Abstract
A blood-based biomarker assay is a non-invasive way to screen that can identify lung cancer at an earlier stage to improve the clinical outcome. MALAT1 is a broadly expressed, long non-coding RNA in human tissues and is overexpressed in numerous human carcinomas. The potential of MALAT1 in the whole blood of lung cancer was evaluated. In the present study, blood samples of patients with lung cancer and healthy volunteers (controls) were recruited and analyzed by quantitative polymerase chain reaction (qPCR) for MALAT1 expression and clinicopathological data. Lung cancer tissues were also analyzed by qPCR. The expression of MALAT1 in the whole blood of lung cancer was lower compared to the control. The area under the receiver operator curve was 0.718 (P<0.001). Relatively, the expression of MALAT1 was stronger in the whole blood of lung cancer with metastasis compared to non-metastasis. Additionally, the whole blood with bone or brain metastasis exhibited a higher expression of MALAT1 compared to the blood with lymph node or pleura metastasis. Subsequently, a lower expression of MALAT1 was detected in metastatic lymph node tissues than that of the carcinoma in situ of the lung. Taken together, these results indicate that MALAT1 as a biomarker to screen lung cancer may represent a host response to lung cancer.
Keywords: MALAT1, lung cancer, quantitative polymerase chain reaction
Introduction
Lung cancer is one of the most common types of cancer. Lung cancer is the leading cause of cancer mortalities worldwide, including China. There have been certain small improvements in the overall survival rate due to the advances in treatment of lung cancer; however, the 5-year survival rate remains low at ∼15% (1) as the majority of lung cancer cases are diagnosed only when they advanced to more malignant stages. Therefore, it is necessary to screen and diagnose individuals at an earlier stage of lung cancer to increase the time to manage the disease and improve clinical outcome.
A blood-based biomarker assay is a non-invasive method to determine the presence of lung cancer during screening or routine clinical visits, which can be useful in identifying subjects that are most likely to have a malignant lesion in the lung that requires further investigation (2). In nasopharyngeal carcinoma (NPC), a blood-based gene signature (LDLRAP1, PHF20 and LUC7L3) that accurately distinguished NPC patients from controls and from patients with other diseases was identified (3). In ovarian cancer, a signature of genes involved in metastasis, invasion and inflammation were identified in native unstimulated blood leukocytes from ovarian cancer patients with a poor prognosis (4). The circulating STC1 mRNA is a promising biomarker in the peripheral blood of non-small cell lung cancer (5). A previous study identified eight genes that can discriminate patients with lung cancer from healthy controls with a high accuracy (6).
MALAT1 is a broadly expressed, long non-coding RNA (∼8.7 kb in humans) in normal human and mouse tissues and is overexpressed in numerous human carcinomas, including cancer of the breast, pancreas, lung, colon, prostate and liver, suggesting that MALAT1 dysregulation is indicated in the development of numerous types of cancers (7,8). In the present study, the expression of MALAT1 is reported in the whole blood of lung cancer and this was lower compared to the control. The expression of MALAT1 was stronger in the whole blood of lung cancer with metastasis compared to non-metastasis. Additionally, the whole blood with bone or brain metastasis represented a higher expression of MALAT1 compared to the blood with lymph node or pleura metastasis. Subsequently, a lower expression of MALAT1 was detected in metastatic lymph node tissues than that of the carcinoma in situ of lung. Taken together, these results indicate that MALAT1 as a biomarker to screen lung cancer may represent a host response to lung cancer. The present study provides a novel perspective on the role of MALAT1 in peripheral whole blood of lung cancer.
Materials and methods
Patients and blood samples
Blood samples of patients with lung cancer and healthy volunteers (controls) were recruited at Tianjin Medical University General Hospital (Tianjin, China) and Hebei General Hospital (Shijiazhuang, China). Consent forms were obtained from all the study participants according to protocols approved by the hospitals' Research Ethics Board.
Gene expression analysis was performed on 105 blood samples (Table I) and 35 tissue samples obtained from patients with tumors confirmed as lung cancer by hospital pathologists and 65 controls. All the samples were collected between November 2011 and January 2013.
Table I.
