Abstract
Host-pathogen protein interactions are fundamental to every microbial infection, yet their identification has remained challenging due to the lack of simple detection tools that avoid abundance biases while providing an open format for experimental modifications. Here, we applied the Nucleic Acid-Programmable Protein Array and a HaloTag-Halo ligand detection system to determine the interaction network of Legionella pneumophila effectors (SidM and LidA) with 10,000 unique human proteins. We identified known targets of these L. pneumophila proteins and potentially novel interaction candidates. In addition, we applied our Click chemistry-based NAPPA platform to identify the substrates for SidM, an effector with an adenylyl transferase domain that catalyzes AMPylation (adenylylation), the covalent addition of adenosine monophosphate (AMP). We confirmed a subset of the novel SidM and LidA targets in independent in vitro pull-down and in vivo cell-based assays, and provided further insight into how these effectors may discriminate between different host Rab GTPases. Our method circumvents the purification of thousands of human and pathogen proteins, and does not require antibodies against or pre-labeling of query proteins. This system is amenable to high-throughput analysis of effectors from a wide variety of human pathogens that may bind to and/or post-translationally modify targets within the human proteome.
Keywords: Nucleic acid programmable protein array (NAPPA), interactome, LidA, SidM, Rab1, AMPylation
INTRODUCTION
Protein molecules orchestrate cellular processes through intricate networks of binding and manipulation events. The study of protein-protein interactions (PPIs) is integral to understanding their molecular mechanism, which in turn guides the development of more efficient therapeutics aimed at the treatment of human diseases caused by protein malfunctions or by pathogens during infection.
The Gram-negative bacterium Legionella pneumophila is the causative agent of Legionnaire’s pneumonia. To survive within alveolar macrophages, the bacterium injects nearly 300 effector proteins directly into the host cell (1, 2). Most L. pneumophila effectors lack significant homology to known proteins, and their biological functions and host cell targets remain unknown. They are, however, key to L. pneumophila virulence and without them L. pneumophila is unable to establish a replication vacuole within the host cell and to persist while utilizing host cell nutrients and membrane components (3).
Small guanine nucleotide binding proteins (GTPases) of the Rab family are key regulators of membrane trafficking in eukaryotic cells and, not surprisingly, the target of some L. pneumophila effector proteins (4–6). LidA, for instance, binds Rab1, a GTPase involved in endoplasmic reticulum (ER) to Golgi membrane trafficking, and assists in the recruitment of ER-derived membranes to the L. pneumophila-containing vacuole (LCV) (7). The effector SidM recruits Rab1 to the LCV, activates it through guanosine diphosphate/triphosphate (GDP/GTP) exchange, and “locks” it in the active conformation via AMPylation (adenylylation), the covalent attachment of adenosine monophosphate (AMP) (7–9). Thus, through the combined efforts of LidA and SidM, L. pneumophila can efficiently exploit Rab1-regulated early secretory vesicle trafficking, thereby promoting its intracellular survival and replication.
Progress towards identifying host-pathogen interactions important for infection by L. pneumophila has been slow mainly due to a lack of screening approaches suitable for the systematic analysis of such a vast number of bacterial effector proteins. Earlier studies that identified human protein targets for L. pneumophila effectors relied primarily on co-precipitation assays, yeast two-hybrid, or gain/loss-of-function studies (7, 8, 10–15). Consequently, we sought to establish a more comprehensive screening approach to efficiently and reliably determine molecular targets of L. pneumophila effectors that are relevant for infection in humans.
Protein microarrays provide a valuable tool for measuring PPIs on a proteomic scale. The fabrication of high quality protein microarrays, however, has its challenges, namely, the need to produce and purify thousands of proteins with good yield and purity. In addition, maintaining protein stability after printing and during storage is a major concern. Previous protein array-based PPI studies required either the covalent labeling of purified query proteins with a fluorophore or the use of anti-tag or protein-specific antibodies which can introduce false negative (because the antibody fails to bind due to steric hindrance within a protein complex) or false positive results (because of non-specific binding) (16). Covalent labeling of proteins adds the concern of protein denaturation and/or biochemical property changes caused by protein purification, storage, or fluorophore cross-linking.
