Abstract
The aims of the present study were to elucidate the transcript levels of DNA methyltransferase (DNMT)1, DNMT3a and DNMT3b by quantitative polymerase chain reaction in patients with primary gastrointestinal diffuse large B-cell lymphoma (PGI-DLBCL), and determine the association of their expression with the clinical parameters and prognostic values of the disease. The results revealed that the expression of DNMT1 in patients with PGI-DLBCL was significantly higher than that in healthy controls (P=0.04), while the expression of DNMT3a and DNMT3b were significantly lower (P<0.001 and P=0.001, respectively). The increased expression of DNMT1 was significantly correlated with shorter overall survival and progression-free survival rates (P=0.018 and P=0.008, respectively). The multivariate analysis demonstrated that the level of DNMT1 was an independent prognostic factor. In conclusion, DNMT1 was identified to be an independent prognostic factor for predicting the survival of patients with PGI-DLBCL; this suggests that it could be used as a marker to indicate the prognosis of PGI-DLBCL.
Keywords: gastrointestinal lymphoma, DNA methyltransferase 1, diffuse large B-cell lymphoma, prognosis
Introduction
Primary gastrointestinal (PGI) lymphoma is an extremely rare disease that accounts for 1–4% of all gastrointestinal malignancies (1). In general, gastrointestinal lymphoma is secondary to widespread nodal diseases. Lymphomas may originate from any section of the gastrointestinal tract, however, the most frequently affected region is the stomach, followed by the small intestine (2). The most commonly diagnosed histopathological subtype of PGI lymphoma is diffuse large B-cell lymphoma (DLBCL), which accounts for ∼50% of all cases (3). Due to its rarity, only a limited number of studies concerning PGI-DLBCL have been published (4–6).
Aberrant DNA methylation, which is characterized by widespread genome hypomethylation, chromosome instability and localized DNA hypermethylation, is considered to be an important event, occurring in early tumor development (7,8). Methylation patterns are predominantly regulated by independent DNA methyltransferases (DNMTs), including DNMT1, DNMT3a and DNMT3b (9,10). DNMT1 mediates maintenance DNA methylation and DNMT3a and DNMT3b are responsible for de novo methylation (9). Previous studies have revealed that DNMT1 and DNMT3b overexpression is associated with unfavorable prognoses in a number of human cancers, including breast and hepatocellular carcinomas, lung cancers, acute myeloid leukemia and epithelial ovarian cancer (11–17).
Increasing evidence suggests that aberrant DNA methylation is significant in the pathogenesis of lymphomas (18–22). However, at present, the prognostic significance of DNMT expression in PGI-DLBCL is yet to be elucidated. The aims of the current study were to determine the transcript levels of DNMT1, DNMT3a and DNMT3b in PGI-DLBCL patients by quantitative polymerase chain reaction (qPCR), and establish their clinical significance.
Materials and methods
Patients and controls
In total, 62 patients with a histopathological diagnosis of PGI-DLBCL, and 30 age- and gender-matched healthy controls were recruited. The study was approved by Tianjin Medical University Cancer Institute and Hospital Ethics Committee, and all the patients provided written informed consent prior to study participation. Fresh samples of cancerous tissues and normal tissues were collected via surgical resection. The samples were frozen in liquid nitrogen for 30 min in cryovials, and subsequently stored at −80°C until further analysis. The diagnosis of PGI-DLBCL was based upon the World Health Organization classification system for hematological malignancies (23). The patients were staged according to the Lugano staging system for gastrointestinal non-Hodgkin's lymphoma (NHL) (24). The diagnostic work-up consisted of the patient's history, their performance status according to the Eastern Cooperative Oncology Group scale, a physical examination, a baseline endoscopy or barium meal examination, gastric mucosal biopsies or a gastrectomy, a complete blood cell count, a biochemical profile, measurement of serum lactate dehydrogenase (LDH), computed tomography scans of the thorax, abdomen and pelvic cavity, and a bone marrow aspiration and biopsy. A low hemoglobin level was defined as <120 g/l, and a high LDH level as >245 U/l. The patients were subsequently grouped according to age, gender, tumor origin, performance status, Lugano staging system outcome, clinical stage, B symptoms, LDH level, International Prognostic Index (IPI) score and chemotherapy response. A retrospective review of the clinical, pathological and treatment results of all patients was conducted and the results were entered into an anonymized database. The clinical characteristics and histological features of the patients with PGI-DLBCL are shown in Table I.
