,3′-diaminobenzidine as a chromogen and a monoclonal mouse anti-rabbit immunoglobulin G antibody (Histostain-SP kit, Invitrogen Life Technologies). The sections were examined using an optical microscope (BX40; Olympus Corporation, Tokyo, Japan). Cells that contained brown-yellow stained granules in the membrane and cytoplasm were considered positive. Next, five hundred cells in five randomly selected fields under high magnification were counted. Presence of <5% of positive cells was classified as (−), then 5–50% as (+), 50–75% as (++) and >75% as (+++).
Statistical analysis
Data are presented as the mean ± standard deviation, and were analyzed by the two-tailed paired t-test with SPSS 18.0 statistical software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of ADAM10 in glioma
RT-PCR confirmed ADAM10 mRNA expression in the glioma and control groups. As demonstrated in Figs. 1 and 2, the relative mRNA expression levels of ADAM10 in the low- and high-grade groups were 21±1.43 and 32±2.7, respectively, compared with the control group (4.63 ±1.23). Statistical analysis demonstrated significant differences between the high- and low-grade groups (P<0.01) and between the low-grade and control groups (P<0.01).
Figure 1.
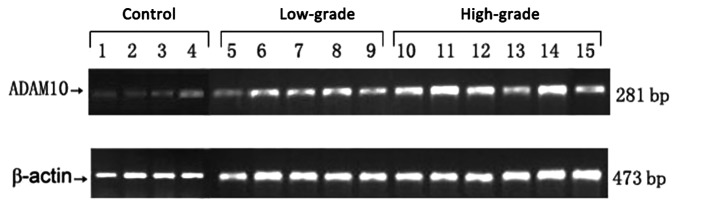
ADAM10 mRNA expression levels detected by RT-PCR. Lanes 1–4 are samples from the non-glioma control group (meningioma); lanes 5–10 are samples from the low-grade group (glioma grades I and II); and lanes 11–16 are samples from the high-grade group (glioma grades III and IV). β-actin serves as an internal control. ADAM10, a-disintegrin and metalloproteinase 10; RT-PCR, reverse transcription-polymerase chain reaction.
Figure 2.
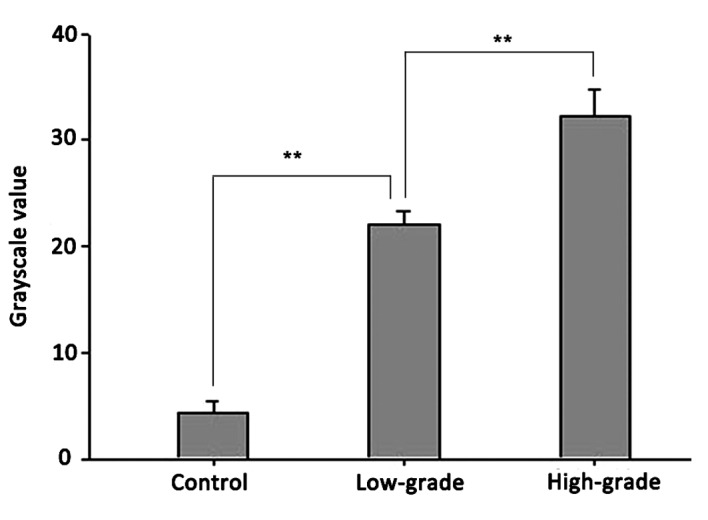
Quantitative histogram of the RT-PCR results. Significant statistical differences existed between the high and low-grade groups and between the low-grade and control groups. **P<0.01. ADAM10, a-disintegrin and metalloproteinase 10; RT-PCR, reverse transcription-polymerase chain reaction.
