Abstract
The advent of genomics in the study of developmental mechanisms has brought a trove of information on gene datasets and regulation during development, where the Zic family of zinc-finger proteins plays an important role. Genomic analysis of the modes of action of Zic3 in pluripotent cells demonstrated its requirement for maintenance of stem cells pluripotency upon binding to the proximal regulatory regions (promoters) of genes associated with cell pluripotency (Nanog, Sox2, Oct4, etc.) as well as cell cycle, proliferation, oncogenesis and early embryogenesis. In contrast, during gastrulation and neurulation Zic3 acts by binding the distal regulatory regions (enhancers, etc) associated with control of gene transcription in the Nodal and Wnt signaling pathways, including genes that act to break body symmetry. This illustrates a general role of Zic3 as a transcriptional regulator that acts not only alone, but in many instances in conjunction with other transcription factors. The latter is done by binding to adjacent sites in the context of multi-transcription factor complexes associated with regulatory elements.
Keywords: Enhancer, Gastrulation, Left-right asymmetry, Neurogenesis, Promoter, Stem cells, Transcription, Zebrafish.
INTRODUCTION
The patterning of the embryo is achieved through a process involving determination of embryonic body axes and defining which cell types develop at each embryonic coordinate. At the core of the mechanism regulating this developmental precision are interconnected gene regulatory networks (GRN) driven by transcription factors (TFs), which control the expression of downstream target genes. It is well established that TFs regulate the tissue-specific transcription of downstream genes by interacting with short (typically 6 – 12 bp) DNA motifs in regulatory elements such as enhancers. DNA looping subsequently brings the TF – enhancer complex close to the target promoter, allowing initiation of transcription [1]. However, the exact mechanism of how binding of TF to regulatory elements is translated into precise spatiotemporal expression of many target genes remains incompletelyunderstood. This is mainly due to limitations of conventional approaches, which focus on the analyses of singular interactions between TFs and cis-regulatory elements [2]. This type of approach fails to answer wider questions including, but not limited to, the variety of genes and/or regulatory elements regulated by any given TF. Next generation sequencing technologies made possible an unbiased analysis of genome-wide TF binding. Embracing these types of approaches, here we review recent progress in the application of genomics to study the role of Zic3 in the molecular mechanisms of developmental regulation.
THE ZIC FAMILY OF TRANSCRIPTION FACTORS
The Zic family proteins are known for their involvement in multiple aspects of embryonic patterning [3]. Their study dates back to almost twenty years ago, when the first gene in the family, murine Zic1, found abundantly in the granule cells of the cerebellum, was cloned [4]. The expression of Zic1 was also found along the dorsal neural tube in the early embryo. subsequent studies identified two other Zic genes, Zic2 and Zic3, similarly expressed in the dorsal neural tube [5]. Comparisons of DNA sequences and gene structures of the three Zic genes revealed their homology to the odd-paired gene of Drosophila, mostly known to specify the anterior-posterior identity of embryonic body segments [6]. Additional vertebrate Zic genes were subsequently identified and characterized [7] making a total of five in frog, chicken, and mammals. Two additional zic genes are present in zebrafish: one arose from gene duplication (zic2b)[8], and another one (zic6)represents a molecular evidence of the early existence of the third pair of Zic genes (Zic3-6) similar to that of the Zic1-4 and Zic2-5 pairs. To date, no evidence exist of the presence of Zic2b in tetrapods, latimeria and sharks, which suggest that it never evolved outside of the teleost lineage; on the other hand Zic6 remains only in teleosts [9].
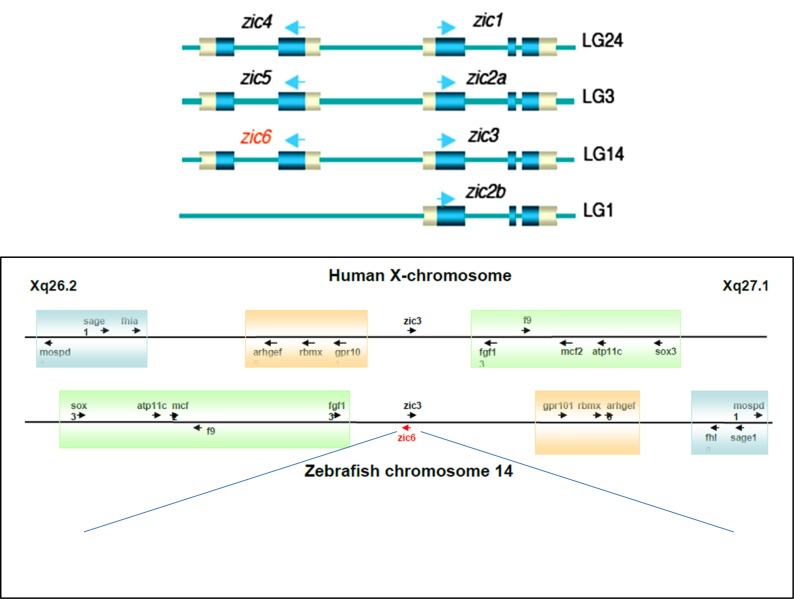
Vertebrate Zic genes are generally located on opposite DNA strands as head-to-head pairs. For instance, Zic1-Zic4 is located in this configuration on human chromosome 3, mouse chromosome 9, and zebrafish chromosome 24; Zic2-Zic5 on human chromosome 13, mouse chromosome 14, and zebrafish chromosome 3; Zic3 on mammalian X chromosome and paired with zic6 on zebrafish chromosome 14 [9a] (Fig. 1). Such close proximity of pairs of genes have been proposed to facilitate the sharing of regulatory regions, which was supported by the similarities in spatiotemporal expression patterns and overlapping functions between pairs of Zic genes [3b, 10]. Nevertheless, all members of the Zic family share a characteristic expression pattern - during gastrulation, Zic genes are expressed in the neural plate and play a key role in the development of the nervous system; later on their expression is detected in the dorsal neural tube and paraxial mesoderm [2, 8, 11]. Interestingly, conservation of this expression pattern extends beyond vertebrates, as demonstrated by characterization of Zic-like genes in amphioxus and ascidians [12]. Moreover, the role of Zic genes during neural development is conserved in all organisms that possess a nervous system [13], suggesting that these genes play an important role in the development and evolution of the nervous system. Comparative analysis across different metazoan phyla revealed that zic genes probably evolved from an ancestral gene of the gli/glis/nk-like family that existed in the last common ancestor of the placozoans, cnidarians, and bilaterians. In these basal metazoans, zic genes are expressed in the endomesodermal tissues and highly neuralized developing tentacles, indicating that their function has likely been conserved since the early stages of metazoan evolution [12b, 14]. However, despite this knowledge, an important question which these evolutionary studies do not answer is whether the molecular mechanism of these ancestral Zic genes is conserved in different tissues. It seems that to answer this question one needs to evaluate a mode of interaction of the Zic proteins with their targets in tissues derived from different germ layers.
Fig. (1).
The pairwise arrangement of zic genes in the zebrafish genome. An additional zic gene in zebrafish, zic6, is located in pair with zic3 on chromosome 14. Although zic6 has been lost in higher vertebrates, the fragment of chromosome 14 containing zic3 retains the syntenic relationship with that of the human X-chromosome
Although members of the Zic family have overlapping functions, loss of function of each individual gene causes a distinct phenotype, suggesting a unique role for each gene [3b, 15]. The roles of Zic family members in development have been addressed in several earlier reviews and readers are directed to those written by Aruga [3c, 16], Grinberg and Millen [15], Merzdorf [3b], and Houtmeyers et al. [17]. This review will focus on Zic3, whose roles in multiple developmental processes have been intensely characterized recently.