Table I.
Clinical characteristics of primary gastrointestinal diffuse large B cell lymphoma patients.
| Characteristic | Patients, n (%) |
|---|---|
| Age, years | |
| ≤60 | 35 (56.5) |
| >60 | 27 (43.5) |
| Gender | |
| Male | 33 (53.2) |
| Female | 29 (46.8) |
| Origin | |
| Stomach | 41 (66.1) |
| Intestinal | 21 (33.9) |
| Performance status | |
| ECOG 0–1 | 45 (72.6) |
| ECOG 2–4 | 17 (27.4) |
| Lugano staging system | |
| I-II | 20 (32.3) |
| IIE-IV | 42 (67.7) |
| Pathological type | |
| Non-GCB | 45 (72.6) |
| GCB | 17 (27.4) |
| LDH | |
| Normal | 22 (35.5) |
| Elevated | 40 (64.5) |
| Hemoglobin | |
| Normal | 30 (48.4) |
| Low | 32 (51.6) |
| B symptoms | |
| Positive | 13 (21.0) |
| Negative | 49 (79.0) |
| IPI | |
| 0–2 | 39 (62.9) |
| 3–5 | 23 (37.1) |
| Therapeutic evaluation | |
| CR | 32 (51.6) |
| PR/SD/PD | 30 (48.4) |
ECOG, Eastern Cooperative Oncology Group; GCB, germinal center B-cell-like; LDH, lactose dehydrogenase; IPI, International Prognostic Index; CR, complete remission; PR, partial remission; SD, stable disease; PD, progressive disease.
RNA, cDNA preparation and qPCR
For the RNA extraction step, ∼25 mg of tumor tissue was pulverized under liquid nitrogen using a pestle and mortar. The RNA was subsequently extracted using RNeasy (Qiagen Inc., Valencia, CA, USA), according to the manufacturer's instructions. Following this, reverse transcription reactions were performed using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's instructions and qPCR was performed using a CM9600 Sequence Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The amplification step was performed in a total volume of 20 µl, with 10 µl kit-supplied QuantiTect™ SYBR® Green RT-PCR Master mix (Applied Biosystems Life Technologies, Foster City, CA, USA), 0.4 µl of each primer (10 µM), 2 µl of cDNA (50 ng RNA) and 7.2 µl ddH2O. The PCR cycling parameters were set as follows: 95°C for 30 sec, followed by 40 cycles of PCR reacting at 95°C for 5 sec, and finally 60°C for 34 sec. β actin was used as an internal standard. The ΔΔCT values were calculated using the differences between the target genes and β actin. The primer sequences are shown in Table II. Each experiment was conducted in triplicate.
Table II.
Primer sequences.
| Gene | Forward sequence (5′→3′) | Reverse sequence (5′→3′) |
|---|---|---|
| DNMT1 | GTGGGGGACTGTGTCTCTGT | TGAAAGCTGCATGTCCTCAC |
| DNMT3a | CAGCTTCCACGTTGCCTTCT | CATCTGCAAGCTGTCTCCCTTT |
| DNMT3b | CCCATTCGAGTCCTGTCATT | GGTTCCAACAGCAATGGACT |
| β actin | CCTGGCACCCAGCACAAT | GGGCCGGACTCGTCATAC |
DNMT, DNA methyltransferase.
