Abstract
Post-translational modification is the most common mechanism of regulating protein function. If phosphorylation is considered a key event in many signal transduction pathways, other modifications must be considered as well. In particular the side chain of lysine residues is a target of different modifications; notably acetylation, methylation, ubiquitylation, sumoylation, neddylation, etc. Mass spectrometry approaches combining highly sensitive instruments and specific enrichment strategies have enabled the identification of modified sites on a large scale. Here we make a comparative analysis of the most representative lysine modifications (ubiquitylation, acetylation, sumoylation and methylation) identified in the human proteome. This review focuses on conserved amino acids, secondary structures preference, subcellular localization of modified proteins, and signaling pathways where these modifications are implicated. We discuss specific differences and similarities between these modifications, characteristics of the crosstalk among lysine post translational modifications, and single nucleotide polymorphisms that could influence lysine post-translational modifications in humans.
Keywords: Acetylation, Lysine post-translational modifications, Methylation, Signaling, Sumoylation, Ubiquitylation.
INTRODUCTION
Post-translational modifications (PTMs) play a key role in regulating a wide range of protein functions, including cellular localization, protein-protein interactions, enzymatic activity, protein turnover and etc. [1]. Moreover, a cross-talk between two or more different PTMs contributes to the fine regulation of cell signaling [2]. Not surprisingly, altered levels of PTMs are implied in several human pathologies and enzymes involved in generating PTMs are a class of drug targets of primary importance [3-6]. Advances in mass spectrometry technology, and the development of novel enrichment strategies [7], in particular the availability of highly specific antibodies, have now made possible the identification of thousands of modified amino acids on proteins. These cutting-edge instrumental and biochemical procedures have allowed the highly accurate characterization of PTMs on a global scale moving the interest from target identification to target characterization [8]. While the phosphorylation of hydroxyl side chains of serine, threonine and tyrosine is the most thoroughly studied form of PTM, the interest towards lysine PTMs is constantly growing, as they have been recognized to play important roles in a variety of cellular functions [9-12] and have been shown to be preferentially located near phosphorylated residues [13].
The ε-amino group of lysine is susceptible to different modifications: acetylation, methylation, ubiquitylation, and ubiquitin-like modifications such as sumoylation, neddhylation, etc. Some PTMs which were initially thought to be restricted to specific cellular targets or cellular functions, have been now recognized as widely occurring [9, 14-16].
Lysine acetylation and methylation have been associated for many years mainly with epigenetic regulation. Reversible acetylation and methylation of histones are indeed key regulators of chromatin remodeling [17]. The enzymes involved in their turnover were therefore named histone acetyltransferase, histone deacetylase and histone methyltransferase, histone demethylase, respectively. Recently, the number of non-histone proteins subjected to lysine acetylation and methylation is growing and the enzymes that catalyze the PTMs are recognized to have a broad spectrum of protein substrates, raising the possibilities of change their name [10, 18, 19].
Lysine acetylation was first identified in histone proteins in the late 1960s [20] and, at the beginning of 2000, several non-histone proteins were found to contain acetylated lysines [21]. However, the acceptance by the scientific community that these modifications could have a functional relevance beyond chromatin regulation occurred only after the publication of high-throughput proteomic studies. In 2006, almost 400 acetylation sites on about 200 proteins were identified by Zhao’s group. Several Non-nuclear acetylated proteins were identified such as metabolic enzymes, chaperones, cytoskeletal and signaling proteins [22]. In 2009, 3600 acetylated lysines were found on 1750 proteins [9], demonstrating that acetylation is a widely observed phenomenon potentially involved in a variety of nuclear and cytoplasmic functions.
Lysine methylation was first identified in a bacterial flagellar protein in 1959 [23] and five years later in histones [24] (for recent reviews see [25, 26]). The identification of methylated lysines in other non-histone proteins has been provided by low-throughput experiments over the years. However, technical limitation for a high-throughput identification of methylated sites (due to lack of effective antibodies) have been overcome only recently [27]. In 2013, Guo’s group identified ~160 lysine-methylated sites on about 130 proteins in HCT116 human colon cancer cells [28] suggesting that methylation could be limited to a small group of proteins, mainly involved in chromatin function. The later identification by Garcia’s group of ~500 lysine methylated sites on about 400 proteins in Hela cells confirmed the primary role of methylation in chromatin organization but also showed that methylation can be involved in a broader range of cellular activities, from nuclear to cytoplasmic compartments [29].
Ubiquitin, a small 76-amino-acid protein, was first discovered in 1975 as the molecular tag to target proteins to proteasomal degradation. In addition to this role, it is widely recognized that ubiquitin modification participates in different signaling pathways including intracellular trafficking, DNA repair, activation of transcription factors and etc. [4, 16]. Ubiquitylation requires an enzymatic cascade involving ubiquitin-activating enzymes (E1), ubiquitin conjugating enzymes (E2) and ubiquitin ligases (E3). The importance of ubiquitylation in cell signaling is supported by the prediction of 16 E1, 53 E2, and 527 E3 enzymes in the human genome [30]. High throughput approach has permitted to identify a growing number of ubiquitylated proteins: from almost 400 ubiquitylated sites on about 200 proteins in 2010 [31], more than 11,000 ubiquitylated sites on about 4,000 proteins in 2011 [32], to reach a figure of about 20,000 unique ubiquitylated sites on more than 5,000 proteins in 2012 [33].