ZIC3 IN HUMAN DISEASE
In 1997, a linkage analysis in five different families with heritable X-linked situs abnormalities identified several different mutations affecting the ZIC3 locus [18]. This established ZIC3 as the first gene associated with left-right patterning defects such as randomization of asymmetry of internal organs (situs ambiguus) or their mirror-image reversal (situs inversus). Additional study of 194 individuals with different forms of X-linked heterotaxy revealed eight different allelic variants in a form of missense or nonsense mutations. Most of them were found in the conserved Zn-finger domain of the ZIC3 [4], which encompasses the 2nd - 5th Zn-fingers [19]. This region seems to be most commonly associated with the disease [18, 20]. More recent screening of 440 unrelated heterotaxy patients revealed eight novel mutations, including five in the N-terminal of ZIC3 [21]. Interestingly, the mutant variants affecting the Zn-finger domain of ZIC3 showed a high degree of aberrant accumulation of ZIC3 in cytoplasm, while in mutations affecting the N-terminal of ZIC3 this defect was less obvious and correlated with severity of disease phenotype [18, 21, 22]. The Zn-finger domain mutations affect the strong atypical nuclear localization signal located in Zn-fingers 2 and 3, which causes mislocalization of ZIC3 to the cytoplasm and prevents activation of target genes [21, 23]. Mutations of ZIC3 also cause a wide spectrum of other disease phenotypes, including congenital heart defects, lumbo-sacral, urogenital and biliary system malformations as well as CNS defects [3a, 18-22, 24]. The complexity of phenotype arising from Zic3 disruption reflects the diverse roles of this TF in regulation of multiple aspects of embryonic development.
ZIC3 AS A TRANSCRIPTION FACTOR
Profiling of Zic3-binding sites using ChIP-chip in mouse ES cells covered 17,000 promoters spanning regions between -5.5kb to +2.5kb of transcription start sites and revealed potential involvement of Zic3 in regulation of some 300 genes, including several linked with pluripotency [25]. Application of next generation sequencing (NGS) allows an unbiased genome-wide assessment of Zic3 binding sites by ChIP-seq, which revealed that a third of Zic3 binding sites were identified within the promoter region (Hong et al., unpublished). A similar approach was applied to study transcriptional activity of Zic3 in the developing zebrafish embryo at 8 hpf, when germ layers are formed and neural induction commenced, and at 24 hpf in the dorsal neural tube during differentiation of primary neurons [26]. This analysis revealed that only a relatively small fraction of Zic3-binding events (8-9%) were associated with promoters. Most of these events were mapped to distant genomic locations. This is in line with an idea that Zic3, similar to other TFs regulates gene activity through long distance regulatory elements [27]. Hence the results of these studies led to the formulation of novel hypotheses regarding Zic3 function.
First, a difference in localization of Zic3 binding sites in stem cells and during embryogenesis possibly reflects changes in the role of this TF during different developmental periods. In stem cells that are in a relatively stable pluripotent state Zic3 often acts as a general TF that binds to the core transcription machinery. This seems to be a common feature among TFs known to regulate ES cell pluripotency in mouse, such as Oct4, Stat3, and Klf4 all of which often bind sites within promoter regions [28]. In contrast, during embryogenesis, when cells actively differentiate in vivo, Zic3 binding to distal elements prevails. Such shift in site-specificity of Zic3 suggests an acquisition of cellular functions specific for differentiating cells. A precise mechanism of this phenomenon remains unknown. At chromatin level it could be due to a decrease in availability of binding sites in promoters or increase in availability of distant binding sites. Both explanations suggest major epigenetic changes taking place during transition from a period of extensive cell proliferation to a period of cell fate determination and differentiation. Epigenetics rearrangements in the form of genome-wide histone methylation pattern changes on gene promoters have been well documented during the midblastula transition [29] and could thus support a model of TF binding site accessibility. Equally important are changes at transcriptome level, which in principle could be both a cause and/or outcome of transcriptional regulation. A shift in Zic3 site-specificity correlates with replacement of maternal transcripts by zygotic ones [30]. Future studies could focus on investigating the relationship between these two events through analysis of epigenome profile and nucleosome occupancy by ChIP-seq or ATAC-seq [31] around the Zic3 binding sites. Other genomics approaches such as variations of the chromatin conformation capture methods - 3C, 4C, and 5C [32], or ChIA-pet [33] could help to determine actual interactions between Zic3-bound regulatory elements and its target. In a larger perspective, a topological map of genomic interaction domains [34] in zebrafish, when available, will greatly facilitate the determination of interactive regulatory domains between different TFs and regulatory elements.
Second, a consensus binding motif of Zic3 in zebrafish is highly similar to that found in mouse ES cells [25] (Hong et al., unpublished). In sharp contrast, most of the surrounding regions appear to be poorly conserved in evolution. It is well documented that the evolution of divergent traits mostly involves modifications of regulatory elements rather than structural or functional changes in effector molecules, as the latter may impose dramatic changes in the GRN controlling development [35]. In accordance with this idea, the binding sites of Zic3 diverge greatly while their core structure and possibly also their binding specificity remain largely conserved across metazoans [14, 26].
Lastly, a large group of Zic3 binding sites are unable to induce a reporter. This can be interpreted as these sites being non-functional or that they perform functions other than enhancers. Analysis of such sites requires experimental output other than an increase in transient expression of reporter during embryogenesis. Possibly such sites could become functional later on or during adulthood. It is also possible that Zic3 requires interacting partners to exert its transcriptional inducing activity at these sites. This possibility is especially attractive since binding motifs of other TFs are often identified in proximity to Zic3 motifs (Winata, unpublished).
Profiling of TF binding sites as well as enhancer studies have demonstrated that multiple TFs binding sites tend to co-localize with enhancers, some even forming large regulatory complexes of up to 50 kb, which previously were termed ‘super enhancers’ [36]. Co-binding of a particular TF with different partners has been shown to cause transcriptional outcome distinct from the one brought about by a single TF. Presence of other TFs’ binding sites nearby Zic3 peaks therefore suggests that Zic3 may act in multi-TF complexes. Among possible candidates for Zic3 binding partners are Gli proteins. These effectors of Hh signaling are structurally similar to Zic [4]. Gli-Zic physical interactions as well as Zic ability to bind Gli consensus motif [37], suggested an interaction with the Hh signaling pathway. This is further supported by the fact that a deficiency of Zic2 has been linked to holoprosencephaly connected to defects in Hh signaling [38]. Finally, genome-wide analysis of Zic3 binding sites showed that almost half of all Zic3 binding sites contain both Zic3 and Gli motifs [26]. This provided additional support for Zic-Gli interaction in regulation of gene activity. Interestingly, the Hh signaling pathway is activated as a result of zygotic transcription, i.e. after a shift towards Zic3 regulation of enhancers. The same could be true regarding other conserved binding sites detected in proximity of Zic3 motifs (Winata, unpublished), which may become functional later on. At least for now, without detailed study of these potential interacting partners, it is difficult to determine the exact nature of their interaction with Zic3.
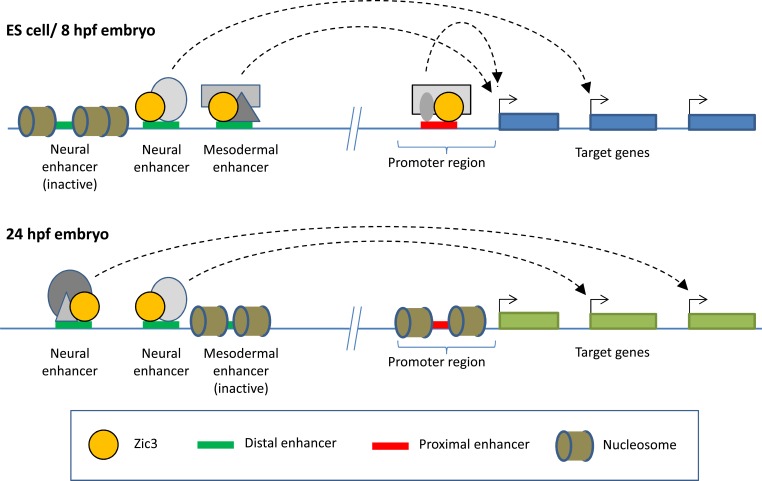
Given a developmental shift from promoter-driven transcriptional regulation by Zic3 to enhancer-driven regulation and possible interaction with some other TFs, a mechanism involving Zic3-mediated transcriptional regulation in different spatiotemporal contexts could be illustrated as in (Fig. 2).