Immunohistochemical staining and scoring
The biopsied tissues were initially fixed in 10% buffered formalin, embedded in paraffin and cut into 4-µm sections. Following this, deparaffinization and heat-induced antigen retrieval steps were performed. The endogenous peroxidase activity was blocked using 0.5% hydrogen peroxide. Subsequent to washing with phosphate-buffered saline (PBS), the sections were incubated with anti-cluster of differentiation (CD)10 (cat. no. ZM0283), -B-cell lymphoma 6 (BCL-6; cat. no. ZM-0011) and -melanoma associated antigen 1 (MUM1; cat. no. ZA-0583) antibodies (ZSGB-BIO, Beijing, China) at 4°C overnight. After washing with PBS again, the secondary antibodies were added to the sections. The nuclei were subsequently stained with hematoxylin, and the color was developed using diaminobenzidine. A positive result was recorded when ≥30% of the cells were stained. The tissues were also analyzed for CD10, BCL-6 and MUM1 immunoreactivity according to the immunophenotypic profile criteria of DLBCL described previously by Hans et al (25). The intensity of the staining and the percentage of positive cells were recorded. The staining intensity was scored between 0 and 3+ as follows: i) 0, Absence of staining; ii) 1+, >25 % of the tumor cells exhibited weak staining; iii) 2+, tumor cells exhibited moderate staining; and iv) 3+, tumor cells demonstrated strong staining. Tumors with a score of 1+, 2+ or 3+ were considered to be positive, while tumors with a score of 0 were considered to be negative. The immunohistochemical analysis was performed independently by at least two histopathologists.
Treatment and response assessment
The patients were treated with four therapeutic modalities; surgery, chemotherapy, Helicobacter pylori (Hp) eradication and radiotherapy. The aim of surgery was to remove the tumor tissue. All patients were treated with 6–8, 21-day cycles of the CHOP [cyclophosphamide (750 mg/m2, day 1), doxorubicin (50 mg/m2, day 1), vincristine (1.4 mg/m2, day 1) and prednisone (100 mg, days 1–5)] or R-CHOP [rituximab (350 mg/m2, day 1), cyclophosphamide (750 mg/m2, day 1), doxorubicin (50 mg/m2, day 1), vincristine (1.4 mg/m2, day 1) and prednisone (100 mg, days 1–5)] regimens. The Hp eradication regimen, which consisted of proton pump inhibitors [omeprazole (20 mg, twice daily), lansoprazole (30 mg, twice daily) or rabeprazole (10 mg, twice daily)] and a combination of antibiotics [amoxicillin (1,000 mg, twice daily), clarithromycin (500 mg, twice daily) and/or metronidazole (200 mg, twice daily)], was administered to eight patients, seven days a month, for three months. In total, six patients received radiotherapy. The responses were assessed by the International Workshop for NHL Standardized Criteria (26).
Statistical analysis
The statistical analysis was performed using SSPS version 19.0 software (SPSS Inc., Chicago, IL, USA). The results are presented as the mean ± standard deviation. The data were analyzed using the non-parametric Mann-Whitney U-test. The significant differences between the survival curves were calculated using the log-rank test. A GraphPad Prism software package was used for the Kaplan-Meier estimate graphs. The Cox proportional hazards regression model was used for the multivariate analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics and histological features
In total, 62 patients with PGI-DLBCL, which included 33 males and 29 females (median age, 54 years; range, 32–76 years), and 30 age- and gender-matched healthy controls were enrolled in the present study. Overall, 20 patients (32.3%) were diagnosed with an advanced Ann Arbor clinical stage (stages IIE-IV), and 45 patients (72.6%) exhibited a good performance status (<2). An elevated serum LDH level was observed in 40 patients (64.5%); low or low-intermediate IPI scores were identified in 39 patients (62.9%). Based upon the results of the immunophenotypical profiles, 17 patients (27.4%) were classified as having a germinal center B-cell-like (GCB) subtype, and 45 patients (72.6%) with a non-GCB subtype. In total, 32 patients (51.6%) achieved complete remission after 6–8 cycles of the CHOP or R-CHOP regimens, 11 (17.8%) were in partial remission and 19 patients (30.6%) exhibited stable or progressive disease.