Following ubiquitin discovery, several proteins related in sequence to ubiquitin and of a similar three-dimensional structure have been identified. Most of these proteins -called ubiquitin-like proteins, Ubls - are conjugated to the ε-amino group of lysine side-chain residue via an enzymatic cascade that resembles ubiquitylation being formed by E1, E2 and E3 enzymes [4, 16]. However, each Ubls requires a distinct set of enzymes. Sumoylation was first discovered in 1996 and among the Ubls is the most investigated for its involvement in a variety of cellular processes [11].
Several studies have already pointed out important features of individual lysine PTMs based on the analysis of the data obtained by high-throughput mass spectrometry experiments [9, 22, 28, 31, 34, 35], or inspecting large databases as performed by Lu and collaborators on lysine acetylation [36]. Very recently a comparative analysis on the most frequent PTMs covering different species has been performed with the aim to identify functional links between PTMs on the basis of their different evolutionary rates [37].
The aim of this paper is not to present a comprehensive review on the biology of lysine PTMs; rather it is to perform a comparative analysis to outline specific differences and similarities between these modifications in humans. So far, a large number of lysine targets have been identified in human cells for four PTMs, i.e. acetylation, ubiquitylation, methylation and sumoylation. This allowed us to re-analyze these PTMs using a variety of bioinformatics tools to highlight similarities and distinctive functions of each modification starting from the analysis of the primary structure.
CONSERVED AMINO ACIDS SURROUNDING THE MODIFIED LYSINES
All human predicted sequences for PTMs (+7, -7) were collected form PhosphositePlus database (www.phospho site.org) [38]. To analyze and visualize the conservation of amino acids in positions close to the modified lysine in the primary structure we utilized the Two sample logo strategy [39]. This tool is here utilized to compare two groups of sequences: surrounding modified lysines vs random collected lysines. The logo visualizes the enrichment (upper) and the depletion (lower) of specific amino acids at positions close to the modified lysine with respect to the reference group, where the size of the symbol is proportional to the difference between the two samples. This comparative analysis permits to highlight the presence of specific conserved patterns for each different type of lysine PTMs.
Fig. 1A shows the Two sample logo for lysine acetylation, methylation, sumoylation and ubiquitylation vs random lysines. e.g., sequences surrounding a modified lysine compared with those surrounding randomly collected lysines.
Fig. (1).
Analysis of conserved patterns in multiple-sequence alignments. A. Two sample logo analysis of acetylated, methylated, ubiquitylated, and sumoylated lysines vs. a random group of lysines extracted from human proteome. Modified sites annotated in the PhosphositePlus database (www.phopshosite.org) [38] can be distinguished in two different groups: low-throughput data and high-throughput data taken from the literature, or high-throughput data generated at Cell Signaling Technology, inc (CST). We have here considered only literature data as for most of the data of the second group mass spectra and the experimental conditions are not available. We collected: 5406 human acetylated lysine sequences from 2728 proteins; 792 human methylated lysine sequences from 558 proteins; 612 human sumoylated lysine sequences from 357 proteins; 23882 human ubiquitylated lysine sequences from 6788 proteins. Human random peptides (+7, -7) were extracted from the Swiss Prot database using a homemade bioperl script and unix text processing commands. Non-redundant sequences were randomized using unix command shuff. Sequence motif analysis was performed using a +7,- 7 residue window around each modified lysine and these data were compared to the same window surrounding random lysine residues extracted from human proteome using Two-Sample logo tool (t-test) [39]. B. Percentage of K/R and D/E upstream and downstream lysine target of acetylation, methylation, ubiquitylation and sumoylation has been calculated. A χ-square test was used to determine the significance of the differences between modified and total lysines (p-value * for 0.05, ** for 0.01, *** for 0.001). C. Solvent accessibility of lysine. Peptides were matched with protein sequences present in PDB database with BlastP. Accessible surface area of modified lysine from the 3-dimensional structure of the protein was calculated using the software STRIDE [49]. The value obtained for each modified lysine was divided by the total surface area of the residue obtained from Chotia [63]. The average value for each type of modified lysine was calculated as percentage of exposition rate and compared with the exposition rate of all lysines present in the PDB database. A χ-square test was used to determine the significance of the differences between modified and total lysines (p-value * for 0.05, ** for 0.01, *** for 0.001).