Fig. (2).
Proposed model of Zic3 regulatory mechanism. In pluripotent ES cells, Zic3 is likely to act as a general TF, binding to basal transcriptional elements near the promoter of pluripotency genes. In the developing embryo, Zic3 binds mainly to distal enhancer elements to regulate tissue-specific expression of target genes. The binding to different enhancer elements is regulated spatiotemporally through epigenetic mechanisms or recruitment by different binding partners represented by grey colored shapes.
ZIC3 IN DIFFERENT CELLULAR AND DEVELOPMENTAL CONTEXT
Analysis of Zic3 interactions in mouse ES cells revealed a role of this TF in inhibiting endodermal differentiation [25, 39]. At the same time when co-expressed with the Oct4, Sox2, and Klf4, Zic3 enhances the reprogramming of human fibroblasts into cells that resemble neural progenitors [40]. This suggests that Zic3, like other members of the Zic family, tends to promote neural fate at the expense of endodermal or mesodermal fates. This idea was further supported by an observation that upon inhibition of the Zic3 function, mesendodermal markers expand [26]. In neural tissue Zic genes seem to control a balance between cell proliferation and differentiation. Their overexpression results in inhibition of neuronal differentiation and reduction of cell proliferation [41].
Zic genes are some of the earliest TFs expressed in the neuroectoderm, where Zic1, Zic2, and Zic3 expression precedes that of the proneural genes [2, 11a, 42]. The zebrafish Zic3 expression is higher in the posterior dorsal neuroectoderm in contrast with the other two genes with higher expression anteriorly [11d]. Analysis in Xenopus and zebrafish have shown that an induction of expression of zic1-3 in dorsal neuroectoderm triggered by inhibition of the ventralizing BMP activity marks the earliest event in determination of the neural fate [11a, 11d, 43]. In zebrafish, zic3 expression in mutants with decreased BMP signaling expands ventrally, showing that BMP activity is necessary for restriction of zic3 expression [11d].
A role of Zic3 during neural induction have been established through studies conducted mostly in Xenopus or zebrafish. The pioneering study by Nakata and colleagues [11a] demonstrated that the overexpression of Zic3 in Xenopus embryo results in an expansion of neural and neural crest derivatives, while ectopic expression of Zic3 in animal cap cells induces the expression of proneural and neural crest genes. However, this is seemingly at odds with a known function of Zic genes in inhibition of neural differentiation. For example, Zic2 was shown to antagonize neural differentiation in the floor plate of Xenopus [44], which suggests that a distinct Zic3 function observed during Xenopus neural induction [11a] is not due to species-specific differences, but rather, differences in developmental stage. Analysis of genome-wide binding sites, combined with functional analysis of Zic3, revealed that Zic3 positively regulates genes essential to maintain neural progenitors during neuroectodermal specification [26]. Some of these are direct targets of Zic3 (dlx4b and msxe), whereas others could be regulated indirectly (msxc, irx1a, irx7). Other targets of Zic3 include several Her genes implicated in the Notch signaling (her4.2, her6, her9), These genes are expressed in the neural plate and its marginal zone containing proliferating progenitors contributing into dorsal neural tube [45]. The identification of neural pre-pattern genes as downstream targets of Zic3, along with the repressive action of Zic3 on proneural genes [26], suggests that Zic3 acts to maintain a certain level of proliferation of neural progenitors resulting in a particular number of neurons. This implies that Zic3 overexpression [11a] may cause an increase in proliferation of neural progenitors, which results in an overall increase of differentiating neurons. In line with this suggestion mouse mutants of Zic1 and/or Zic2 exhibit reduced cell proliferation and enhanced expression of motor neuron marker Wnt7a in the cerebellum [46]. Therefore, Zic3 seems to act by maintaining an undifferentiated state of neural progenitors by positive regulation of neural fate repressors and, possibly, negatively regulating proneural genes. In contrast, a loss of Zic3 function caused an increase of neural differentiation markers, such as neurog1 and her9, indicating the repressive action of Zic3 on neural differentiation. Interestingly, binding sites of Zic3 were also found within 100kb of neurog1, neurod4, and ncam1a promoters, which suggest that Zic3 could also directly regulate genes involved in neural differentiation [26]. In particular, this mode of action is consistent with Zic3 action in parallel to Notch, which is supported by changes on expression of her9 as well as association of her4.2 and her6 with Zic3 binding peaks [26].
Zic proteins promote differentiation of neural crest cells. This cell lineage originates from precursor cells located during gastrulation at the periphery of the neural plate. Together with precursors of roof plate (see below), they converge at the dorsal neural tube as a result of neurulation. subsequently, neural crest cells undergo epithelial to mesenchymal transition, delaminate from the roof plate, and migrate out of the neural tube to differentiate into various cell types [47]. Zic genes are known to be involved in migration and differentiation of the neural crest. Overexpression of Zic1, Zic2, Zic3 and Zic5 in Xenopus embryos resulted in hyperplastic neural crest tissue and expansion of neural crest markers [7a, 11a, 44, 48]. Loss of Zic2 and Zic5 functions in mouse resulted in a decrease of neural crest cells and malformations of the structures they contribute towards [10a, 49]. Zic genes were also expressed in the chick neural crest [50]. In zebrafish, neural crest markers such as foxd3 and pax3a were identified as downstream targets of Zic3 at 24 hpf [26]. These genes involved in neural crest induction and migration [51] were down-regulated upon zic3 knockdown. This suggested that Zic3 positively regulates their transcription. Although this result was derived from observations at 24 hpf, i.e. later than the time of neural crest specification and migration from the dorsal neural tube [52], zic3 is constantly expressed in neural crest cells starting from gastrula. Its role in neural crest migration can therefore be extrapolated based on this evidence.
Upon migration of the neural crest cells out of a neural tube, the roof plate becomes the most dorsal cell lineage [53]. Zic3 negatively regulates several proneural bHLH genes, such as neurog1, neurod4 and her9 [26]. This may prevent differentiation of the roof plate cells to maintain these as signaling glia. In the zebrafish, Zic1 and Zic4 control the expression of roof plate determinant lmx1b [54], which is also a target of Zic3 [26]. Zic6 have been implicated in regulation of cell adhesion in the dorsal neural tube during elongation of the roof plate [55]. Hence Zic genes regulate multiple aspects of roof plate development.
It is accepted that cell specification in the dorsal spinal cord depends mostly on Gli3-independent Wnt signaling. Hence it comes as no surprise that several genes of the Wnt signaling pathway expressed in the dorsal neural tube are targets of Zic3 [11d, 26, 56]. This developmental regulation may play a role in a major morphogenetic rearrangement that prospective roof plate cells undergo between 24 hpf and 36 hpf. Being initially polarized along the medio-lateral axis, these cellsrearrange polarity along the dorso-ventral axis [55] and a deficiency in the Zic genes affects this process (I.K., unpublished). lgl2 and dlg2 are Zic3 targets expressed in the roof plate where they regulate cell polarity at the level of cell adhesion [26, 57]. Hence it is possible that Zic3 regulation of lgl2 and dlg2 plays an essential part in regulation of cell adhesion necessary for reorientation of the prospective roof plate cells and their stretching morphogenesis [57e].