mRNA expression of DNMT1, DNMT3a and DNMT3b in patients with PGI-DLBCL
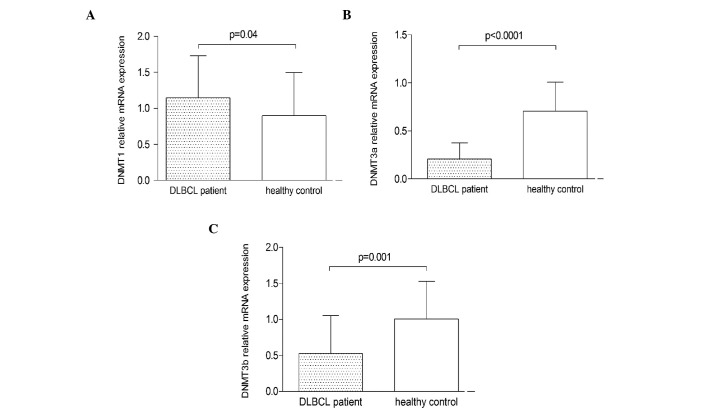
The mRNA expression of DNMT1, DNMT3a and DNMT3b was determined using qPCR. The expression of DNMT1 mRNA in PGI-DLBCL patients was significantly higher than that of the healthy controls (P=0.04), while the expression of DNMT3a and DNMT3b mRNA was significantly lower (P<0.0001 and P=0.001, respectively) (Fig. 1).
Figure 1.
Relative mRNA expression of DNMT1, DNMT3a and DNMT3b in PGI-DLBCL patients and healthy controls. The mRNA expression was detected using quantitative polymerase chain reaction. The bars indicate the relative mRNA levels (mean ± standard deviation). (A) The levels of DNMT1 mRNA from PGI-DLBCL patients were significantly higher than those of healthy controls (P=0.04), while the expression of (B) DNMT3a and (C) DNMT3b mRNA was lower (P<0.0001 and P=0.001, respectively). DNMT, DNA methyltransferase; PGI-DLBCL, primary gastrointestinal diffuse large B cell lymphoma.
Association of DNMT1 expression with the clinicopathological characteristics of PGI-DLBCL
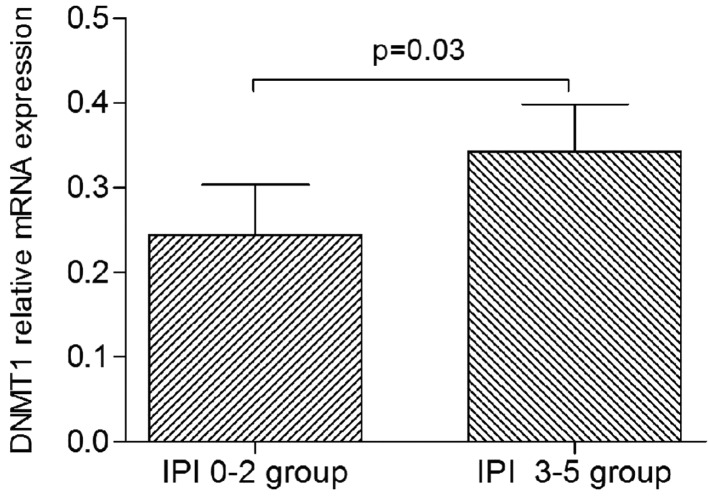
The patients with PGI-DLBCL were divided into subgroups according to specific clinical parameters, such as the Lugano stage (stages I-II and stages IIE-IV subgroups), the IPI score (low IPI, 0–2; and high IPI, 3–5 subgroups), and the DLBCL type (non-GCB and GCB subgroups). It was revealed that DNMT1 expression was significantly higher in the high IPI subgroup than in the low IPI subgroup (P=0.03; Fig. 2). By contrast, there were no significant differences in the expression of DNMT3a and DNMT3b between any of the subgroups (data not shown).
Figure 2.