A feature that strikingly arises is the great variability in the amino acid conservation among lysine modifications. Sumoylation differs from all the other PTMs because the substrate recognition mechanism is highly primary-structure dependent: there is an enrichment of a glutamic acid in +2 position (63.9% of modified residues present a glutamic acid in +2 position). This primary-structure dependence for sumoylation, originally proposed after the identification of the first sumoylated sites [40] and subsequently supported by proteomic studies (see for example ref. [35]) is confirmed by our analysis covering ~600 sumo modified sites in humans. The core sequence is ΨKxE (Ψ, any hydrophobic residue) where the E at +2 is prominent. From this analysis, the so-called PDSM (Phosphorylation-Dependent Sumoylation Motif) motif (YKxExxSP) is recognizable. This is a SUMO motif followed by one proline-directed phosphorylation site whose phosphorylation plays an important role in promoting sumoylation [41]. The high degree of primary structure conservation in sumoylation suggests that the recognition mechanism is quite similar for the enzymes responsible for this modification. As mentioned above, sumoylation is promoted by the cascade of E1, E2 and E3 enzymes resembling the ubiquitin pathways, and the specificity in substrate modification is driven by different combinations of E2 and E3 enzymes. However, in contrast to ubiquitin cascade, only one E2 enzyme, Ubc9, has been identified. Moreover, Ubc9 is frequently able to sumoylate substrates in vitro without the presence of the ligase [11]. The Ubc9 dependence for substrate recognition in lysine sumoylation is evident in the reported two sample logo.
All the other modifications, i.e. acetylation, methylation and ubiquitylation, display a lower degree of conservation (at each position the degree of conservation is lower than 10%) consistent either with a reduced primary structure specificity [34], or with the recognition of several different motifs. This latter hypothesis is supported by an analysis on the primary structure conservation at the phosphorylation sites [42]. When all the Ser/Thr phosphosites are analyzed in a Two sample logo vs Ser/Thr proteome, a conservation below 15% is observed at all the considered positions with the only exception at the +1 position. This result is expected from the diversity of kinases involved in phosphorylating these sites. Infact, if only the subset of phosphosites produced by a single kinase is considered, the conservation of amino acids surrounding the target site is much stronger [42]. Moreover it cannot be excluded the possibility that some modifications can be non-enzymatically produced in cells. For example non-enzymatic lysine acetylation of proteins in the presence of physiological concentrations of acetyl-CoA occurs in vitro [43, 44]. Moreover, the demonstration that acetyl-CoA levels are correlated with acetylation levels in vivo, suggests that non-enzymatic acetylation could occur also in cells [45].
Despite the low level of amino acid conservation, the Two sample logos deserves some additional comments: methylation seems to recognize specific positions at N-terminus with respect to the target lysine, while ubiquitylation seems to recognize residues both at the N- and C-terminus close to the modified site. For acetylation we observe a similar amino acid conservation on both sides.
These observations disclose striking differences in primary structure conservation that may have several biological implications. We can indeed suppose that mutations or modifications of residues close to the acetylated, methylated or ubiquitylated lysine could have a limited effect on these PTMs. On the contrary, these changes for sumoylated sites could abolish this modification or create new sumoylation sites if critical positions such as -1 and/or +2 are affected.
These differences should be also be kept in mind when bioinformatics methods are chosen to predict lysine modified sites. Fig. 1 clearly shows that computational approaches based on primary protein structure to predict lysine modifications could be useful mainly for the identification of putative sumoylated sites.
Other considerations can be done analyzing the electrostatic and the hydrophobic characters of the amino acids surrounding modified lysine.
Lysine PTMs can be clustered according to the preference for acidic or basic residues in the +7, -7 residue window centered on the target site. Figs. 1A and B show that acetylation and methylation display an enrichment of basic residues at several positions close to the modified site (with particular reference to the downstream positions respect to the modified site for acetylation, and to the upstream positions for methylation, see Fig. 1B). The logos published by Choudhary and collaborators on 3600 acetylated sites [9] show an enrichment in lysines similar to our two sample logo calculated on 20,000 modified sites. Sumoylation and ubiquitylation display instead an enrichment of acidic residues at positions surrounding the target site with glutamic preference for sumoylation (downstream from the modified lysine and mainly at position +2), and preference for aspartic residues for ubiquitylation (both upstream and downstream from the modified lysine) (see Figs. 1A and B). The preference for acidic residues that are downstream from the sumoylated sites was first suggested by Sharrock’s group in 2006 [46] and here confirmed by our analysis on a more representative sample. The enrichment in acidic residues at positions close to the ubiquitylated sites has been previously observed in murine sequences [33] and human sequences [47].
According to these differences, some considerations can be offered regarding the potential crosstalk between lysine PTMs and phosphorylation. Ser/Thr protein kinases are historically divided into three main groups based on local structural determinants close to the target site: proline-directed kinases that require a Pro at the + 1 position of the modified site (notably the mitogen activated kinases and the cyclin dependent kinases), basophilic kinases that phosphorylate residues with clustered positive charged (PKA, Akt, PKC, etc), and the few but highly pleiotropic acidophilic kinases that phosphorylate residues specified by negatively charged side chains (CK2, CK1, and others) [48]. On the basis of the above observations (see Fig. 1A and B) we may expect interplay especially between acetylation/methylation and basophilic kinases on one side, and between ubiquitylation/sumoylation and acidophilic kinases on the other. Moreover sumoylation sites display an enrichment of proline at several positions close to the modified site suggesting a cross-talk of this modification with the proline-directed kinases (see the above mentioned PDSM motif [41]).