Subsequent stages of dorso-ventral patterning of the neural tube involve both Gli3-dependent and -independent mechanisms that mediate Wnt action at intermediate and ventral levels. In the ventral neural tube Wnts expressed in the floor plate contribute into development of motor neurons [58]. The mechanisms by which Wnts pattern the neural tube in a Gli3-independent manner lack a few important details. It was proposed that Wnts acting in parallel with Bmps directly control the expression of homeodomain and basic helix-loop-helix (bHLH) TFs [59]. But in absence of a mechanism for delivery of Wnts expressed dorsally into the ventral neural tube this model remains incomplete. This is of particular importance since, unlike some other morphogens, the hydrophobic Wnts do not diffuse efficiently and act only at a short distance from Wnt producing cells [60]. In Drosophila the long-distance transport of the Wnt-related Wg is achieved by specialized cell extensions (cytonemes) [61] or transcytosis [62]. Morphogens are often secreted from highly polarized cells such as the roof plate. As a matter of fact the roof plate is tightly aligned with stem-like cells prior to, during and after stretching morphogenesis of the roof plate. Such elongation of the roof plate allows a long distance transport of Wnts across most of the neural tube [55]. Furthermore, it has been shown that the secreted Frizzled –related proteins enhance the diffusion of Wnt ligands to expand their signaling range [63]. Since Zic3 negatively regulates sfrp1a in the roof plate [26], this could be a mechanism to restrict a spread of Wnt signaling to a vicinity of a small apical footprint of the roof plate cell. It seems that the long-distance Wnt signaling could be regulated by the in-built transcriptionally regulated molecular systems that prevent Wnt spread in the extracellular space by blocking the soluble Wnt-binding modulators. Given a well-known role of Wnts as oncogenes and an activation of Zic expression in brain tumors, the regulation of Zic3 and its targets in tumors should be explored further in search for anti-cancer therapy.
ZIC3 IN GASTRULATION AND LEFT-RIGHT (L-R) PATTERNING
Zic3 is distinguished from other Zic family members by its involvement in the L-R patterning [3c]. In vertebrates, the L-R axis is established in the early mesoderm by means of left-sided Nodal signaling which induces the expression of Pitx2, a key TF which directs the development of left-sided structures such as heart and determines the directionality of gut looping [64]. In the mammalian embryo, a leftward fluid flow caused by ciliary rotation in the embryonic node [65] maintains a left-sided localization of Sonic hedgehog morphogen and retinoic acid known for their role in regulating developmental processes [66]. These signals are necessary for the establishment of Nodal signaling at the left portion of the lateral plate mesoderm. In zebrafish, the Kupffer’s vesicle is a structure equivalent to the mouse node [67]. Nodal cilia rotation in the Kupffer’s vesicle causes localization of Ca2+ ions, which induces Notch and BMP4 on the left lateral plate mesoderm. These subsequently activate Pitx2 expression [67b]. A similar mechanism acts to establish the left side localization of Nodal in the neural plate resulting in asymmetry of the brain [68]. In frogs, cortical rotation of the embryo during fertilization induces left-sided processing of the Vg1 protein, which in turn results in left-sided Xnr1 expression. This subsequently directs L-R axis specification through Pitx2 activation [69]. Zic3 expression in the mesoderm is induced by Xbra, a TF regulating notochord development [70]. Overexpression of Zic3 in the right-side embryonic mesoderm results in right-sided expansion of left side markers Pitx2 and Xnr1, culminating in defective heart and gut looping [70]. This indicates that Zic3 acts as a determinant of the left-sided signaling pathway. In mouse, targeted deletion of Zic3 resulted in congenital heart defects and pulmonary reversal or isomerism [3a, 24a]. The expression pattern of Nodal and Pitx2 in these mutants was randomized similar to the Xenopus overexpression study. More recently, Cast et al. [71] showed that Zic3 loss-of-function (LOF) causes laterality defects in Xenopus and zebrafish in support of the conserved role of Zic3 in regulating L-R specification in vertebrates.
Despite its role as a determinant of L-R asymmetry, Zic3 is not expressed unilaterally [11a, 11d, 70]. Moreover, organs for which laterality is affected by Zic3 LOF do not express Zic3, raising a question as to how Zic3 confers L-R patterning. Overexpression of Zic3 in the right hand-side blastomeres of the Xenopus embryo, and not those at the left side, resulted in L-R axis disruption, suggesting that Zic3 acts depending on the spatial context [70]. More recent studies suggested that an action of Zic3 in L-R asymmetry is an early developmental event, in which Zic3 was shown to regulate the formation of the dorsal organizer, and therefore the midline structures [72], through its suppression of the canonical Wnt signaling [26]. Defects of the midline structures are associated with aberrations in L-R patterning [73] and mutations of genes in the Nodal signaling pathway (NODAL, ACVRIIB, FOXH1, and LEFTYA) known to regulate midline development have been identified in patients with heterotaxy [74]. Furthermore, Cast et al. [71] demonstrated that upon Zic3 LOF defects in convergence-extension (C-E) correlate with subsequent defects in L-R patterning. Therefore, an involvement of Zic3 in C-E could be sufficient to ensure proper L-R patterning later on. However, considering that Zic3 expression persists in mesoderm after establishing embryonic midline, it is possible that Zic3 regulates L-R specification through a combination of interaction with proteins involved in early midline development and direct regulation of components of L-R specification. Genomic study in zebrafish suggested that this is indeed the case [26]. While Zic3 downstream targets include genes acting in the Nodal and canonical Wnt pathways that regulate early midline development, Zic3 also regulates genes directly implicated in L-R patterning, which include members of the non-canonical Wnt (or planar cell polarity) signaling pathway, such as dvl2, invs, and vangl2, known to regulate ciliogenesis in the mouse node and zebrafish Kupffer’s vesicle [75]. Disruptions in ciliogenesis cause human left-right patterning disorders linked to mutations in genes encoding motor proteins responsible for cilia function [76]. Taken together, Zic3 is required at two stages of L-R patterning through its regulation of midline development as well as ensuring the proper formation and function of the Kupffer’s vesicle.
ZIC3 AND GLOBAL REGULATION OF DEVELOPMENT
The role of Zic3 in multiple, disparate aspects of development reflects its ‘mosaic pleiotropism’ [35b, 77]. This property is exemplified by its involvement in the patterning of at least two different germ layers (ectoderm and mesoderm), or its role in activating different pathways at different developmental stages. The ability of TF to perform multiple functions in different spatiotemporal context could be achieved through interactions with different partners which confers spatiotemporal specifity of its function [35b]. In the case of Zic3, the presence of this mechanism is supported by the identification of binding sites of different TFs located nearby Zic3 binding sites, as well as the evidence of possible physical interactions between Zic3 and Gli proteins.
Within the wider context, comparative studies of metazoans showed that the conserved Zic protein is repeatedly utilized in developmental processes during evolution. This is compatible with an idea of evolutionary ‘bricolage’ [78], which manifests itself as redeployment of existing sets of molecules during evolution of new GRNs that acquire novel developmental functions [35b]. The novel features generated from redeployment of a conserved TF often results in changes in the sequences of cis-regulatory elements (CREs) in the form of addition or deletion of a TF binding site, or the modification to the strength of its regulatory effects through changes in the number of binding sites (reviewed in [35b]). The case of Zic3 illustrates this principle – a majority of Zic3 binding sites are surrounded by poorly conserved regions, which may suggest distinct compositions of multiprotein complexes binding to the target CREs, resulting in evolutionary diversification. It is possible that an additional round of genome duplication in teleosts further contributed into relaxing selection pressure on CREs as it led to even greater diversification of regulatory elements. This could be seen not only due to the genome-wide shift in a mode of Zic3 binding. It also correlates with a shift in Zic3 functionality, which is evident due to a difference in GO enrichment of associated genes during the developmental stages studied. Importantly, the recognition motif of Zic3 involved in two GRNs remains the same, which highlights the importance of its pleiotropism.