Characteristics of the patients and control.
| Characteristics | Lung cancer | Control | P-value |
|---|---|---|---|
| No. of samples | 105 | 65 | |
| Age, median (range) | 51 (39–65) | 49 (35–63) | 0.75 |
| Gender | |||
| Male | 63 | 35 | 0.43 |
| Female | 42 | 30 |
Blood collection, RNA isolation and RNA quality control
Peripheral whole blood (10 ml) was collected from patients in EDTA Vacutainer tubes and RNA was extracted with the AxyPrep™ Blood Total RNA Miniprep kit (Axygen Scientific, Inc., Union City, CA, USA) in compliance with the manufacturer's instructions. Total RNA was assessed spectrophotometrically using a nucleic acid analyzer (Beckman Coulter, Inc., Brea, CA, USA). RNA quality was determined by the absorbance at 260/280 nm quota and quantity was measured as ng/µl.
cDNA synthesis and quantitative polymerase chain reaction (qPCR) gene expression
One microgram of total RNA was transcribed into cDNA in a 20 µl reaction volume using reverse transcriptase for first-strand cDNA synthesis. Following reverse transcription of RNA, all the qPCR reactions were performed using an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems Life Technologies, Foster City, CA, USA) with the designed primers for target genes and an internal control gene, GAPDH. Each sample for each gene was run in triplicate.
Statistical analysis
The gene expression levels in the whole blood of lung cancer patients were compared to those in the control, or in the lung cancer tissues compared to the metastatic lymph node tissues with the use of the Wilcoxon test. The associations between the gene expression levels and potential explanatory variables, including lymph node metastasis, were evaluated with the independent sample t-test. In addition, receiver operating characteristic (ROC) analysis was generated by plotting the sensitivity against the false positive rate (1-specificity) as the discrimination between lung cancer patients and controls. Area under the curve (AUC) was calculated. All the statistical analyses were performed using SPSS, version 16.0 for Windows software (SPSS, Inc., Chicago, IL, USA). Two-sided P-values were calculated and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of MALAT1 in the whole blood of lung cancer and control patients
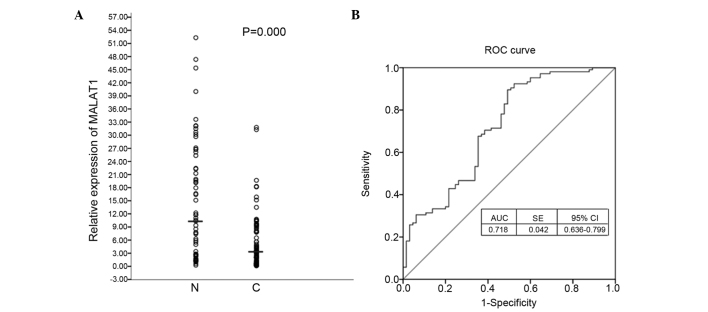
The expression level of the MALAT1 gene was measured in the whole blood of lung cancer and control patients using reverse transcription-qPCR. The expression of MALAT1 in the whole blood of lung cancer patients (n=105) was lower compared to the control (n=65) (Fig. 1A). ROC curves show sensitivity versus specificity in discriminating between lung cancer and control patients (Fig. 1B). The AUC was calculated as 0.718 (P<0.001). MALAT1 has the potential to distinguish between cancer cases and controls with a cutoff value (10.3444) for the clinical test. The correlations between MALAT1 expression and these clinicopathological features were analyzed and the expression of MALAT1 was associated with metastasis (lymph node, pleura, bone and brain) (Fig. 2). The expression of MALAT1 was stronger in the whole blood of lung cancer patients with metastasis compared to weak expression in non-metastasis. In addition, the whole blood with bone or brain metastasis represented higher expression of MALAT1 compared to the blood with lymph node or pleura metastasis.
Figure 1.
MALAT1 expression in the whole blood of lung cancer and control. (A) The expression level of the MALAT1 gene in the whole blood of lung cancer and healthy volunteers (controls) was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The difference was subject to statistical analyses by Wilcoxon test and the median of MALAT1 relative expression is shown. (N, normal control; C, lung cancer.) (B) Receiver operating characteristic (ROC) curves based on RT-qPCR data. The area under the curve (AUC) values and corresponding 95% confidence interval (CI) are reported. SE, standard error.
Figure 2.