The third aspect concerns the hydrophobic residues. Fig. 1A shows that methylated and sumoylated sites show a preference for hydrophobic residues at position -1 respect to the modified site. The enrichment of leucine at -1 position in methylated sites has been shown in a recent proteomic study on 119 methylated sites [28] and here confirmed by our two sample logo generated on ~800 methylated residues. Acetylated and in particular ubiquitylated sites show an enrichment in hydrophobic residues at several positions close to the modified lysine. Lysines surrounded by hydrophobic residues may be less solvent exposed in the protein structure and therefore their modifications could be responsible for important conformational changes. This idea has been examined by screening the solvent accessibility of modified lysines on crystal structures deposited in PDB database. Fig. 1C compares the average value of the solvent accessibility between total lysines found in the PDB structures and modified lysines; on the average, all the modified lysines are more exposed to solvent with respect to the total lysines (Fig. 1C) as expected assuming that lysine to be modified should be exposed to solvent. However no significant differences among the different PTMs are observed.
POSITION OF MODIFIED LYSINES IN SECONDARY STRUCTURES
Next, we checked if the targets of PTMs display preference for secondary structures. We utilized STRIDE, a program that recognizes secondary structural elements in proteins from known atomic coordinates [49]. The distribution in protein secondary structures between modified lysines and total lysines was compared. Fig. 2A shows that lysines are selected for a specific PTM also on the basis of the secondary structure. Acetyl and ubiquityl modifications shows a similar trend for secondary structure preference: enrichment in α-helix, no significant difference in turn, reduction in β-sheets and in coiled regions. The preference for a-helix and the reduction in coiled regions of acetylated and ubiquitylated lysine is in agreement with previous observations (see for acetylation [9, 22, 36] and for ubiquitylation [31, 34]).
Fig. (2).
Secondary structure analysis. Peptides containing a modified lysine were matched with protein sequences present in PDB database with BlastP. The secondary structure assignment was calculated according STRIDE [49]. Based upon this data four structures are predicted: helix (H+G+I), beta-strands (E+B), turns (T) and coil (the rest of the letters). A χ-square test was used to determine the significance of the differences between modified and total lysines (p-value * for 0.05, ** for 0.01, *** for 0.001). A. Lysine residues for which a predicted structure exists are mapped and the comparison between all lysines present in the database and modified lysines are shown. B. Serine, threonine and tyrosine for which a predicted structure exists are mapped and the comparison between all serine, threonine and tyrosine present in the database and modified residues are shown. Phosphorylated sites were extracted from PhosphositePlus database (www.phopshosite.org) [38] excluding not published data as in (Fig. 1). We collected 86036 human phosphorylated serine sequences from 14869 proteins; 40332 human phosphorylated threonine sequences from 12333 proteins; 33951 human phosphorylated tyrosine sequences from 11463 proteins.
Sumo and methyl modifications seem to show a reduction in ordered structures (α-helix and β-sheets) and an enrichment in coiled sequences when compared to all lysines, even if a higher number of identified sites are necessary to have statistically significant differences.
Preferences for secondary structure in lysine targeting have been here compared with preferences exhibited by phosphorylation (affecting Ser/Thr and Tyr residues), the most well studied PTM. In this case, we have the same type of modification, phosphorylation, that affects different residues on the same functional group. Fig. 2B shows the distribution in protein secondary structures of phosphorylated vs total Ser, Thr and Tyr residues. We observed a significant reduction of phosphorylated amino acids in β-sheet, and an enrichment for residues localized in α-helix statistically significant only for phosphor-tyrosines. Preference for residues in turn is observed in all these modifications but the difference with the reference set is statistically significant only for Ser-phosphorylation. This comparative analysis confirms the complexity and the great variability among PTMs in secondary structure preference.
A separate discussion should be given for coiled regions that can be mostly considered unstructured/high mobile regions. While acetylation and ubiquitylation modifications show a significant reduction, sumoylation shows a significant enrichment for lysine in coiled regions when compared with total lysines (Fig. 2A). This feature of sumoylation is shared with Ser and Thr phosphorylation that display an increase of the percentage of the modified residue localized in coiled regions (Fig. 2B) when compared with the control. One limit of this analysis is based on the fact that this assignment is based on PDB structures where the coiled regions, that are highly mobile, are often not visible. Therefore a potential underestimation for coiled preferences has to be taken into consideration. A further analysis has been done utilizing IUPred, a web server for the prediction of unstructured/disordered regions in proteins based on primary structure. Fig. 3 shows an increased preference for lysines localized in disordered regions for sumoylation and methylation when compared to the reference set. On the contrary acetylation shows no preference between ordered/disordered regions, while ubiquitylation shows a significant enrichment in ordered regions. This particular feature of ubiquitylated sites was early suggested on a small number of sequences [50] and here confirmed on a larger dataset.
Fig. (3).