In this context, it is worthwhile to mention competence, an actively acquired ability to respond to an inductive signal [79]. Competence could be dictated by the epigenetic state of chromatin in responding cells due to the developmental regulation of accessibility of DNA regions, i.e. enhancers and promoters. An analysis of developmental regulation of genetic activity by Zic3 revealed an important genome-wide switch from regulation of the promoter-driven cellular functions during pluripotency state towards enhancer-driven regulation of functions associated with progressing development – cell migration, commitment and determination during gastrulation, as well as cell differentiation in the dorsal neural tube [25, 26] (Fig. 3). Given a role of Zic genes in brain tumors [80], it is easy to imagine that under pathological conditions which involve dedifferentiation, a reversal from enhancer-driven regulation towards promoter-driven general cellular activities such as cell proliferation may take place. When supported by experimental evidence this emerging knowledge may help to formulate a novel paradigm of searching druggable targets.
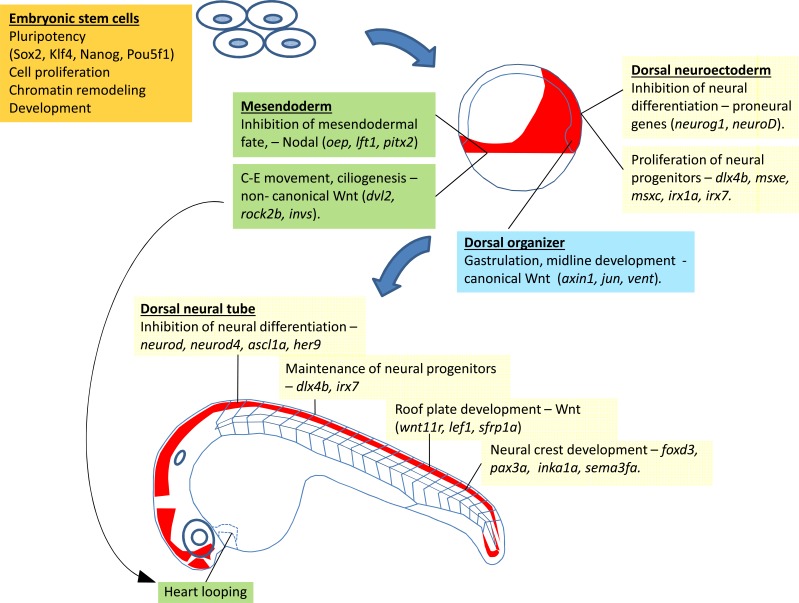
Fig. (3).
Summary of the multiple roles of Zic3 in different spatiotemporal contexts in pluripotent cells and during zebrafish development. Expression of zic3 is indicated with red shade. Functions of Zic3 within a specific expression domain, as well as the relevant downstream target genes (direct and indirect) are denoted in colored boxes.
CONCLUSION
Since its first characterization two decades ago, the roles of Zic3 in various aspects of embryonic development have been increasingly recognized. Importantly, it provides an example of the multiple utilization of a single TF in various developmental processes. Genomic studies using ChIP-seq has enabled elucidation of developmental changes in the molecular regulatory mechanism involving a mode of interaction of Zic3 with regulatory regions and determination of a large number of direct and indirect targets of Zic3 in various spatiotemporal contexts. Future characterizations of genetic and epigenetic factors, which determine spatiotemporal specificity of Zic3 action will further illuminate the molecular mechanism of differential Zic3 deployment across different developmental stages and cell types, as well as provide invaluable insights into the general mechanism of regulation of pleiotropic factors in development.
ACKNOWLEDGEMENTS
Authors are thankful to S. Mathavan for critical discussions and David Racine for expert English editing. CLW lab is supported by the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 316125 (Fishmed). VK lab is funded by the Agency for Science Technology and Research of Singapore.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
References
- 1. (a) Ong C.T, Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b)Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 2013;25(3):387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech. Dev. 1998;75(1-2):43–51. doi: 10.1016/S0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 3.(a) Ware S. M, Harutyunyan K. G, Belmont J. W. Zic3 is critical for early embryonic patterning during gastrulation. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;3(1-2):776–85. doi: 10.1002/dvdy.20668. [DOI] [PubMed] [Google Scholar]; (b) Merzdorf C.S. Emerging roles for zic genes in early development. Dev. Dyn. 2007;236(4):922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]; (c)Aruga J. The role of Zic genes in neural development. Mol. Cell. Neurosci. 2004;26(2):205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Aruga J, Yokota N, Hashimoto M, Furuichi T, Fukuda M, Mikoshiba K. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J. Neurochem. 1994;63(5):1880–1890. doi: 10.1046/j.1471-4159.1994.63051880.x. [DOI] [PubMed] [Google Scholar]
- 5.Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman V.M, Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem. 1996;271(2):1043–1047. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- 6.Benedyk M.J, Mullen J.R, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8(1):105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- 7.(a)Nakata K, Koyabu Y, Aruga J, Mikoshiba K. A novel member of the Xenopus Zic family, Zic5, mediates neural crest development. Mech. Dev. 2000;99(1-2):83–91. doi: 10.1016/S0925-4773(00)00480-9. [DOI] [PubMed] [Google Scholar]; (b) Ishiguro A, Inoue T, Mikoshiba K, Aruga J. Molecular properties of Zic4 and Zic5 proteins: functional diversity within Zic family. Biochem. Biophys. Res. Commun. 2004;324(1):302–307. doi: 10.1016/j.bbrc.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 8.Toyama R, Gomez D.M, Mana M.D, Dawid I.B. Sequence relationships and expression patterns of zebrafish zic2 and zic5 genes. Gene Expr. Patterns. 2004;4(3):345–350. doi: 10.1016/j.modgep.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.(a)Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dynamics : an official publication of the American Association of Anatomists. 2004;231(2):449–59. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]; (b) Keller M.J, Chitnis A.B. Insights into the evolutionary history of the vertebrate zic3 locus from a teleost-specific zic6 gene in the zebrafish, Danio rerio. Dev. Genes Evol. 2007;217(7):541–547. doi: 10.1007/s00427-007-0161-4. [DOI] [PubMed] [Google Scholar]
- 10.(a)Inoue T, Hatayama M, Tohmonda T, Itohara S, Aruga J, Mikoshiba K. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 2004;270(1):146–162. doi: 10.1016/j.ydbio.2004.02.017. [DOI] [PubMed] [Google Scholar]; (b) Nyholm M.K, Wu S.F, Dorsky R.I, Grinblat Y. The zebrafish zic2a-zic5 gene pair acts downstream of canonical Wnt signaling to control cell proliferation in the developing tectum. Development. 2007;134(4):735–746. doi: 10.1242/dev.02756. [DOI] [PubMed] [Google Scholar]; (c) Ohtsuka M, Kikuchi N, Yokoi H, Kinoshita M, Wakamatsu Y, Ozato K, Takeda H, Inoko H, Kimura M. Possible roles of zic1 and zic4, identified within the medaka Double anal fin (Da) locus, in dorsoventral patterning of the trunk-tail region (related to phenotypes of the Da mutant) Mech. Dev. 2004;121(7-8):873–882. doi: 10.1016/j.mod.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 11.(a) Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. USA. 1997;94(22):11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nagai T, Aruga J, Takada S, GA1/4nther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 1997;182(2):299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]; (c) Rohr K.B, Schulte-Merker S, Tautz D. Zebrafish zic1 expression in brain and somites is affected by BMP and hedgehog signalling. Mech. Dev. 1999;85(1-2):147–159. doi: 10.1016/S0925-4773(99)00044-1. [DOI] [PubMed] [Google Scholar]; (d) Grinblat Y, Sive H. zic Gene expression marks anteroposterior pattern in the presumptive neurectoderm of the zebrafish gastrula. Developmental dynamics : an official publication of the American Association of Anatomists. 2001;222(4):688–93. doi: 10.1002/dvdy.1221. [DOI] [PubMed] [Google Scholar]
- 12.(a) Imai K.S, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129(11):2723–2732. doi: 10.1242/dev.129.11.2723. [DOI] [PubMed] [Google Scholar]; (b) Wada S, Saiga H, Hrzic N. HrzicN, a new Zic family gene of ascidians, plays essential roles in the neural tube and notochord development. Development. 2002;129(24):5597–5608. doi: 10.1242/dev.00156. [DOI] [PubMed] [Google Scholar]; (c) Gostling N.J, Shimeld S.M. Protochordate Zic genes define primitive somite compartments and highlight molecular changes underlying neural crest evolution. Evol. Dev. 2003;5(2):136–144. doi: 10.1046/j.1525-142X.2003.03020.x. [DOI] [PubMed] [Google Scholar]