Immunohistochemical and western blot analyses confirmed ADAM10 protein expression in the controls, and low- and high-grade gliomas. In the western blot assay, two bands for ADAM10 were observed, in which the 100-kDa band is the ADAM10 protein precursor, while the 75-kDa band is its active form. The 100-kDa ADAM10 protein precursor is non-functional and thus was excluded from the analysis. Analysis of the mean grayscale values of the mature 75 kDa band indicated that there was a trend of increasing ADAM10 protein expression correlated with increasing pathological grade (Figs. 3 and 4; Table II). In addition, a significant increase in ADAM10 protein levels was observed in the low-grade glioma group compared with the non-glioma control group (P<0.05), while the ADAM10 protein levels in the high-grade glioma group were significantly higher compared with the control non-glioma (P<0.01) and low-grade glioma groups (P<0.05).
Figure 3.
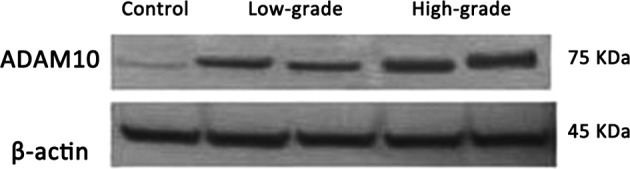
Western blot analysis results for ADAM10 (75 kDa) protein expression. From left to right, the samples are from the non-glioma control, low-grade glioma and high-grade glioma groups. β-actin (45 kDa, lower panel) serves as an internal control. ADAM10, a-disintegrin and metalloproteinase 10.
Figure 4.
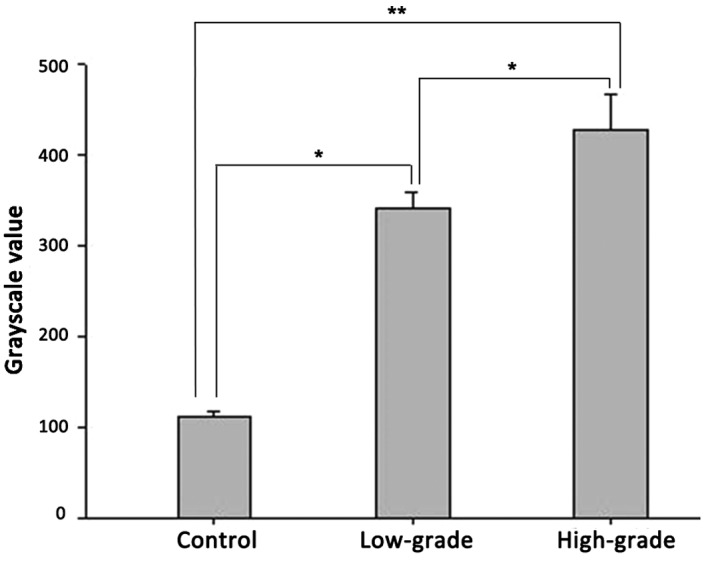
Quantitative histogram of the western blot results. As shown, there were statistically significant differences among 3 groups. *P<0.05 and **P<0.01. ADAM10, a-disintegrin and metalloproteinase 10.
Table II.
Immunohistochemical staining for a-disintegrin and metalloproteinase 10 in different groups.
| Tissues | Number of positive cells (n) | Total number of cells (n) | Percentage of cells (%) |
|---|---|---|---|
| Control | 47 | 500 | 9.4 |
| Low-grade | 269 | 500 | 53.8 |
| High-grade | 382 | 500 | 76.4 |
Localization of ADAM10 protein in tissues
As observed under the light microscope, ADAM10 protein was found to be localized on the cell membrane and blood vessel walls within tumor tissues (Fig. 5). Cells which contained brown-yellow granules in the membrane and cytoplasm were considered to be positive for ADAM10 protein expression. The positive rate of ADAM10 protein expression in the control non-glioma group was only 9.4% of cells, while 53.8% of cells in the low-grade and 76.4% of cells in the high-grade glioma groups were deemed positive for ADAM10 protein expression. Therefore, these observations indicated a malignancy-dependent pattern for ADAM10 mRNA and protein expression.