DNMT1 expression in the IPI subgroups. Patients with primary gastrointestinal diffuse large B cell lymphoma were divided into low IPI (0–2) and high IPI (3–5) subgroups. DNMT1 expression was significantly higher in the high IPI subgroup (p=0.03) compared with the low IPI subgroup. DNMT1, DNA methyltransferase 1; IPI, International Prognostic Index.
DNMT1 is a negative independent prognostic factor for patients with PGI-DLBCL
The patients were divided into high-expression (above the median) and low-expression (below the median) groups depending on mRNA expression levels. The results demonstrated that the overall survival (OS) and progression-free survival (PFS) rates of the high-DNMT1 group were significantly shorter compared with those of the low-DNMT1 group (P=0.038 and P=0.012, respectively; Fig. 3). The univariate analysis revealed that DNMT1 expression, Lugano staging, pathological type and IPI scores were factors associated with the prognosis of patients with PGI-DLBCL (Table III). The multivariate analysis showed that in addition to the IPI score, DNTM1 expression was also an independent predictive factor for mortality and disease progression (Table IV).
Figure 3.
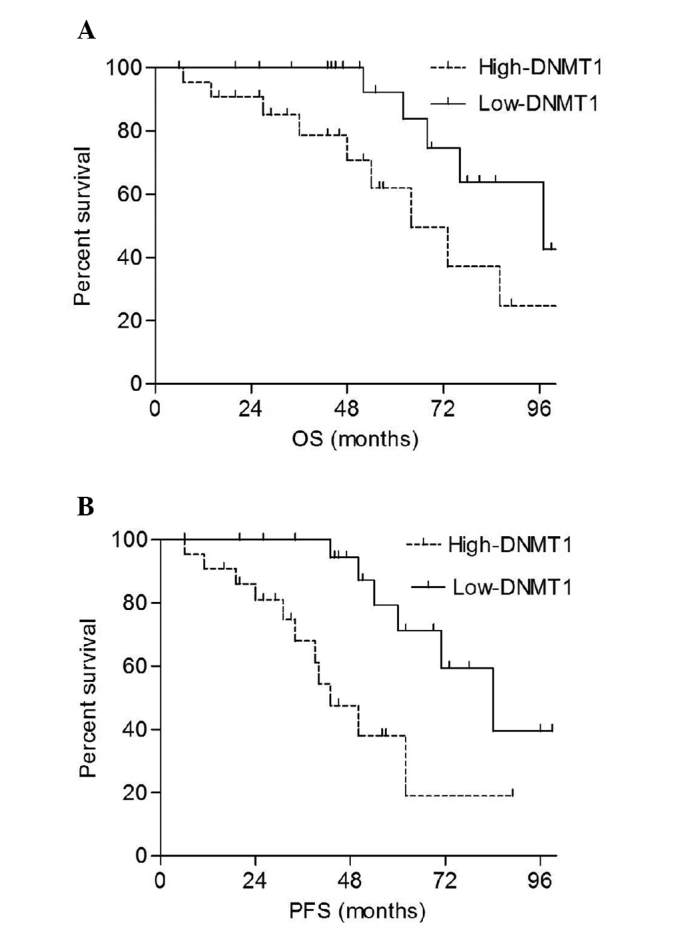
Effect of DNMT1 expression on OS and PFS in patients with primary gastrointestinal diffuse large B cell lymphoma. The (A) OS and (B) PFS rates of the high-DNMT1 group were significantly shorter than those of the low-DNMT1 group (P=0.038 and P=0.012, respectively). DNMT1, DNA methyltransferase 1; OS, overall survival; PFS, progression-free survival.
Table III.