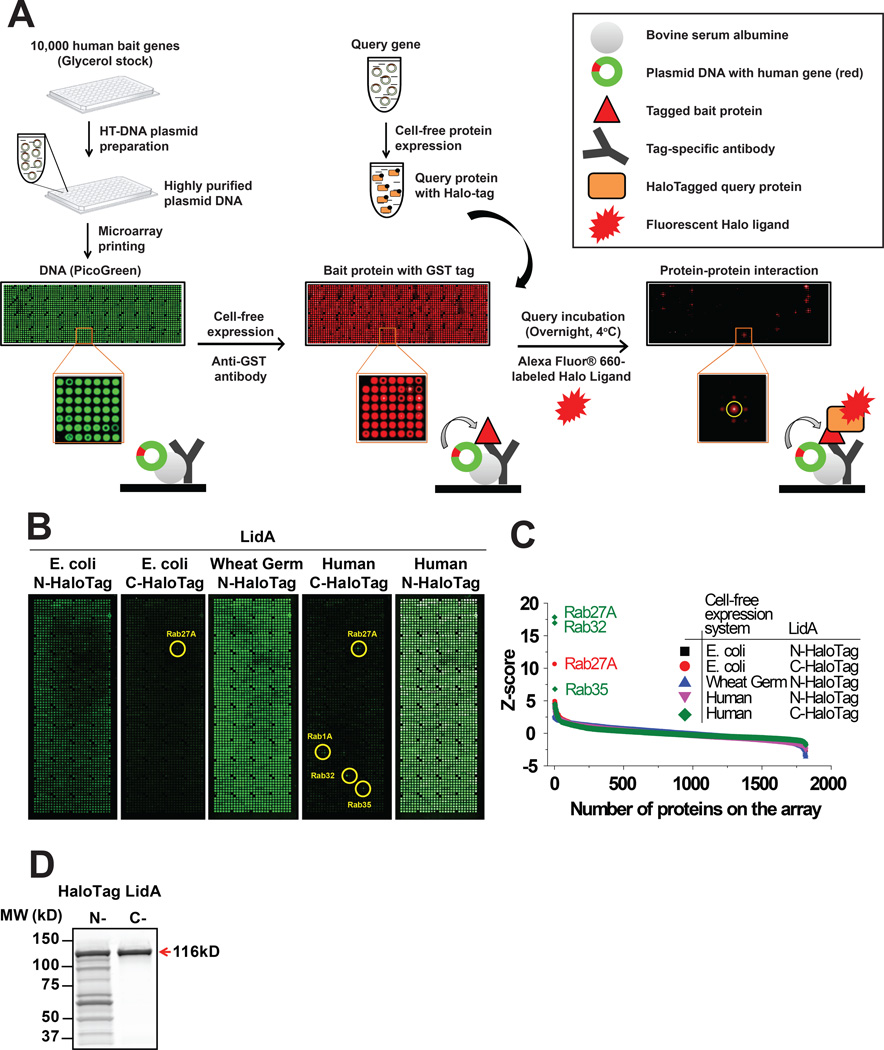
To address these issues, we looked to Nucleic Acid-Programmable Protein Arrays (NAPPA), where thousands of unique genes encoding proteins of interest are printed on an aminosilane-coated slide. Proteins are then freshly synthesized at the time of assay through in vitro transcription/translation (IVTT) and displayed in situ using co-spotted anti-tag antibodies (Figure 1) (17, 18). In the present approach, instead of using a tag that requires detection by anti-tag antibodies, we introduced the HaloTag (Promega) at the C-terminus of the bacterial query protein. HaloTag is a modified haloalkane dehalogenase designed to covalently bind to synthetic Halo-ligands (haloalkanes) (19). Once applied to NAPPA, binding of HaloTag query protein to its interactor(s) can be specifically detected among thousands of proteins using an Alexa660-labeled Halo-ligand (Figure 1) (20).
Figure 1.