Distribution of modified amino acids in ordered and disordered regions of the proteins. Peptides containing a modified amino acid were matched with protein sequences using BlastP. Reference set: all human protein sequences extracted from Swissprot database. All the protein sequences were analyzed using IUPred [64] and the values for specific amino acid were extracted. Residues with a value above 0.5 were considered disordered. The percentage of disorder/order for all residues and for modified residues was calculated and displayed. A χ-square test was used to determine the significance of the differences between modified and total lysines (p-value * for 0.05, ** for 0.01, *** for 0.001). A. Protein disorder prediction for total and modified lysines. B. Protein disorder prediction for total and modified serine, threonine and tyrosine.
Applying the same disorder/order prediction program to phosphorylation (Fig. 3B) we observe that, regardless of the type of residue, this PTM invariably displays a significant enrichment for residues localized in disordered region when compared with the reference set displaying a behavior similar to methylation and sumoylation. Differences between Ser, Thr and Tyr target sites may be due to the hydrophilic character of the amino acid considered (increasing from Tyr, Thr to Ser) (Fig. 3B).
These observations could give us new insights into the mechanisms of substrate selection for lysine PTMs. We can indeed expect that enzymes involved in lysine ubiquitylation recognize specific secondary/tertiary structures. In contrast, a dynamic structure (disorder region) could be necessary to adapt peptide structure to the active site of the enzyme involved in lysine methylation and sumoylation. This especially applies to sumoylation since a crystallographic analysis of a complex between Ubc9 and a c-terminal domain of RanGAP1 reveals that lysine adopts an extended conformation that is critical for its interaction with the Ubc9 enzyme [51].
Moreover the modification of a lysine in an ordered or disordered region could have a different functional significance: modification of lysines in a folded domain can interfere with the protein activity [52, 53], while modification of lysines in disordered regions, can regulate signaling [54] and protein interaction networks [55].
SUBCELLULAR AND FUNCTIONAL CLASSIFICATION OF MODIFIED PROTEINS
To disclose functional differences among lysine PTMs, we analyzed subcellular localization (Fig. 4) and molecular functions (Fig. S1 (192.2KB, pdf) ) of modified proteins. Subcellular localization analysis shows that sumoylated and methylated proteins exhibit a preferential nuclear localization. Acetylated and ubiquitylated proteins are more similarly distributed between cytoplasmic and nuclear compartments. Moreover, all the modified proteins show a wide distribution in the subcellular compartments (Fig. 4).
Fig. (4).
>Subcellular localization and functional analysis. Subcellular localization of lysine modified proteins assigned using GeneCoDis3 webserver [65, 66]. Only results with p values less than 0.01 are shown.
An analysis of the molecular functions of the modified proteins shown in Fig. S1 (192.2KB, pdf) is in agreement with the subcellular localization analysis. Overrepresented categories of molecular functions of sumoylated and methylated proteins are mostly linked to nuclear functions while acetylated and ubiquitylated proteins cover a wide range of biological functions (Fig. S1 (192.2KB, pdf) ).
OVERLAP OF MODIFICATIONS ON LYSINE RESIDUE
The modification of a lysine chain could induce different biological effects according to the type of modification [56]. Therefore we wondered what is the extent of overlap between lysine PTMs. Fig. 5A shows that 94% of the total modified lysines are modified by only one type of PTM, about 6% may be modified by two different competing PTMs, and about 0.2% may be modified by three different, competing PTMs. We also find four sequences where the same lysine is potentially susceptible to all four PTMs (Table 1). Three proteins, p53, histone H3, and SUMO-1 activating enzyme subunit 2 (SAE2), can be modified by four different competing PTMs (Table 1).
Fig. (5).
Determination of the overlapping degree between different PTMs. A. On the left, Venn diagram showing the overlap between lysine PTMs is shown. On the right, the percentage of unique lysines (susceptible only to one modification) and of lysines that can be modified by two or more PTMs are shown. B. Two sample logos between ubiquitylated and acetylated lysines, ubiquitylated and methylated lysines, ubiquitylated and sumoylated lysines, acetylated and methylated lysines, and acetylated and sumoylated lysines are shown.
Table 1.
Sequences modified by four competing PTMs.
| Protein | Site | Modification | Sequence |
|---|---|---|---|
| H3 | K24 | (a,m,s,u) | PRKQLATKAARKSAP |
| P53 | K386 | (a,m,s,u) | RHKKLMFKTEGPDSD |
| SAE2 | K271 | (a,m,s,u) | RYLLTMDKLWRKRKP |
| ACTA1/ACTA2 | K70 | (a,m,u) | KRGILTLKYPIEHGI |
| ACTB | K68 | (m,s) | KRGILTLKYPIEHGI |
| ACTC1 | K70 | (a,m,u) | KRGILTLKYPIEHGI |
| ACTG1 | K68 | (m) | KRGILTLKYPIEHGI |
| ACTG2 | K69 | (a,m,u) | KRGILTLKYPIEHGI |
H3 (histone H3); p53; SAE2 (SUMO-1 activating enzyme subunit 2); ACTA1/ACTA2 (actin α 1/α 2); ACTB [16](actin β1); ACTC1 (actin, alpha, cardiac muscle 1); ACTG1 (Actin, cytoplasmic 2); ACTG2 (actin, gamma 2). For the references see www.phosphosite.org.