Univariate analysis revealing the significance of different prognostic variables for primary gastrointestinal diffuse large B cell lymphoma.
| Overall survival | Progression-free survival | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| DNMT1 expression (low vs. high) | 5.87 (2.31–12.68) | 0.015 | 3.78 (1.24–8.62) | 0.012 |
| ECOG (0–1 vs. −4) | 3.24 (1.67–8.90) | 0.072 | 2.56 (1.24–9.78) | 0.145 |
| Lugano stage (I-II vs. IIE-IV) | 2.13 (0.45–7.89) | 0.032 | 3.02 (0.24–8.34) | 0.024 |
| Pathological type (non-GCB vs. GCB) | 3.56 (0.83–10.76) | 0.027 | 1.56 (0.45–6.76) | 0.032 |
| LDH (normal vs. elevated) | 0.67 (0.06–1.18) | 0.124 | 0.92 (0.06–3.54) | 0.294 |
| IPI (0–2 vs. 3–5) | 3.45 (0.23–11.92) | 0.016 | 2.30 (0.23–10.34) | 0.031 |
| Treatment (surgery+R-CHOP vs. surgery+CHOP) | 2.01 (0.54–5.72) | 0.569 | 1.03 (0.78–8.45) | 0.756 |
| Therapeutic evaluation (SD vs. PR/SD/PD) | 2.69 (0.35–6.97) | 0.187 | 1.45 (0.56–12.46) | 0.357 |
HR, hazard ratio; DNMT1, DNA methyltransferase 1; ECOG, Eastern Cooperative Oncology Group; GCB, germinal center B-cell-like; LDH, lactose dehydrogenase; IPI, International Prognostic Index; R-CHOP, rituximab administered with cyclophosphamide, doxorubicin, vincristine and prednisone; CHOP, cyclophosphamide, doxorubicin, vincristine and prednisone; SD, stable disease; PR, partial remission; PD, progressive disease.
Table IV.
Multivariate analysis revealing the significance of independent prognostic variables for primary gastrointestinal diffuse large B cell lymphoma.
| Overall survival | Progression-free survival | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| DNMT1 expression (low vs. high) | 4.32 (1.25–8.23) | 0.018 | 4.02 (1.42–12.63) | 0.008 |
| Lugano stage (I-II vs. IIE-IV) | 1.34 (0.69–6.90) | 0.082 | 1.56 (0.54–7.78) | 0.160 |
| Pathological type (non-GCB vs. GCB) | 1.13 (0.45–4.56) | 0.143 | 1.02 (0.24–3.34) | 0.237 |
| IPI (0–2 vs. 3–5) | 2.96 (0.58–5.76) | 0.017 | 2.56 (0.83–5.76) | 0.020 |
HR, hazard ratio; DNMT1, DNA methyltransferase 1; GCB, germinal center B-cell-like; IPI, International Prognostic Index.
Discussion
The processes that underlie the pathogenesis of PGI-DLBCL are yet to be elucidated. Previous studies have hypothesized that abnormal DNA methylation is an important event that occurs early in tumor development (27,28). DNA methylation is regulated by DNMTs, predominantly DNMT1, DNMT3a and DNMT3b (9,10). Therefore, we hypothesize that the dysregulated expression and function of DNMT1, DNMT3a and DNMT3b is significant in the pathogenesis of PGI-DLBCL.
Increasing evidence indicates that high DNMT1 and/or DNMT3b expression has a modulating effect upon the clinical outcome of cancer patients. A previous study revealed that high DNMT1 expression was associated with a subgroup of gastric cancer patients with poorer outcomes following platinum/fluorouracil-based neoadjuvant chemotherapy (29). Another study identified that the level of DNMT1 mRNA was significantly higher in gastric cancer patients with methylated Runx3, compared with those with unmethylated Runx3 (30). Furthermore, increased expression of DNMT1 and DNMT3b have been detected in the tumor tissues of patients with lung cancer, and were also associated with poorer prognoses (31–34). In a study concerning pancreatic cancer patients, those with a higher DNMT1 expression exhibited a lower overall survival rate (35). In addition, DNMT1 and DNMT3b were markedly upregulated in malignant glioma tumor tissues (36), and DNMT3b overexpression was identified to be an independent prognostic factor for predicting the reduced survival of DLBCL patients (37).