Optimization of the high-throughput NAPPA interaction assay for L. pneumophila effectors. (A) Flow scheme of Nucleic Acid Programmable Protein Array (NAPPA) fabrication and protein interaction assay. Plasmid cDNA of ~10,000 human genes was printed on aminosilane-coated slides at a density of ~2,000 genes per slide. DNA immobilization was validated with PicoGreen staining (green); display of recombinant tagged bait proteins was examined with appropriate anti-tag antibody (red) after IVTT. Binding of HaloTag-query protein to its interactor on NAPPA was detected using Alexa660-labeled Halo-ligand. (B) Comparison of protein interaction assay using the query protein LidA produced from E. coli-, wheat germ-, and human HeLa cell-based IVTT systems as well as N-terminal and C-terminal HaloTag constructs. Circles indicate the location of protein spots with enhanced signal. (C) Quantitative comparison of interactors for LidA sorted by their Z-score calculated from (B). (D) In-gel fluorescence analysis of in vitro translated LidA with N-terminal and C-terminal HaloTag.
First, we systematically tested the influence of different IVTT systems (bacterial, wheat germ and human) and the location of HaloTag in recombinant query proteins on PPI screening assays. We then screened ~10,000 human proteins for both known and novel interaction partners of the two L. pneumophila effectors LidA and SidM. We specifically asked i) whether our approach can recapitulate the proteins identified to date as targets, and ii) whether those are indeed representative of the major protein families targeted by these effectors, or if there are additional proteins or protein families that have been missed. In addition, we probed the array for host substrates of SidM-mediated AMPylation by combining NAPPA with a Click chemistry-based nonradioactive AMPylation assay (21). Our findings add to our understanding of L. pneumophila pathogenesis and demonstrate the versatility and potential of our improved NAPPA platform for host-pathogen interactome mapping.
MATERIALS AND METHODS
Strains and plasmids
The strains and plasmids used in this study are listed in Table 1. L. pneumophila strains were grown and maintained as described (22, 23). L. pneumophila strains Lp02 (thyA hsdR rpsL) and Lp03 (Lp02 dotA3 (T4SS-)) are thymidine-auxotroph derivatives of Philadelphia-1 (24, 25). Halo-LidA and SidM were constructed using the Flexi vector system (Promega). A DNA fragment containing the lidA open reading frame (lpg0940) was amplified by PCR using the forward primer MMO63 containing a SgfI restriction site (5’-ACTTGCGATCGCCATGGCAAAAGATAACAAATC-3’) and the reverse primer MMO64 containing a PmeI restriction site (5’-AATCGTTTAAACTGATGTCTTGAATGGAGATAAAG-3’). The sidM open reading frame (lpg2464) was similarly PCR-amplified using forward primer MMO490 containing an SgfI site (5'-CTAAGCGATCGCCATGAGCATAATGGGGAGAAT-3') and reverse primer MMO491 containing a PmeI site (5'- ACAGGTTTAAACTTTTATCTTAATGGTTTGTCTTTC-3'). PCR fragments were digested with the specified enzymes and cloned into pFN22k or pFC15k (Promega), encoding an N-terminal or C-terminal HaloTag, respectively. To generate the previously described inactive mutant of SidM (9), the aspartates at positions 110 and 112 were substituted with alanine using site-directed mutagenesis (26). Plasmid pGEX6p1-sidM was used as a template for PCR amplification with mutation-directing primers MMO325 (5'-GCCACTGAGTATAGTGCGCTAGCTGCCTTTGTTATTGTT-3') and MMO326 (5'-AACAATAACAAAGGCAGCTAGCGCACTATACTCAGTGGC-3') to generate plasmid pGEX6p1-sidMD110/112A. These primers were also used to generate pmCherry-C1-sidMD110/112A by site-directed mutagenesis. Plasmids for production and purification of recombinant His-tagged LidA and SidM in Escherichia coli were constructed by recombining pDONR221 plasmids containing the lidA or sidM open reading frame (gift of R. Isberg, Tufts University) with pDEST17 (Life Technologies), encoding an N-terminal His6 fusion, using LR clonase (Life Technologies). Plasmids for production of GFP-tagged Rab1B, Rab8B, Rab10, Rab27A, and OCEL1 were constructed by recombining pDONR221 plasmids containing each individual gene (DNASU DNA plasmid repository, http://dnasu.org) with pcDNA6.2/N-EmGFP-DEST (Life Technologies) using LR Clonase (Life Technologies). For the expression of mCherry-tagged LidA, a DNA fragment containing the lidA open reading frame was amplified by PCR using the forward primer MMO660 containing an EcoRI restriction site (5’-CGGAATTCTATGGCAAAAGATAAC-3’) and the reverse primer MMO642 containing a BamHI restriction site (5’-CGGTGGATCCCGTGATGTCTTGAATGG-3’). The PCR fragment was digested with the specified enzymes and cloned into pmCherry-C1 at the respective cut sites to generate an N-terminal mCherry fusion.