Figure 5.
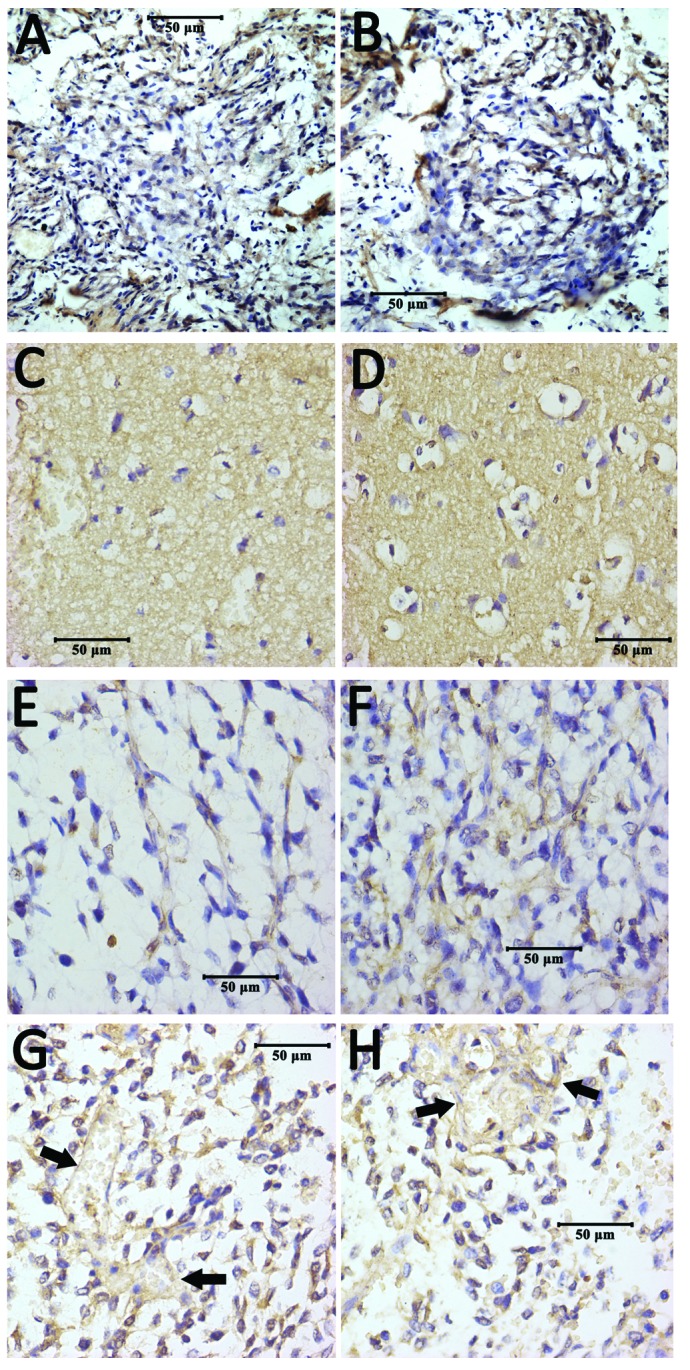
Immunohistochemical staining of ADAM10 protein. (A and B) Representative images from the control group, in which there were no ADAM10 positive cells visible. (C and D) Images from the low-grade group, in which the structure of many tumor cells had been destroyed and the cells appeared vacuous with non-specific background staining; in (D), partial positive staining is observed, which is scattered in tumor cell membranes and in the cytoplasm. (E and F) Representative images from the high-grade group. A relatively greater number of tumor cells with positive staining are present in (E), which is even more apparent compared with (F), where positive staining is demonstrated to be further increased in the perinuclear membrane or the cytoplasm of tumor cells. (G and H) Images from high-grade gliomas, in which blood vessel walls display positive staining (arrows), and the nuclei are stained by hematoxylin. ADAM10, a-disintegrin and metalloproteinase 10.