The present study revealed that DNMT1 mRNA expression in PGI-DLBCL patients was higher than that of the healthy controls, and that it was increased in the high IPI subgroup compared with the low IPI subgroup. The Kaplan-Meier analysis showed that the high-DNMT1 subgroup exhibited shorter OS and PFS rates compared with the low-DNMT1 subgroup. This suggests that DNMT1 is a negative predictive factor for PGI-DLBCL. The Cox regression analysis revealed that DNMT1 was an independent prognostic factor, which suggests that DNMT1 expression may be a suitable prognostic marker for patients with PGI-DLBCL. These findings are consistent with those of other previous studies (28–36). Although previous studies have reported that the levels of DNMT3a and DNMT3b were significantly higher in cancer patients (31,33,35,36), the present study indicated that the expression of DNMT3a and DNMT3b in PGI-DLBCL patients was markedly lower than that of the healthy controls. This may be due to the small number of samples used in the present study. Future studies with larger patient cohorts are therefore required in order to address this inconsistency.
In conclusion, the results of the present study suggest that DNMT1, DNMT3a and DNMT3b may be involved in the etiology of PGI-DLBCL. In addition, the results indicate that DNMT1 is a negative and independent prognostic parameter for patients with PGI-DLBCL, and, therefore, may be used as a biomarker in order to guide prognosis.
References
- 1.Wang T, Gui W, Shen Q. Primary gastrointestinal non-Hodgkin's lymphoma: clinicopathological and prognostic analysis. Med Oncol. 2010;27:661–666. doi: 10.1007/s12032-009-9265-1. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann R, Panahon AM, Barcos MP, Walsh D, Stutzman L. Gastrointestinal involvement in non-Hodgkin's lymphoma. Cancer. 1980;46:215–222. doi: 10.1002/1097-0142(19800701)46:1<215::AID-CNCR2820460136>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 3.van Krieken JH, Otter R, Hermans J, et al. Malignant lymphoma of the gastrointestinal tract and mesentery. A clinico-pathologic study of the significance of histologic classification. NHL Study Group of the Comprehensive Cancer Center West. Am J Pathol. 1989;135:281–289. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F, Yang G, Xia B. Increased expression of the spindle checkpoint protein BubR1 is associated with high cell proliferation in primary gastrointestinal diffuse large B cell lymphoma. Cell Biochem Biophys. 2013;66:747–752. doi: 10.1007/s12013-013-9519-6. [DOI] [PubMed] [Google Scholar]
- 5.Ding D, Pei W, Chen W, Zuo Y, Ren S. Analysis of clinical characteristics, diagnosis, treatment and prognosis of 46 patients with primary gastrointestinal non-Hodgkin lymphoma. Mol Clin Oncol. 2014;2:259–264. doi: 10.3892/mco.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Shen W, Cao J, et al. Treatment of gastrointestinal diffuse large B cell lymphoma in China: a 10-year retrospective study of 114 cases. Ann Hematol. 2012;91:1721–1729. doi: 10.1007/s00277-011-1375-0. [DOI] [PubMed] [Google Scholar]
- 7.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol. 2003;109:37–45. doi: 10.1016/S1521-6616(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 10.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 11.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Saito Y, Kanai Y, Nakagawa T, et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 13.Oue N, Kuraoka K, Kuniyasu H, et al. DNA methylation status of hMLH1, p16(INK4a), and CDH1 is not associated with mRNA expression levels of DNA methyltransferase and DNA demethylase in gastric carcinomas. Oncol Rep. 2001;8:1085–1089. doi: 10.3892/or.8.5.1085. [DOI] [PubMed] [Google Scholar]
- 14.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4122. [PubMed] [Google Scholar]