Table 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| E. coli strains | ||
| GC5 | F- Φ80lacZM15 (lacZYA-argF)U169 recA1 endA1 hsdR1 7(rK mK+) phoA supE44 thi-1 gyrA96 relA1 tonA |
Genesee |
| BL21(DE3) | FompT hsdSB(rB mB) gal dcm (DE3) | Novagen |
| L. pneumophila strains | ||
| Lp02 | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph; Smr |
(24) |
| Lp03 | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph, lacking dotA3; Smr |
(27) |
| Plasmids | ||
| pCPDnHalo |
E.coli cell-free expression vector; encodes HaloTag alone; Ampr |
This study |
| pJFT6_nHalo | Wheat germ cell-free expression vector for N-temrinal HaloTag fusion; Ampr |
This study |
| pJFT7_nHalo | Mammalian cell-free expression vector for N-terminal HaloTag fusion; Ampr |
(28) |
| pJFT7_cHalo | Mammalian cell-free expression vector for C-terminal HaloTag fusion; Ampr |
(28) |
| pANT7_cGST | T7-based mammalian expression vector; encodes C-terminal GST tag; Ampr |
(29) |
| pLDNT7_nFLAG | T7-based mammalian expression vector; encodes N-terminal FLAG tag; Ampr |
(30) |
| pFN22k–lidA | Encodes LidA with N-terminal HaloTag; Kanr | This study |
| pFC15k–lidA | Encodes LidA with C-terminal HaloTag; Kanr | This study |
| pFN22k–sidM | Encodes SidM with N-terminal HaloTag; Kanr | This study |
| pFC15k–sidM | Encodes SidM with C-terminal HaloTag; Kanr | This study |
| pGEX-6p-1-sidM | Encodes SidM with N-terminal GST tag; Ampr | (7) |
| pGEX-6p1-sidMD110/ 112A | Encodes SidMD110/112A with N-terminal GST tag; Ampr | This study |
| pDEST17-lidA | Encodes His6-LidA; Ampr | This study |
| pDEST17-sidM | Encodes His6-SidM; Ampr | This study |
| pcDNA6.2/N-EmGFP- DEST |
Encodes GFP; Camr, Ampr | Life Technologies |
| pcDNA6.2/N-EmGFP- DEST-rab1B |
Encodes GFP- Rab1B (human); Ampr | This study |
| pcDNA6.2/N-EmGFP- DEST-rab8B |
Encodes GFP- Rab8B (human); Ampr | This study |
| pcDNA6.2/N-EmGFP- DEST-rab10 |
Encodes GFP- Rab10 (human); Ampr | This study |
| pcDNA6.2/N-EmGFP- DEST-rab27A |
Encodes GFP- Rab27A (human); Ampr | This study |
| pcDNA6.2/N-EmGFP- DEST-ocel1 |
Encodes GFP- OCEL1 (human); Ampr | This study |
| pmCherry-C1 | Encodes mCherry; Kanr | Clontech |
| pmCherry-lidA | Encodes mCherry-LidA; Kanr | This study |
| pmCherry-sidMD110/112A | Encodes mCherry-SidMD110/112A; Kanr | This study |
Abbreviations: Smr, streptomycin resistance; Kanr, kanamycin resistance; Ampr, ampicillin resistance; Camr, chloramphenicol resistance.