Discussion
ADAM10, which is secreted as a precursor protein, is a member of the ADAM transmembrane protein family and has four potential functional domains: proteolytic, adhesion, fusion and intracellular signaling domains (13,15,16,18,24,25). Previous studies have reported that a number of ADAMs are involved in the biological processes of malignant tumor cells, including proliferation and invasive growth (26–30). In addition, ADAM10 has been found to be highly expressed in local secretory cells in prostate cancer and expressed in the basal cells of benign glands, while aberrant ADAM10 expression has been demonstrated to correlate with osteosarcoma progression (17,31,32). A previous study has demonstrated that ADAM10 promotes the proliferation of gastric cancer cells by mediating ectodomain shedding and, therefore, activating epidermal growth factor receptor ligands (25). Other studies have demonstrated that ADAM10 plays a role in the hydrolysis of placental type IV collagen, an important extracellular matrix component, further indicating that ADAM10 is involved in cancer invasion and metastasis (33).
In the present study, ADAM10 expression was assessed in different grades of gliomas at the transcriptional and translational levels, while the association between glioma malignancy and ADAM10 expression was investigated. ADAM10 expression was measured in controls (meningioma) using RT-PCR and western blot analysis. The present study demonstrated that ADAM10 expression was significantly higher with increasing malignancy grade. The statistical analysis revealed that there were significant differences among the high- and low-grade groups and the control in a malignancy-dependent pattern, indicating that ADAM10 expression is closely associated with glioma malignancy. In accordance with previous studies on the role of ADAM family members in other malignancies, the present study revealed that ADAM10 may also be involved in glioma growth and invasiveness. The semi-quantitative RT-PCR and western blot analysis demonstrated that the expression level of ADAM10 is highest in high-grade gliomas, and relatively higher in low-grade gliomas compared with the controls. In the control benign brain tumors (meningiomas), ADAM10 was not significantly expressed, as confirmed by the immunohistochemical assay (Table II). Although the specific mechanism of ADAM10 in glioma biology remains unclear, immunohistochemical staining for ADAM10 may provide new insights. As Fig. 5 shows, positive staining was mainly observed on the cell membrane, further confirming that ADAM10 is a transmembrane protein. The results of the present study demonstrated that the higher the tumor grade, the higher the ADAM10 expression level, and thus the more aggressive the tumor. In accordance with existing evidence (12–20,28,31,33,34), one hypothesis is that ADAM10 promotes the invasive growth of glioma cells by hydrolyzing extracellular matrix collagen. This hydrolysis may transmit intercellular signals and promote subsequent ectodomain EGFR ligand shedding, thus stimulating intracellular relevant genes to increase tumor cell proliferation. However, this hypothesis requires further verification.
The present study also revealed that ADAM10 protein was present in the blood vessel walls of glioma tissues but the implication of this finding is unclear at present. However, a previous study suggested that ADAM10 modulates the connections between vessel wall cells by regulating vascular endothelial cadherin, a cell-adhesion glycoprotein that regulates blood vessel permeability, implying a pathological mechanism for the edema formation that can occur in glioma (32). In addition, a previous study reported that ADAM10 may be associated with peripheral migration of glioma cells (34). Based on these findings, ADAM10 may be involved in the regulation of tumor vascular permeability, which is likely to be one of the mechanisms responsible for the formation of evident peritumoral edema in gliomas.