- 15.Xing J, Stewart DJ, Gu J, Lu C, Spitz MR, Wu X. Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer. 2008;98:1716–1722. doi: 10.1038/sj.bjc.6604343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayette S, Thomas X, Jallades L, et al. High DNA methyltransferase DNMT3B levels: poor prognostic marker in acute myeloid leukemia. PLoS One. 2012;7:e51527. doi: 10.1371/journal.pone.0051527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X, Song Z, Fu Y, et al. Clinicopathological significance and prognostic value of DNA methyltransferase 1, 3a, and 3b expressions in sporadic epithelial ovarian cancer. PLoS One. 2012;7:e40024. doi: 10.1371/journal.pone.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaknovich R, Melnick A. Epigenetics and B-cell lymphoma. Curr Opin Hematol. 2011;18:293–299. doi: 10.1097/MOH.0b013e32834788cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor KH, Briley A, Wang Z, Cheng J, Shi H, Caldwell CW. Aberrant epigenetic gene regulation in lymphoid malignancies. Semin Hematol. 2013;50:38–47. doi: 10.1053/j.seminhematol.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 20.De S, Shaknovich R, Riester M, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9:e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pike BL, Greiner TC, Wang X, et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98:1912–1920. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomonaga M. Outline and direction of revised WHO classification of tumors of haematopoietic and lymphoid tissues. Rinsho Ketsueki. 2009;50:1401–1406. (In Japanese) [PubMed] [Google Scholar]
- 24.No authors listed: 5th International Conference on Malignant Lymphoma, Part 2. Proceedings. Lugano, Switzerland, June 9–12, 1993. Ann Oncol. 1994;5(Suppl 2):S1–S163. [PubMed] [Google Scholar]
- 25.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 26.Cheson BD, Horning SJ, Coiffier B, et al. Report of an international workshop to standardize response criteria for non-Hodgkin's lymphomas. NCI Sponsored International Working Group. J Clin Oncol. 1999;17:1244. doi: 10.1200/JCO.1999.17.4.1244. [DOI] [PubMed] [Google Scholar]
- 27.Fukushige S, Horii A. DNA methylation in cancer: a gene silencing mechanism and the clinical potential of its biomarkers. Tohoku J Exp Med. 2013;229:173–185. doi: 10.1620/tjem.229.173. [DOI] [PubMed] [Google Scholar]
- 28.Sincic N, Herceg Z. DNA methylation and cancer: ghosts and angels above the genes. Curr Opin Oncol. 2011;23:69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 29.Mutze K, Langer R, Schumacher F, et al. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer. 2011;47:1817–1825. doi: 10.1016/j.ejca.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 30.Chen W, Gao N, Shen Y, Cen JN. Hypermethylation downregulates Runx3 gene expression and its restoration suppresses gastric epithelial cell growth by inducing p27 and caspase3 in human gastric cancer. J Gastroenterol Hepatol. 2010;25:823–831. doi: 10.1111/j.1440-1746.2009.06191.x. [DOI] [PubMed] [Google Scholar]
- 31.Gao P, Yang X, Xue YW, Zhang XF, Wang Y, Liu WJ, Wu XJ. Promoter methylation of glutathione S-transferase pi1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma and its correlation with DNA methyltransferase 1 expression. Cancer. 2009;115:3222–3232. doi: 10.1002/cncr.24369. [DOI] [PubMed] [Google Scholar]
- 32.Xing J, Stewart DJ, Gu J, Lu C, Spitz MR, Wu X. Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer. 2008;98:1716–1722. doi: 10.1038/sj.bjc.6604343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary non-small cell lung cancer. Cancer. 2006;107:1042–1049. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- 34.Vallbohmer D, Brabender J, Yang D, et al. DNA methyltransferases messenger RNA expression and aberrant methylation of CpG islands in non-small-cell lung cancer: association and prognostic value. Clin Lung Cancer. 2006;8:39–44. doi: 10.3816/CLC.2006.n.031. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Gao J, Man XH, Li ZS, Gong YF. Significance of DNA methyltransferase-1 and histone deacetylase-1 in pancreatic cancer. Oncol Rep. 2009;21:1439–1447. doi: 10.3892/or_00000372. [DOI] [PubMed] [Google Scholar]
- 36.Kreth S, Thon N, Eigenbrod S, et al. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amara K, Ziadi S, Hachana M, Soltani N, Korbi S, Trimeche M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101:1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]