Production and purification of recombinant proteins
Proteins used in NAPPA PPI assays were produced as described below. For NAPPA AMPylation assays, untagged SidM and SidMD110/112A were produced and purified as previously described (31). Briefly, protein production in E. coli BL21 (DE3) was induced with 0.2 mM isopropyl-β-dithiogalactopyranoside (IPTG) at 20°C overnight. GST-SidM was purified from the soluble fraction of the bacterial lysate using Glutathione Sepharose 4B slurry (GE Healthcare) pre-equilibrated in PBS supplemented with 1 mM MgCl2 and 1 mM β-mercaptoethanol (β-ME). Protein-bound beads were washed with the equilibration buffer and incubated overnight with PreScission protease (GE Healthcare) to remove the GST tag.
Fabrication of high-density NAPPA human arrays
All 10,000 sequence-verified, full-length human ORFs in T7-based mammalian expression vectors (pANT7-cGST and pLDNT7_nFLAG) were obtained from DNASU. To determine the quality of NAPPA arrays, the printed plasmid cDNA was stained with PicoGreen and the expression and display of human proteins in situ was examined using human HeLa lysate-based cell-free expression system (Thermo Fisher Scientific) and monoclonal mouse anti-GST antibody (1:200 dilution) (Sigma Aldrich) (32).
Protein-protein interaction assays using NAPPA and magnetic beads
The NAPPA array was blocked for 1 hr at 23°C using Superblock solution (Pierce), and then covered with a hybridization chamber (Grace Bio-Labs, Inc.). Protein expression was performed by injection of 160 µL human cell-free expression mix into the chamber and incubation for 1.5 hr at 30°C and 0.5 hr at 15°C. After washing three times with PBST (PBS, 0.2% Tween), the resulting NAPPA was further blocked with PPI blocking buffer (1×PBS, 1% Tween 20 and 1% BSA, pH7.4) for 2 hr at 4°C. In parallel, LidA and SidM proteins were produced by incubation of 9 µg of DNA in 180 µL human cell-free expression system for 2 hr at 30°C. The NAPPA was incubated with unpurified LidA or SidM in human HeLa lysate for 16 hr at 4°C. The unbound molecules were washed away using PPI washing buffer (PBS, 5mM MgCl2, 0.5% Tween20, 1% BSA and 0.5% DTT, pH7.4) three times. Detection was executed using 12.5 µM Alexa660-conjugated Halo-ligand (Promega) by incubation for 2 hr at 4°C. After washing three times, the array was dried with brief centrifugation at 1,000 rpm for 1 min, and scanned using a Tecan PowerScanner. The fluorescent signal was quantified using Array-Pro Analyzer (Media Cybernetics).