The key biological function of ADAM10 and other ADAM family members is their enzymatic activity as metalloproteinases (11,13–15,18,24,26,34). More specifically, ADAM10 is involved in the intramembrane proteolysis process, whereby it mediates ectodomain shedding of various membrane-bound receptors, adhesion molecules, growth factors and cytokines (35). For instance, ADAM10 is involved in regulating the ectodomain shedding of Notch, human epidermal growth factor receptor 2, CD44, interleukin-6 receptor, amyloid precursor protein and certain cadherins (18). Considering the infiltrative and invasive growth characteristics of gliomas (3,4), the expression of ADAM10 is expected to correlate with not only the pathological grades, but also the aggressiveness of gliomas to a certain extent. In other words, the more malignant the glioma is, the more invasive it may be. Thus, the malignancy-dependent pattern of ADAM10 expression in gliomas may indicate that ADAM10 plays an important role in glioma invasion and may subsequently be used to predict clinical intractability and poor outcome.
In conclusion, the present study demonstrated that ADAM10 expression is closely associated with glioma grade in a malignancy-dependent pattern, indirectly indicating an important biological role for ADAM10 in glioma growth and development, particularly the invasiveness of gliomas. The expression of ADAM10 in the blood vessel walls of tumors may be associated with the formation of peritumoral edema. Further evidence is required to prove these hypotheses in future studies; however, the present study indicates that ADAM10 may be a promising target for glioma therapy.
Acknowledgements
This study was supported by grants from the Liaoning Provincial Natural Science Foundation of China (No. 2013021075 to Bo Qiu) and the Fund for Scientific Research of the First Hospital of China Medical University (No. fsfh1304 to Bo Qiu).
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group: Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Qiu B, Zhang D, Wang Y, et al. Interleukin-6 is overexpressed and augments invasiveness of human glioma stem cells in vitro. Clin Exp Metastasis. 2013;30:1009–1018. doi: 10.1007/s10585-013-9599-0. [DOI] [PubMed] [Google Scholar]
- 4.Qiu B, Zhang D, Tao J, Tie X, Wu A, Wang Y. Human brain glioma stem cells are more invasive than their differentiated progeny cells in vitro. J Clin Neurosci. 2012;19:130–134. doi: 10.1016/j.jocn.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Stylli SS, Howes M, MacGregor L, Rajendra P, Kaye AH. Photodynamic therapy of brain tumours: evaluation of porphyrin uptake versus clinical outcome. J Clin Neurosci. 2004;11:584–596. doi: 10.1016/j.jocn.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Stylli SS, Kaye AH, MacGregor L, Howes M, Rajendra P. Photodynamic therapy of high grade glioma - long term survival. J Clin Neurosci. 2005;12:389–398. doi: 10.1016/j.jocn.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Park DM, Rich JN. Biology of glioma cancer stem cells. Mol Cells. 2009;28:7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 8.Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coniglio SJ, Segall JE. Review: molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol. 2013;32:372–380. doi: 10.1016/j.matbio.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Kaczarek E, Zapf S, Bouterfa H, Tonn JC, Westphal M, Giese A. Dissecting glioma invasion: interrelation of adhesion, migration and intercellular contacts determine the invasive phenotype. Int J Dev Neurosci. 1999;17:625–641. doi: 10.1016/S0736-5748(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 11.Stamenkovic I. Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol. 2003;200:448–464. doi: 10.1002/path.1400. [DOI] [PubMed] [Google Scholar]