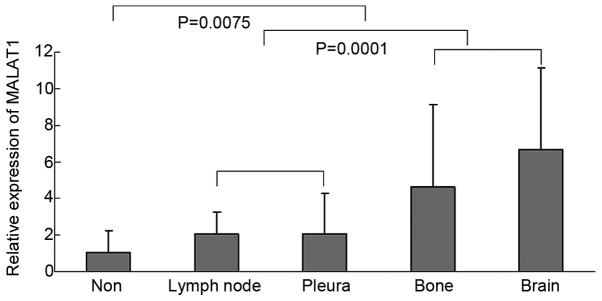
Correlation between MALAT1 expression and metastatic features. The correlation was analyzed among the metastasis (lymph node, pleura, bone and brain). The difference was evaluated with the independent sample t-test.
Expression of MALAT1 in lung cancer and metastatic lymph node tissues
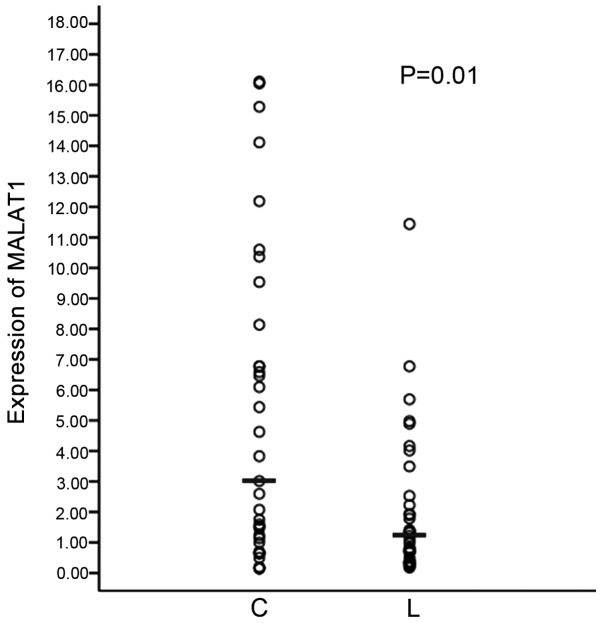
There is a small section of circulating tumor cells (CTCs) that are released into blood from carcinoma in situ in the peripheral whole blood of cancer patients and the expression of MALAT1 is lowest in the blood with lymph node metastasis compared to metastases of other sections. Whether CTCs with a low MALAT1 expression metastasize to lymph nodes was questioned. Subsequently, the expression of MALAT1 in lung cancer and metastatic lymph nodes tissues was examined. As shown in Fig. 3, in the metastatic lymph node tissues, the expression of MALAT1 is lower than that of the carcinoma in situ of lung.
Figure 3.
MALAT1 expression in lung cancer and metastatic lymph nodes tissues. The expression level of the MALAT1 gene was detected using reverse transcription-quantitative polymerase chain reaction. The difference was subject to statistical analyses by Wilcoxon test and the median of MALAT1 relative expression is shown. C, lung cancer; L, lymph nodes.
Discussion
MALAT1 lacks open reading frames and does not translate MALAT1 in vitro to yield peptides, suggesting that MALAT1 functions as a long non-coding RNA (∼8.7 kb in humans) (7,9,10). The long form of MALAT1 is subsequently localized to nuclear speckles (11). MALAT1 is overexpressed in cancer tissues. In a previous study of cervical cancer, the inhibition of MALAT1 in CaSki human cervical cancer cells suppressed cell proliferation and invasion (12). Another study identified that MALAT1 enhanced cell motility of lung adenocarcinoma cells (13). Lai et al (14) reported that MALAT1 overexpression predicted tumor recurrence of hepatocellular carcinoma following liver transplantation. Certain studies have reported that the long non-coding RNAs may be detected in cancers and provide novel tumor biomarkers (15–17).
In the present study, the expression of MALAT1 in the whole blood of lung cancer patients and controls was explored. The expression of MALAT1 in the blood of lung cancer was lower than that of the control. The expression of MALAT1 in blood is contrary to that in tissues. In tissues, the expression of MALAT1 in cancer was higher compared to normal tissues (data not shown). Additionally, the expression of MALAT1 was detected in carcinoma in situ of lung and metastatic lymph node tissues. The expression of MALAT1 in carcinoma in situ was higher compared to that of the metastatic lymph node tissues. These results demonstrate that MALAT1 may be able to identify lung cancer using whole blood.