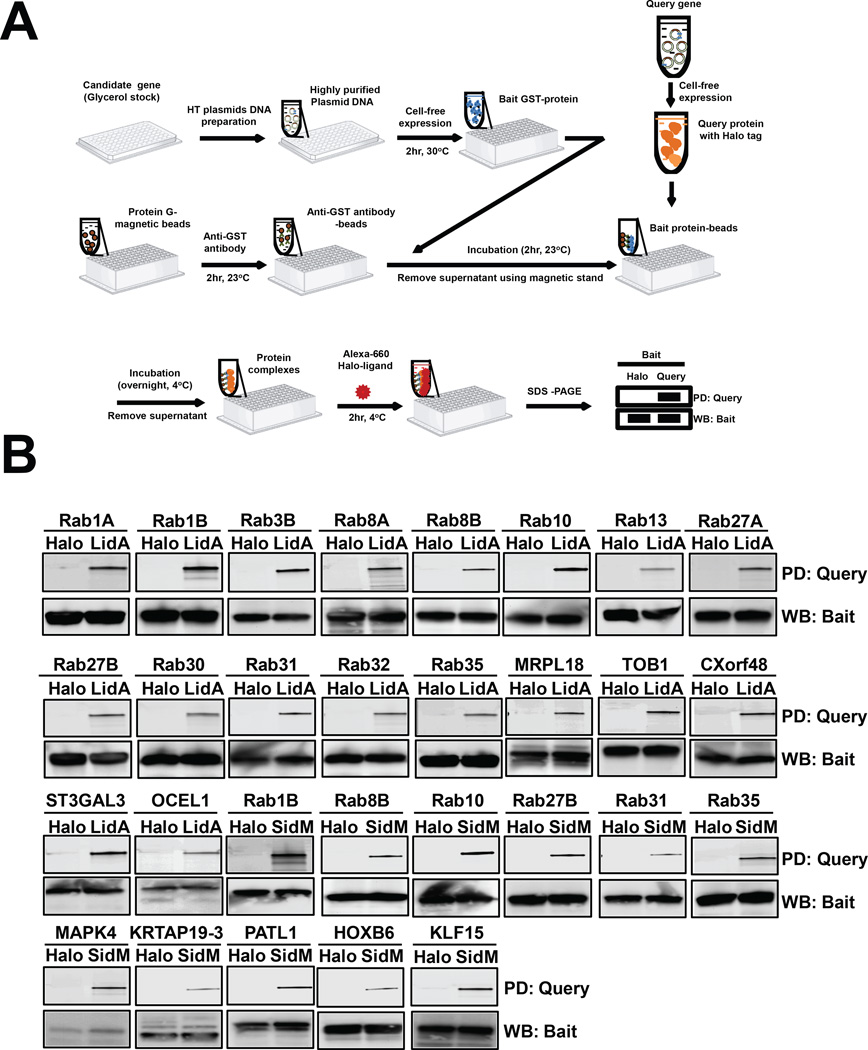
A schematic illustration of the bead-based pull-down assay is shown in Figure 3A. The query protein (LidA or SidM) containing a HaloTag was prepared as described above. To perform the pull-down assay, 25 µL of query HaloTagged proteins in HeLa lysate were added to a 96-well plate containing the bait protein-coated beads and incubated at 4°C overnight in an Eppendorf Thermomixer® R mixer incubator at 1400 rpm. After washing with PPI washing buffer, detection was performed by adding 25 µL of Alexa660-labeled Halo-ligand with incubation for 2 hr at 4°C. After that, the beads with protein complexes were washed three times with PBST and the supernatant was removed. To elute the query and bait proteins from the beads, 20 µL 1×SDS loading buffer containing 10% 2-mercaptoethanol was injected into each well of 96-well plate and treated for 5 min at 95 °C. Then the protein was separated on a 4–15% Tris-Criterion™ Precast Gel (Bio-rad) at 200 V for 35 min. After washing three times with deionized water, the protein gels were scanned in an Amersham Bioscience Typhoon 9400 variable mode imager at 635 nm.
Figure 3.
Confirmation of target candidates using in vitro bead-based pull-down assay. (A) Flow scheme of the high throughput pulldown assay. (B) Immunoblot analysis of the pulldown. Top panel (pulldown, PD) shows that the query proteins (HaloTag-LidA or -SidM), but not the Tag, were precipitated by GST-bait-coated beads. Bottom panel (western blot, WB) shows detection of the GST-bait protein by immunoblot using anti-GST antibody.
Subsequent to in-gel fluorescence scanning, the expressed GST-protein in SDS-PAGE gel was examined by western blot. Briefly, the protein-in-gel was transferred to a nitrocellulose membrane using Trans-Blot® SD Semi-Dry Transfer Cell (Bio-rad) at 20 V for 1 hr. After blocking with 5% milk for 1 hr, the membrane was incubated with 1:2000 dilution of mouse monoclonal anti-GST antibody overnight. After washing with PBST, the membrane was incubated with 1:5000 dilution of HRP conjugated sheep anti-mouse secondary antibody for 1 hr at room temperature. The detection was performed using chemiluminescence with SuperSignal West Femto Luminol/Enhancer Solution (Pierce).