- 13.Lindgens D, Holstein T.W, Technau U. Hyzic, the Hydra homolog of the zic/odd-paired gene, is involved in the early specification of the sensory nematocytes. Development. 2004;131(1):191–201. doi: 10.1242/dev.00903. [DOI] [PubMed] [Google Scholar]
- 14.Layden M.J, Meyer N.P, Pang K, Seaver E.C, Martindale M.Q. Expression and phylogenetic analysis of the zic gene family in the evolution and development of metazoans. Evodevo. 2010;1(1):12. doi: 10.1186/2041-9139-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinberg I, Millen K.J. The ZIC gene family in development and disease. Clin. Genet. 2005;67(4):290–296. doi: 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- 16.Aruga J, Mikoshiba K. Role of BMP, FGF, calcium signaling, and Zic proteins in vertebrate neuroectodermal differentiation. Neurochem. Res. 2011;36(7):1286–1292. doi: 10.1007/s11064-011-0422-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houtmeyers R, Souopgui J, Tejpar S, Arkell R. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell. Mol. Life Sci. 2013;70(20):3791–3811. doi: 10.1007/s00018-013-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebbia M, Ferrero G.B, Pilia G, Bassi M.T, Aylsworth A, Penman-Splitt M, Bird L.M, Bamforth J.S, Burn J, Schlessinger D, Nelson D.L, Casey B. X-linked situs abnormalities result from mutations in ZIC3. Nat. Genet. 1997;17(3):305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 19.Ware S.M, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, Towbin J, Belmont J.W. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am. J. Hum. Genet. 2004;74(1):93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MA(c)garbanA(c) A, Salem N, Stephan E, Ashoush R, Lenoir D, Delague V, Kassab R, Loiselet J, Bouvagnet P. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur. J. Hum. Genet. 2000;8(9):704–708. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- 21.Cowan J, Tariq M, Ware S.M. Genetic and functional analyses of ZIC3 variants in congenital heart disease. Hum. Mutat. 2014;35(1):66–75. doi: 10.1002/humu.22457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessels M.W, Kuchinka B, Heydanus R, Smit B.J, Dooijes D, de Krijger R.R, Lequin M.H, de Jong E.M, Husen M, Willems P.J, Casey B. Polyalanine expansion in the ZIC3 gene leading to X-linked heterotaxy with VACTERL association: a new polyalanine disorder? J. Med. Genet. 2010;47(5):351–355. doi: 10.1136/jmg.2008.060913. [DOI] [PubMed] [Google Scholar]
- 23.Hatayama M, Tomizawa T, Sakai-Kato K, Bouvagnet P, Kose S, Imamoto N, Yokoyama S, Utsunomiya-Tate N, Mikoshiba K, Kigawa T, Aruga J. Functional and structural basis of the nuclear localization signal in the ZIC3 zinc finger domain. Hum. Mol. Genet. 2008;17(22):3459–3473. doi: 10.1093/hmg/ddn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Purandare S.M, Ware S.M, Kwan K.M, Gebbia M, Bassi M.T, Deng J.M, Vogel H, Behringer R.R, Belmont J.W, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129(9):2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]; (b) Ware S.M, Harutyunyan K.G, Belmont J.W. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev. Dyn. 2006;235(6):1631–1637. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]; (c) Casey B, Devoto M, Jones K.L, Ballabio A. Mapping a gene for familial situs abnormalities to human chromosome Xq24-q27.1. Nat. Genet. 1993;5(4):403–407. doi: 10.1038/ng1293-403. [DOI] [PubMed] [Google Scholar]
- 25.Lim L.S, Hong F.H, Kunarso G, Stanton L.W. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells. 2010;28(11):1961–1969. doi: 10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- 26.Winata C.L, Kondrychyn I, Kumar V, Srinivasan K.G, Orlov Y, Ravishankar A, Prabhakar S, Stanton L.W, Korzh V, Mathavan S. Genome wide analysis reveals Zic3 interaction with distal regulatory elements of stage specific developmental genes in zebrafish. PLoS Genet. 2013;9(10):e1003852. doi: 10.1371/journal.pgen.1003852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Sanyal A, Lajoie B.R, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wederell E.D, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, Ingham M, Hirst M, Robertson G, Marra M.A, Jones S, Hoodless P.A. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36(14):4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Carroll J.S, Liu X.S, Brodsky A.S, Li W, Meyer C.A, Szary A.J, Eeckhoute J, Shao W, Hestermann E.V, Geistlinger T.R, Fox E.A, Silver P.A, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Vega V.B, Ng H.H. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:203–209. doi: 10.1101/sqb.2008.73.026. [DOI] [PubMed] [Google Scholar]
- 29.(a) Lindeman L.C, Winata C.L, Aanes H, Mathavan S, Alestrom P, Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 2010;54(5):803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]; (b) Lindeman L.C, Andersen I.S, Reiner A.H, Li N, Aanes H, A~strup O, Winata C, Mathavan S, MA1/4ller F, Alestrom P, Collas P. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell. 2011;21(6):993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]; (c) Vastenhouw N.L, Zhang Y, Woods I.G, Imam F, Regev A, Liu X.S, Rinn J, Schier A.F. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraldez A.J, Mishima Y, Rihel J, Grocock R.J, Van Dongen S, Inoue K, Enright A.J, Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 31.Buenrostro J.D, Wu B, Chang H.Y, Greenleaf W.J. ATACseq: A Method for Assaying Chromatin Accessibility Genome- Wide. Curr. Prot. Mol. Biol. 2015;109(21 29):1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]; (b) Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu K.S, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38(11):1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]; (c) Dostie J, Zhan Y, Dekker J. Chromosome conformation capture carbon copy technology. Curr. Prot. Mol. Biol. 2007;Chapter 21, Unit 21 14. doi: 10.1002/0471142727.mb2114s80. [DOI] [PubMed] [Google Scholar]
- 33.Fullwood M.J, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell. Biochem. 2009;107(1):30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon J.R, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu J.S, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.(a)Wittkopp P.J, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012;13(1):59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]; (b)Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 36.Whyte W.A, Orlando D.A, Hnisz D, Abraham B.J, Lin C.Y, Kagey M.H, Rahl P.B, Lee T.I, Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J. Biol. Chem. 2001;276(3):2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- 38.(a) Brown S.A, Warburton D, Brown L.Y, Yu C.Y, Roeder E.R, Stengel-Rutkowski S, Hennekam R.C, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 1998;20(2):180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]; (a) Sanek N.A, Grinblat Y. A novel role for zebrafish zic2a during forebrain development. Dev. Biol. 2008;317(1):325–335. doi: 10.1016/j.ydbio.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Quinn M.E, Haaning A, Ware S.M. Preaxial polydactyly caused by Gli3 haploinsufficiency is rescued by Zic3 loss of function in mice. Hum. Mol. Genet. 2012;21(8):1888–1896. doi: 10.1093/hmg/dds002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim L.S, Loh Y.H, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng H.H, Stanton L.W. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell. 2007;18(4):1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Declercq J, Eggermont K, Agirre X, Prosper F, Verfaillie C.M. Zic3 induces conversion of human fibroblasts to stable neural progenitor-like cells. J. Mol. Cell Biol. 2012;4(4):252–255. doi: 10.1093/jmcb/mjs015. [DOI] [PubMed] [Google Scholar]