The mechanistic basis for the alterations in whole blood MALAT1 in the presence of lung cancer is not clear. The whole blood consists of several cell subtypes and subsets of differentiated immune cells, such as neutrophils, lymphocytes and monocytes, which may have different mRNA expression profiles (18). The blood-based biomarkers represent a host response to tumor. The changes in MALAT1 expression in blood show the body's systemic response to the presence of lung cancer, including different clinical characteristics. Controls (healthy patients) have a strong immune system compared to lung cancer. MALAT1 may also be a marker of the cells in immune system. There is a previous study reporting that certain genes that are involved in cancer metastasis and invasion were significantly downregulated in the native unstimulated blood leukocytes from ovarian cancer patients with a poor prognosis (4). Two previous studies showed that the distinct whole-blood RNA expression signatures identified can predict the severity of castration-resistant prostate cancer (19,20).
The expression of MALAT1 in the whole blood of lung cancer patients with metastasis was stronger compared to non-metastasis, which showed that MALAT1 promotes the tumor metastasis and additionally, the whole blood with lymph node metastasis represented a lower expression of MALAT1 compared to bone or brain metastasis. In addition to immunocytes, the peripheral blood of cancer patients contains a small section of CTCs, which are released into blood from carcinoma in situ. The CTCs escape from the primary tumor and settle down at a secondary site to cause metastasis. CTCs spread from the primary tumor and colonize in the lymph node. The settlement is affected by host and molecular characteristics. The expression of MALAT1 is lower in metastatic lymph node tissues than that of the carcinoma in situ of the lungs. The CTCs with the signature of MALAT1 expression are not suitable to locate in the lymph nodes. This may suggest that the immune system, particularly for inactive ones, expressed less of the MALAT1 gene in accordance with the expression of MALAT1 in the whole blood of lung cancer.
Acknowledgements
The present study was supported by grants from the Tianjin Education Committee Foundation (no. 20110101), the National Natural Science Foundation of China (no. 81201645), Doctoral Program Foundation of Institutions of Higher Education (no. 20121202120008) to Dr FJ Guo; the National Natural Science Foundation of China (no. 30973383) to Dr ZG Li; and the Key Project from the National Natural Science Foundation of China (no. 30430300), National 863 Program (no. 2006AAOZA401), National 973 Program (no. 2010CB529405) and China-Sweden Cooperative Foundation (no. 09ZCZDSF04100) to Dr QH Zhou.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PLoS One. 2012;7:e46045. doi: 10.1371/journal.pone.0046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaatar AM, Lim CR, Bong CW, et al. Whole blood transcriptome correlates with treatment response in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2012;31:76. doi: 10.1186/1756-9966-31-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaksson HS, Sorbe B, Nilsson TK. Whole blood RNA expression profiles in ovarian cancer patients with or without residual tumors after primary cytoreductive surgery. Oncol Rep. 2012;27:1331–1335. doi: 10.3892/or.2012.1680. [DOI] [PubMed] [Google Scholar]
- 5.Du YZ, Gu XH, Li L, Gao F. The diagnostic value of circulating stanniocalcin-1 mRNA in non-small cell lung cancer. J Surg Oncol. 2011;104:836–840. doi: 10.1002/jso.21948. [DOI] [PubMed] [Google Scholar]
- 6.Rotunno M, Hu N, Su H, et al. A gene expression signature from peripheral whole blood for stage I lung adenocarcinoma. Cancer Prev Res (Phila) 2011;4:1599–1608. doi: 10.1158/1940-6207.CAPR-10-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 8.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 9.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 13.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 15.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arita T, Ichikawa D, Konishi H, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 17.Sun Y, Wang Z, Zhou D. Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med Hypotheses. 2013;81:319–321. doi: 10.1016/j.mehy.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Passtoors WM, Beekman M, Deelen J, et al. Gene expression analysis of mTOR pathway: Association with human longevity. Aging Cell. 2013;12:24–31. doi: 10.1111/acel.12015. [DOI] [PubMed] [Google Scholar]
- 19.Olmos D, Brewer D, Clark J, et al. Prognostic value of blood mRNA expression signatures in castration-resistant prostate cancer: A prospective, two-stage study. Lancet Oncol. 2012;13:1114–1124. doi: 10.1016/S1470-2045(12)70372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross RW, Galsky MD, Scher HI, et al. A whole-blood RNA transcript-based prognostic model in men with castration-resistant prostate cancer: A prospective study. Lancet Oncol. 2012;13:1105–1113. doi: 10.1016/S1470-2045(12)70263-2. [DOI] [PubMed] [Google Scholar]