We have analyzed primary structure of sequences where lysines could be modified by two different PTMs and the two sample logo has been generated for each pair (Fig. 5B). Comparing the two sample logo of Fig. 5B and Fig. 1A, lysines that can be acetylated or ubiquitylated maintain the acetylation hallmark in the positions close to the lysine (in particular in the window n-2, n+2). This would means that acetyltransferase enzymes are more dependent on primary structure recognition than enzymes involved in ubiquitylation. Lysines that are sumoylated or acetylated and sumoylated or ubiquitylated maintain the predominance of the classical sumo motif (ΨKxE), confirming the Stringent primary structure requirement for these enzymes. Lysines that are ubiquitylated or methylated and acetylated or methylated display a selection in primary structure that seems to be a compromise between the two patterns (see Fig. 5B and Fig. 1A).
GENETIC POLYMORPHISMS THAT INFLUENCE LYSINE PTMs
Recently, the concept of personalized medicine has attracted much attention for the possibility to have a more precise, predictable medicine customized for each individual patient. In this area the knowledge of human genetic polymorphisms could provide a basis for understanding the differences in disease susceptibility, prognosis and/or treatment response (for review see [57]). It has been evaluated that about 90% of human genetic variations are caused by single nucleotide polymorphisms (SNPs) that can occur in approximately every 300 base pairs along human chromosomes [58]. A non-synonymous SNP that occurs in the coding region causes an amino acid substitution. Missense mutation is the type of variation most frequently related to human disease. Amino acid substitution may change the physiochemical nature of the wild-type amino acid, but it can also significantly affect PTMs. In this case it could directly abrogate or create new PTMs, or if it occurs in the flanking residues, it could change the affinity of the modifying enzyme for the target site. The potential involvement of SNPs on protein phosphorylation has already been analyzed [59, 60]. In particular, Ren et al. computationally observed that 70% of the reported non-synonymous SNPs could potentially influence protein phosphorylation: ~25% by a direct change of the phosphorylation site and the other by affecting the protein kinase specificity [59]. Very recently a similar analysis has been performed on protein lysine acetylation by Suo and colleagues [61]. Using an acetylation prediction system the authors suggest that about 50% of the variations in amino acid could potentially affect lysine acetylation [61].
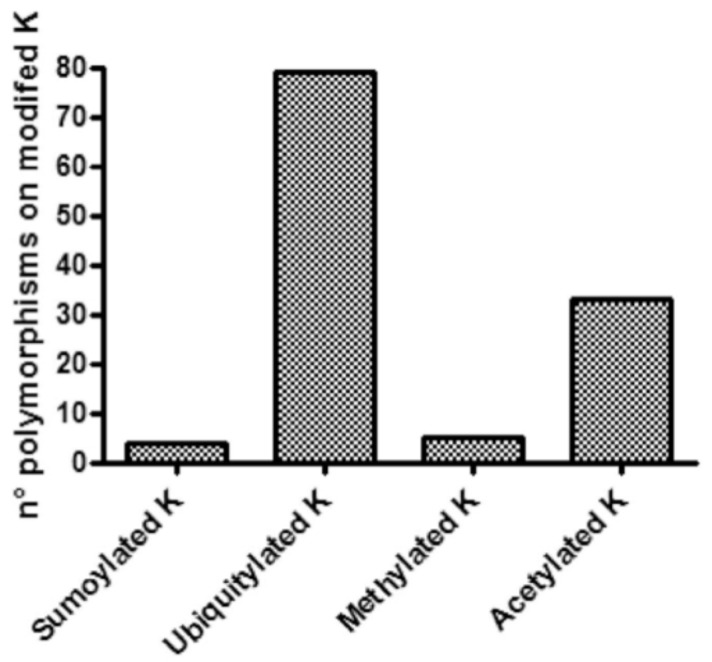
We can easily detect the impact of a genetic variant on lysine PTMs when the affected amino acid is itself the target of the modification. The information about human genetic variations was extracted from the Swiss Variant database [62]. Fig. 6 shows the number of human polymorphisms that changes the lysine targets of PTMs (in ~40% of the acetylation, and ~50% of the ubiquitylation cases, the substitution of the target lysine is associated with a disease). By inspecting the published protein structures the impact of the presence or absence of a lysine modification in some cases could be easily predicted. Fig. 7 shows the structures of two proteins for which a mutant lacking the lysine modified by ubiquitylation has been described. K16 of the ribosyldihyd- ronicotinamide dehydrogenase, a cytosolic flavoenzyme that catalyzes the two-electron reduction of quinones to hydroquinones, is prone to ubiquitylation (see www.phospho site.org for references) and it is absent in the natural variant VAR_021399, K→R. The position of this residue being close to the active site (Fig. 7A) suggests that its ubiquitylation could affect the catalytic activity of the enzyme. The mutation in this case does not change the physiochemical property of the wild-type amino acid but prevents its ubiquitylation and therefore it is expected that it could have some functional consequences.
Fig. (6).
Number of SNPs affecting lysines subjected to PTMs.
Fig. (7).