- 12.Bulstrode H, Jones LM, Siney EJ, et al. A-Disintegrin and Metalloprotease (ADAM) 10 and 17 promote self-renewal of brain tumor sphere forming cells. Cancer Lett. 2012;326:79–87. doi: 10.1016/j.canlet.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Huovila AP, Turner AJ, Pelto-Huikko M, Kӓrkkӓinen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 17.Zhao R, Ni D, Tian Y, Ni B, Wang A. Aberrant ADAM10 expression correlates with osteosarcoma progression. Eur J Med Res. 2014;19:9. doi: 10.1186/2047-783X-19-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armanious H, Gelebart P, Anand M, Belch A, Lai R. Constitutive activation of metalloproteinase ADAM10 in mantle cell lymphoma promotes cell growth and activates the TNFα/NFκB pathway. Blood. 2011;117:6237–6246. doi: 10.1182/blood-2010-10-313940. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Liu S, Liu K, et al. A disintegrin and metalloprotease (ADAM)10 is highly expressed in hepatocellular carcinoma and is associated with tumour progression. J Int Med Res. 2014;42:611–618. doi: 10.1177/0300060513505500. [DOI] [PubMed] [Google Scholar]
- 20.Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW, Liu TY. Increase of disintergin metalloprotease 10 (ADAM10) expression in oral squamous cell carcinoma. Cancer Lett. 2007;245:33–43. doi: 10.1016/j.canlet.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 21.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–229. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- 22.Qiu B, Li X, Sun X, et al. Overexpression of aquaporin-1 aggravates hippocampal damage in mouse traumatic brain injury models. Mol Med Rep. 2014;9:916–922. doi: 10.3892/mmr.2014.1899. [DOI] [PubMed] [Google Scholar]
- 23.Qiu B, Sun X, Zhang D, Wang Y, Tao J, Ou S. TRAIL and paclitaxel synergize to kill U87 cells and U87-derived stem-like cells in vitro. Int J Mol Sci. 2012;13:9142–9156. doi: 10.3390/ijms13079142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfsberg TG, White JM. ADAMs in fertilization and development. Dev Biol. 1996;180:389–401. doi: 10.1006/dbio.1996.0313. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimura T, Tomita T, Dixon MF, Axon AT, Robinson PA, Crabtree JE. ADAMs (a disintegrin and metalloproteinase) messenger RNA expression in Helicobacter pylori-infected, normal, and neoplastic gastric mucosa. J Infect Dis. 2002;185:332–340. doi: 10.1086/338191. [DOI] [PubMed] [Google Scholar]
- 26.Rocks N, Paulissen G, El Hour M, et al. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–379. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Wildeboer D, Naus S, Amy Sang QX, Bartsch JW, Pagenstecher A. Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J Neuropathol Exp Neurol. 2006;65:516–527. doi: 10.1097/01.jnen.0000229240.51490.d3. [DOI] [PubMed] [Google Scholar]
- 28.Kodama T, Ikeda E, Okada A, et al. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am J Pathol. 2004;165:1743–1753. doi: 10.1016/S0002-9440(10)63429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Abaco GM, Ng K, Paradiso L, Godde NJ, Kaye A, Novak U. ADAM22, expressed in normal brain but not in high-grade gliomas, inhibits cellular proliferation via the disintegrin domain. Neurosurgery. 2006;58:179–186. doi: 10.1227/01.NEU.0000192363.84287.8B. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X, Jiang F, Katakowski M, et al. Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer Sci. 2007;98:674–684. doi: 10.1111/j.1349-7006.2007.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCulloch DR, Akl P, Samaratunga H, Herington AC, Odorico DM. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10:314–323. doi: 10.1158/1078-0432.CCR-0846-3. [DOI] [PubMed] [Google Scholar]
- 32.Schulz B, Pruessmeyer J, Maretzky T, et al. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circ Res. 2008;102:1192–1201. doi: 10.1161/CIRCRESAHA.107.169805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millichip MI, Dallas DJ, Wu E, Dale S, McKie N. The metallo-disintegrin ADAM10 (MADM) from bovine kidney has type IV collagenase activity in vitro. Biochem Biophys Res Commun. 1998;245:594–598. doi: 10.1006/bbrc.1998.8485. [DOI] [PubMed] [Google Scholar]
- 34.Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci. 2009;29:4605–4615. doi: 10.1523/JNEUROSCI.5126-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang CL, Jiang FQ, Xu F, Jiang GX. ADAM10 overexpression confers resistance to doxorubicin-induced apoptosis in hepatocellular carcinoma. Tumour Biol. 2012;33:1535–1541. doi: 10.1007/s13277-012-0405-4. [DOI] [PubMed] [Google Scholar]