- 41.Aruga J, Tohmonda T, Homma S, Mikoshiba K. Zic1 promotes the expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev. Biol. 2002;244(2):329–341. doi: 10.1006/dbio.2002.0598. [DOI] [PubMed] [Google Scholar]
- 42.Gamse J.T, Sive H. Early anteroposterior division of the presumptive neurectoderm in Xenopus. Mech. Dev. 2001;104(1-2):21–36. doi: 10.1016/S0925-4773(01)00358-6. [DOI] [PubMed] [Google Scholar]
- 43.Marchal L, Luxardi G, ThomA(c) V, Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc. Natl. Acad. Sci. USA. 2009;106(41):17437–17442. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393(6685):579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- 45.(a) Ekker M, Akimenko M.A, Allende M.L, Smith R, Drouin G, Langille R.M, Weinberg E.S, Westerfield M. Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol. Biol. Evol. 1997;14(10):1008–1022. doi: 10.1093/oxfordjournals.molbev.a025707. [DOI] [PubMed] [Google Scholar]; (b) Thisse B, Pflumio S, FA1/4rthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier X.Q, Thisse C. Expression of the zebrafish genome during embryogenesis. ZFIN direct data submission. 2001. ; (c) Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill J.P, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/S0091-679X(04)77027-2. [DOI] [PubMed] [Google Scholar]; (d)Woda J.M, Pastagia J, Mercola M, Artinger K.B. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130(2):331–342. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lecaudey V, Anselme I, Dildrop R, RA1/4ther U, Schneider-Maunoury S. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J. Comp. Neurol. 2005;492(3):289–302. doi: 10.1002/cne.20765. [DOI] [PubMed] [Google Scholar]; (f) Lecaudey V, Anselme I, Rosa F, Schneider-Maunoury S. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development. 2004;131(13):3121–3131. doi: 10.1242/dev.01190. [DOI] [PubMed] [Google Scholar]
- 46.Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J. Neurosci. 2002;22(1):218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisen J.S, Weston J.A. Development of the neural crest in the zebrafish. Dev. Biol. 1993;159(1):50–59. doi: 10.1006/dbio.1993.1220. [DOI] [PubMed] [Google Scholar]
- 48.Kuo J.S, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125(15):2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- 49.Elms P, Siggers P, Napper D, Greenfield A, Arkell R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev. Biol. 2003;264(2):391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Warner S.J, Hutson M.R, Oh S.H, Gerlach-Bank L.M, Lomax M.I, Barald K.F. Expression of ZIC genes in the development of the chick inner ear and nervous system. Dev. Dyn. 2003;226(4):702–712. doi: 10.1002/dvdy.10262. [DOI] [PubMed] [Google Scholar]
- 51.(a) Stewart R.A, Arduini B.L, Berghmans S, George R.E, Kanki J.P, Henion P.D, Look A.T. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 2006;292(1):174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]; (b)Bang A.G, Papalopulu N, Goulding M.D, Kintner C. Expression of Pax-3 in the lateral neural plate is dependent on a Wnt-mediated signal from posterior nonaxial mesoderm. Dev. Biol. 1999;212(2):366–380. doi: 10.1006/dbio.1999.9319. [DOI] [PubMed] [Google Scholar]
- 52.(a)Raible D.W, Wood A, Hodsdon W, Henion P.D, Weston J.A, Eisen J.S. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev. Dyn. 1992;195(1):29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]; (b) Kimmel C.B, Ballard W.W, Kimmel S.R, Ullmann B, Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 53.(a) Krispin S, Nitzan E, Kalcheim C. The dorsal neural tube: a dynamic setting for cell fate decisions. Dev. Neurobiol. 2010;70(12):796–812. doi: 10.1002/dneu.20826. [DOI] [PubMed] [Google Scholar]; (b) Krispin S, Nitzan E, Kassem Y, Kalcheim C. Evidence for a dynamic spatiotemporal fate map and early fate restrictions of premigratory avian neural crest. Development. 2010;137(4):585–595. doi: 10.1242/dev.041509. [DOI] [PubMed] [Google Scholar]
- 54.(a)Chizhikov V.V, Millen K.J. Mechanisms of roof plate formation in the vertebrate CNS. Nat. Rev. Neurosci. 2004;5(10):808–812. doi: 10.1038/nrn1520. [DOI] [PubMed] [Google Scholar]; (b)Chizhikov V.V, Millen K.J. Control of roof plate development and signaling by Lmx1b in the caudal vertebrate CNS. J. Neurosci. 2004;24(25):5694–5703. doi: 10.1523/JNEUROSCI.0758-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Elsen G.E, Choi L.Y, Millen K.J, Grinblat Y, Prince V.E. Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Dev. Biol. 2008;314(2):376–392. doi: 10.1016/j.ydbio.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kondrychyn I, Teh C, Sin M, Korzh V. Stretching morphogenesis of the roof plate and formation of the central canal. PLoS One. 2013;8(2):e56219. doi: 10.1371/journal.pone.0056219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujimi T.J, Mikoshiba K, Aruga J. Xenopus Zic4: conservation and diversification of expression profiles and protein function among the Xenopus Zic family. Dev. Dyn. 2006;235(12):3379–3386. doi: 10.1002/dvdy.20906. [DOI] [PubMed] [Google Scholar]
- 57.(a) Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 2003;5(1):53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]; (b) Harris T.J, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 2004;167(1):135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sabherwal N, Tsutsui A, Hodge S, Wei J, Chalmers A.D, Papalopulu N. The apicobasal polarity kinase aPKC functions as a nuclear determinant and regulates cell proliferation and fate during Xenopus primary neurogenesis. Development. 2009;136(16):2767–2777. doi: 10.1242/dev.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Sonawane M, Carpio Y, Geisler R, Schwarz H, Maischein H.M, Nuesslein-Volhard C. Zebrafish penner/lethal giant larvae 2 functions in hemidesmosome formation, maintenance of cellular morphology and growth regulation in the developing basal epidermis. Development. 2005;132(14):3255–3265. doi: 10.1242/dev.01904. [DOI] [PubMed] [Google Scholar]; (e) Sonawane M, Martin-Maischein H, Schwarz H, NA1/4sslein-Volhard C. Lgl2 and E-cadherin act antagonistically to regulate hemidesmosome formation during epidermal development in zebrafish. Development. 2009;136(8):1231–1240. doi: 10.1242/dev.032508. [DOI] [PubMed] [Google Scholar]
- 58.(a)Agalliu D, Takada S, Agalliu I, McMahon A.P, Jessell T.M. Motor neurons with axial muscle projections specified by Wnt4/5 signaling. Neuron. 2009;61(5):708–720. doi: 10.1016/j.neuron.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b)Liu A, Majumdar A, Schauerte H.E, Haffter P, Drummond I.A. Zebrafish wnt4b expression in the floor plate is altered in sonic hedgehog and gli-2 mutants. Mech. Dev. 2000;91(1-2):409–413. doi: 10.1016/S0925-4773(99)00308-1. [DOI] [PubMed] [Google Scholar]; (c) Ungar A.R, Kelly G.M, Moon R.T. Wnt4 affects morphogenesis when misexpressed in the zebrafish embryo. Mech. Dev. 1995;52(2-3):153–164. doi: 10.1016/0925-4773(95)00386-F. [DOI] [PubMed] [Google Scholar]
- 59.Ulloa F, MartA- E. Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 2010;239(1):69–76. doi: 10.1002/dvdy.22058. [DOI] [PubMed] [Google Scholar]
- 60.Logan C.Y, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 61.Roy S, Hsiung F, Kornberg T.B. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332(6027):354–358. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.(a) Gallet A, Staccini-Lavenant L, ThA(c)rond P.P. Cellular trafficking of the glypican Dally-like is required for full-strength Hedgehog signaling and wingless transcytosis. Dev. Cell. 2008;14(5):712–725. doi: 10.1016/j.devcel.2008.03.001. [DOI] [PubMed] [Google Scholar]; (b) Marois E, Mahmoud A, Eaton S. The endocytic pathway and formation of the Wingless morphogen gradient. Development. 2006;133(2):307–317. doi: 10.1242/dev.02197. [DOI] [PubMed] [Google Scholar]; (c) Strigini M, Cohen S.M. Wingless gradient formation in the Drosophila wing. Curr. Biol. 2000;10(6):293–300. doi: 10.1016/S0960-9822(00)00378-X. [DOI] [PubMed] [Google Scholar]