Ribosyldihydronicotinamide dehydrogenase (NQO2) (PDB entry: 1QR2) and piruvate kinase (PK) (PDB entry:1A49) models. A. NQO2 structure. The side chain of K16 prone to ubiquitylation and FADH2 molecule are shown in sticks. B. PK structure. The side chain of K348 prone to ubiquitylation and ATP molecule are shown in sticks. All the images were generated with PyMOL Software (Scroedinger, LLC).
K348 of the mitochondrial pyruvate kinase is prone to ubiquitylation (see www.phosphosite.org for references) and a natural variant (VAR_011456, K→N) has been linked to a pyruvate kinase deficiency in red blood cells. K348 is near to the active site (Fig. 7B) and the addition of a ubiquitin moiety in this position could interfere with the catalytic activity. In this case an amino acid with a positively charged side chain has been substituted with a polar one but we can expect that also the lack of ubiquitylation will be functionally relevant.
On the contrary, it is more difficult to predict the outcome when amino acid substitution takes place at positions surrounding the target lysine: considering the low primary structure conservation for most of the lysine PTMs (refer to discussion above and see Fig. 1) we can hypothesize that non-synonymous SNPs could have a modest effect for most of PTMs. This is not the case of sumoylation that is highly primary-structure dependent (see Fig. 1); in this regard, we may expect that the loss of the glutamic acid at +2 and/or the hydrophobic amino acid at -1 will be deleterious for this PTM. Likewise the appearance of these aminoacids (at +2 and -1 of a lysine) may generate new “unnatural” sumoylation sites.
CONCLUSION
Lysine is the target of different competing PTMs. Here, we made a comparative survey of lysine PTMs that have been extensively identified in humans, i.e. methylation, ubiquitylation, acetylation and sumoylation. Our analysis accounted for the primary amino acid structure, solvent accessibility, secondary structure and order/disorder prediction, cellular localization and biological functions, highlighting specific differences and similarities between these lysine PTMs. Future analysis will be extended to other lysine PTMs when a sufficient amount of data becomes available.
ACKNOWLEDGEMENTS
This work was supported by Associazione Italiana per la Ricerca sul Cancro, AIRC (grant number IG10312) (to L.A.P.).
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
SUPPLEMENTARY MATERIALS
Supplementary material is available on the publisher’s web site along with the published article.
LIST OF ABBREVIATIONS
- PTMs =
Post-translational Modifications
- SNPs =
Single Nucleotide Polymorphisms
REFERENCES
- 1.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4(6):565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rust H L, Thompson P R. Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem. Biol. 2011;6(9):881–892. doi: 10.1021/cb200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen P. Protein kinases--the major drug targets of the twenty-first centuryκ. Nat. Rev. Drug. Discov. 2002;1(4):309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinasesκ. Cell. 2010;143(5):686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Venugopal B, Evans T R. Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr. Med. Chem. 2011;18(11):1658–1671. doi: 10.2174/092986711795471284. [DOI] [PubMed] [Google Scholar]
- 6.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316(2):113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Olsen J V, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteom. 2013;12(12):3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toppo S, Pinna LA, Salvi M. Matching up Phosphosites to Kinases: A Survey of Available Predictive Programs. Curr. Bioinform. 2010;5(2):11. [Google Scholar]
- 9.Choudhary C, Kumar C, Gnad F, Nielsen M L, Rehman M, Walther T C, Olsen J V, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Wen H, Shi X. Lysine methylation: beyond histones. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):14–27. doi: 10.1093/abbs/gmr100. [DOI] [PubMed] [Google Scholar]
- 11.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 12.Reyes-Turcu F E, Ventii K H, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma K, D'Souza R C, Tyanova S, Schaab C, Wisniewski J R, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell. Rep. 2014;8(5):1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 14.Lanouette S, Mongeon V, Figeys D, Couture JF. The functional diversity of protein lysine methylation. Mol. Syst. Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 16.van der Veen A G, Ploegh H L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 17.Barth T K, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 2010;35(11):618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Aka J A, Kim G W, Yang X J. K-acetylation and its enzymes: overview and new developments. Handb. Exp. Pharmacol. 2011;206:1–12. doi: 10.1007/978-3-642-21631-2_1. [DOI] [PubMed] [Google Scholar]
- 19.Allis C D, Berger S L, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Gershey E L, Vidali G, Allfrey V G. Chemical studies of histone acetylation The occurrence of epsilon-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 1968;243(19):5018–5022. [PubMed] [Google Scholar]
- 21.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylationκ. EMBO J. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S C, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin N V, White M, Yang X J, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Ambler R P, Rees M W. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56–57. doi: 10.1038/184056b0. [DOI] [PubMed] [Google Scholar]
- 24.Murray K. The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 25.Moore K E, Gozani O. An unexpected journey: Lysine methylation across the proteome. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbagrm.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paik W K, Paik D C, Kim S. Historical review: the field of protein methylation. Trends Biochem. Sci. 2007;32(3):146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Bock I, Dhayalan A, Kudithipudi S, Brandt O, Rathert P, Jeltsch A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics. 2011;6(2):256–263. doi: 10.4161/epi.6.2.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee K A, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil S A, Silva J C, Vemulapalli V, Bedford M T, Comb M J. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell. Proteomics. 2014;13(1):372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X J, Arnaudo A M, Garcia B A. Large-scale global identification of protein lysine methylation in vivo. Epigenetics. 2013;8(5):477–485. doi: 10.4161/epi.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta. 2006;1764(12):1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G, Paige J S, Jaffrey S R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010;28(8):868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner S A, Beli P, Weinert B T, Nielsen M L, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics. 2011;10(10):M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner S A, Beli P, Weinert B T, Scholz C, Kelstrup C D, Young C, Nielsen M L, Olsen J V, Brakebusch C, Choudhary C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics. 2012;11(12):1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen M L. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics. 2011;10(3):M110 003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matic I, Schimmel J, Hendriks I A, van Santen M A, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal A C. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 2010;39(4):641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Lu Z, Cheng Z, Zhao Y, Volchenboum SL. Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation. PLoS One. 2011;6(12):e28228. doi: 10.1371/journal.pone.0028228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minguez P, Parca L, Diella F, Mende D R, Kumar R, Helmer-Citterich M, Gavin A C, van Noort V, Bork P. Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. 2012;8:599. doi: 10.1038/msb.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucl. Acids Res. 2012;40(Database issue):D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacic V, Iakoucheva L M, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22(12):1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 40.Johnson E S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 41.Hietakangas V, Anckar J, Blomster H A, Fujimoto M, Palvimo J J, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci.; U S A. 2006;103(1):45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvi M, Cesaro L, Pinna LA. Variable contribution of protein kinases to the generation of the human phosphoproteome: a global weblogo analysis. Biomol. Concepts. 2010;1(2):185–196. doi: 10.1515/bmc.2010.013. [DOI] [PubMed] [Google Scholar]
- 43.Paik W K, Pearson D, Lee H W, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta. 1970;213(2):513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 44.Weinert B T, Iesmantavicius V, Moustafa T, Scholz C, Wagner S A, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinert B T, Iesmantavicius V, Wagner S A, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013;51(2):265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Yang S H, Galanis A, Witty J, Sharrocks A D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25(21):5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim W, Bennett E J, Huttlin E L, Guo A, Li J, Possemato A, Sowa M E, Rad R, Rush J, Comb M J, Harper J W, Gygi S P. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinna L A, Ruzzene M. How do protein kinases recognize their substratesκ. Biochim. Biophys. Acta. 1996;1314(3):191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 49.Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucl. Acids Res. 2004;32:W500–502. doi: 10.1093/nar/gkh429. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagai T, Azia A, Toth-Petroczy A, Levy Y. Intrinsic disorder in ubiquitination substrates. J. Mol. Biol. 2011;412(3):319–324. doi: 10.1016/j.jmb.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Bernier-Villamor V, Sampson D A, Matunis M J, Lima C D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108(3):345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Zhang J, Lin Y, Lei Q, Guan K-L, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO reports. 2011;12(6):534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimazu T, Hirschey M D, Hua L, Dittenhafer-Reed K E, Schwer B, Lombard D B, Li Y, Bunkenborg J, Alt F W, Denu J M, Jacobson M P, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uversky V N, Oldfield C J, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 55.Haynes C, Oldfield C J, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky V N, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X J, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31(4):449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laing R E, Hess P, Shen Y, Wang J, Hu S X. The role and impact of SNPs in pharmacogenomics and personalized medicine. Curr. Drug. Metab. 2011;12(5):460–486. doi: 10.2174/138920011795495268. [DOI] [PubMed] [Google Scholar]
- 58.Collins F S, Brooks L D, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8(12):1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 59.Ren J, Jiang C, Gao X, Liu Z, Yuan Z, Jin C, Wen L, Zhang Z, Xue Y, Yao X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics. 2010;9(4):623–634. doi: 10.1074/mcp.M900273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryu G M, Song P, Kim K W, Oh K S, Park K J, Kim J H. Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucl. Acids Res. 2009;37(4):1297–1307. doi: 10.1093/nar/gkn1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suo S B, Qiu J D, Shi S P, Chen X, Huang S Y, Liang R P. Proteome-wide analysis of amino acid variations that influence protein lysine acetylation. J. Proteome Res. 2013;12(2):949–958. doi: 10.1021/pr301007j. [DOI] [PubMed] [Google Scholar]
- 62.Yip Y L, Scheib H, Diemand A V, Gattiker A, Famiglietti L M, Gasteiger E, Bairoch A. The Swiss-Prot variant page and the ModSNP database: a resource for sequence and structure information on human protein variants. Hum. Mutat. 2004;23(5):464–470. doi: 10.1002/humu.20021. [DOI] [PubMed] [Google Scholar]
- 63.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105(1):1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 64.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 65.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucl. Acids Res. 2012;40:W478–483. doi: 10.1093/nar/gks402. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucl. Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucl. Acids Res. 2013;41:D377–386. doi: 10.1093/nar/gks1118. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.