- 63.Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development. 2009;136(24):4083–4088. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- 64.(a) Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol. Cell. 2001;7(1):137–149. doi: 10.1016/S1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]; (b) Ryan A.K, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la PeA a J, Sabbagh W, Greenwald J, Choe S, Norris D.P, Robertson E.J, Evans R.M, Rosenfeld M.G, IzpisA a Belmonte J.C. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394(6693):545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 65.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95(6):829–837. doi: 10.1016/S0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435(7039):172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 67.(a) Essner J.J, Vogan K.J, Wagner M.K, Tabin C.J, Yost H.J, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418(6893):37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]; (b) Essner J.J, Amack J.D, Nyholm M.K, Harris E.B, Yost H.J. Kupffer ?(tm)s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132(6):1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 68.Sampath K, Rubinstein A.L, Cheng A.M, Liang J.O, Fekany K, Solnica-Krezel L, Korzh V, Halpern M.E, Wright C.V. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395(6698):185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- 69.(a) Yost H.J. Left-right development in Xenopus and zebrafish. Semin. Cell Dev. Biol. 1998;9(1):61–66. doi: 10.1006/scdb.1997.0191. [DOI] [PubMed] [Google Scholar]; (b)Hyatt B.A, Lohr J.L, Yost H.J. Initiation of vertebrate left-right axis formation by maternal Vg1. Nature. 1996;384(6604):62–65. doi: 10.1038/384062a0. [DOI] [PubMed] [Google Scholar]
- 70.Kitaguchi T, Nagai T, Nakata K, Aruga J, Mikoshiba K. Zic3 is involved in the left-right specification of the Xenopus embryo. Development. 2000;127(22):4787–4795. doi: 10.1242/dev.127.22.4787. [DOI] [PubMed] [Google Scholar]
- 71.Cast A.E, Gao C, Amack J.D, Ware S.M. An essential and highly conserved role for Zic3 in left-right patterning, gastrulation and convergent extension morphogenesis. Dev. Biol. 2012;364(1):22–31. doi: 10.1016/j.ydbio.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimi T.J, Hatayama M, Aruga J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/I -catenin signaling pathway. Dev. Biol. 2012;361(2):220–231. doi: 10.1016/j.ydbio.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 73.(a) Bisgrove B.W, Essner J.J, Yost H.J. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127(16):3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]; (b) Danos M.C, Yost H.J. Role of notochord in specification of cardiac left-right orientation in zebrafish and Xenopus. Dev. Biol. 1996;177(1):96–103. doi: 10.1006/dbio.1996.0148. [DOI] [PubMed] [Google Scholar]
- 74.(a) Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach S.D, Molinari L, Niesh S.R, Jefferies J.L, Craigen W.J, Towbin J.A, Belmont J.W, Ware S.M. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum. Mol. Genet. 2009;18(5):861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin J.A, Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am. J. Med. Genet. 1999;82(1):70–76. doi: 10.1002/(SICI)1096-8628(19990101)82:1<70::AID-AJMG14>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]; (c)Kosaki K, Bassi M.T, Kosaki R, Lewin M, Belmont J, Schauer G, Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am. J. Hum. Genet. 1999;64(3):712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d)Bamford R.N, Roessler E, Burdine R.D, Saplako Ylu U, dela Cruz J, Splitt M, Goodship J.A, Towbin J, Bowers P, Ferrero G.B, Marino B, Schier A.F, Shen M.M, Muenke M, Casey B. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat. Genet. 2000;26(3):365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 75.(a)Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Matthews L, Gardner A, Schuster G, Vien L, Harrison W, Elder F.F, Penman-Splitt M, Overbeek P, Strachan T. Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat. Genet. 1998;20(2):149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]; (b) Otto E.A, Schermer B, Obara T, O ?(tm)Toole J.F, Hiller K.S, Mueller A.M, Ruf R.G, Hoefele J, Beekmann F, Landau D, Foreman J.W, Goodship J.A, Strachan T, Kispert A, Wolf M.T, Gagnadoux M.F, Nivet H, Antignac C, Walz G, Drummond I.A, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 2003;34(4):413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Okada Y, Takeda S, Tanaka Y, IzpisA a Belmonte J.C, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121(4):633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]; (d) Hashimoto M, Shinohara K, Wang J, Ikeuchi S, Yoshiba S, Meno C, Nonaka S, Takada S, Hatta K, Wynshaw-Boris A, Hamada H. Planar polarization of node cells determines the rotational axis of node cilia. Nat. Cell Biol. 2010;12(2):170–176. doi: 10.1038/ncb2020. [DOI] [PubMed] [Google Scholar]; (e) Wang G, Cadwallader A.B, Jang D.S, Tsang M, Yost H.J, Amack J.D. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer ?(tm)s vesicle in zebrafish. Development. 2011;138(1):45–54. doi: 10.1242/dev.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f)Watanabe D, Saijoh Y, Nonaka S, Sasaki G, Ikawa Y, Yokoyama T, Hamada H. The left-right determinant Inversin is a component of node monocilia and other 9+0 cilia. Development. 2003;130(9):1725–1734. doi: 10.1242/dev.00407. [DOI] [PubMed] [Google Scholar]
- 76.(a) Olbrich H, Haffner K, Kispert A, Volkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, Zariwala M, Noone P.G, Knowles M, Mitchison H.M, Meeks M, Chung E.M, Hildebrandt F, Sudbrak R, Omran H. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30(2):143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]; (b) Pennarun G, Escudier E, Chapelin C, Bridoux A.M, Cacheux V, Roger G, ClA(c)ment A, Goossens M, Amselem S, Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 1999;65(6):1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c)Sutherland M.J, Ware S.M. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am. J. Med. Genet. Part C, Seminars in Med. Genet. 2009;151C(4):307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- 77.Hadorn E. Patterns of biochemical and developmental pleiotropy. Cold Spring Harb. Symp. Quant. Biol. 1956;21:363–373. doi: 10.1101/SQB.1956.021.01.030. [DOI] [PubMed] [Google Scholar]
- 78.(a)Duboule D, Wilkins A.S. The evolution of ?~bricolage ?(tm) Trends Genet. 1998;14(2):54–59. doi: 10.1016/S0168-9525(97)01358-9. [DOI] [PubMed] [Google Scholar]; (b)Jacob F. Evolution and tinkering. Science. 1977;196(4295):1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]; (c)Morange M. Evolutionary developmental biology its roots and characteristics. Dev. Biol. 2011;357(1):13–16. doi: 10.1016/j.ydbio.2011.03.013. [DOI] [PubMed] [Google Scholar]; (d)Wilkins A.S. Genetic networks as transmitting and amplifying devices for natural genetic tinkering; Novartis Foundation Symposium; 2007. pp. 71–86. [DOI] [PubMed] [Google Scholar]; (e)Wilkins A.S. Between ?odesign ?? and ?obricolage ??: genetic networks, levels of selection, and adaptive evolution. Proc. Natl. Acad. Sci. USA. 2007;104(suppl. 1):8590–8596. doi: 10.1073/pnas.0701044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waddington C.H. Organizers and Genes. Cambridge, United Kingdom: Cambridge University Press; 1940. [Google Scholar]
- 80.Aruga J, Nozaki Y, Hatayama M, Odaka Y.S, Yokota N. Expression of ZIC family genes in meningiomas and other brain tumors. BMC Cancer. 2010;10:79. doi: 10.1186/1471-2407-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]