Nucleotide exchange assay
Following production by IVTT, the NAPPA GTPase protein array was treated with Tris-EDTA buffer (50mM Tris pH 7.5, 50mM NaCl, 0.1mM DTT, 1% BSA, 5mM EDTA) for 1 hr at room temperature, with the aim of removing GDP and GTP contributed by the HeLa cell-based IVTT system during protein synthesis. After that, the solution was removed by centrifugation of the slide for 2 min at 1,000rpm at 4 °C. The array was then covered with a sixteen-well chamber using Whatman Chip Clip™ holder (GE Healthcare). Each well received either 60µl 1mM GDP or GTPγS, or buffer (50mM Tris pH 7.5, 50mM NaCl, 0.1mM DTT, 5mM EDTA) for the nucleotide-free GTPase proteins. The loading of nucleotides was executed for 2 hr at room temperature. The reaction was stopped using PPI blocking buffer containing 5mM MgCl2. Nucleotide loading was confirmed using a fluorescence competition assay in which 1µM FL-GDP or FL-GTP was mixed with 1mM GDP or GTPγS and loaded to respective arrays.
AMPylation assay using NAPPA and magnetic beads
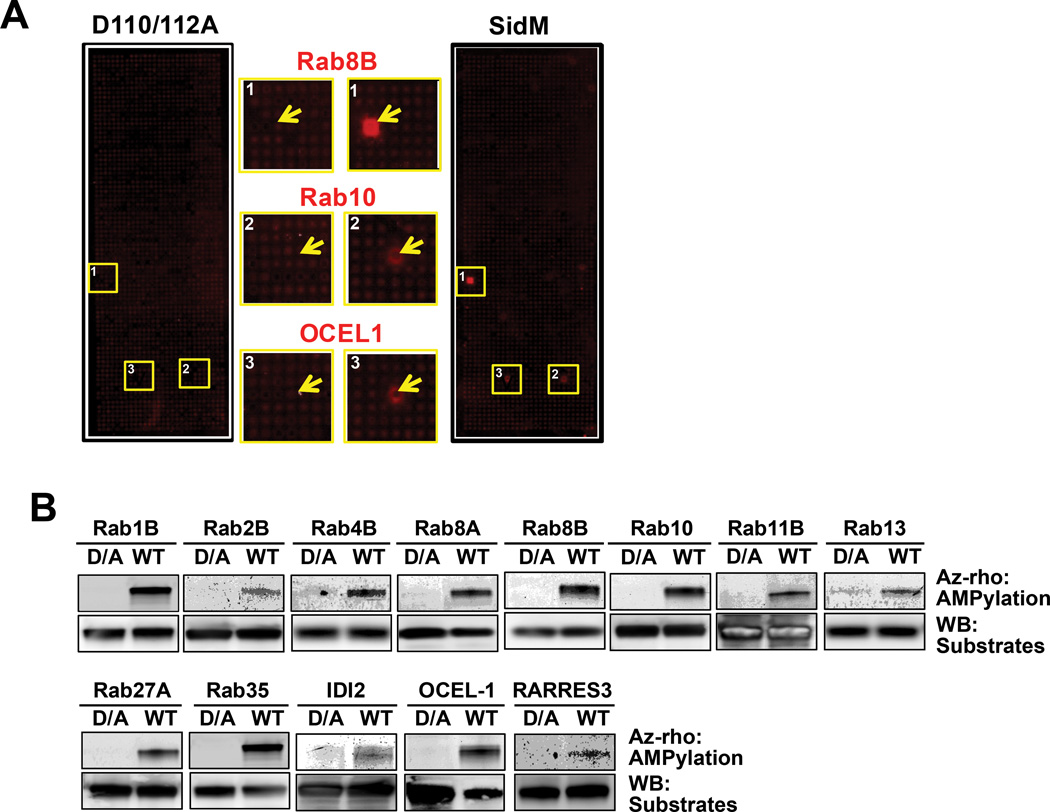
To detect potential substrates of SidM by AMPylation, the NAPPA array was incubated with 100 µg/mL purified SidM or SidMD110/112A and 250 µM N6pATP in 160 µL AMPylation solution (20 mM HEPES pH 7.4, 100 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA, 1 mM DTT) for 2 hr at 23°C. After washing with PBST three times, the detection was executed using 160 µL click reagents (250 µM az-rho, 1 mM TCEP, 0.1 mM TBTA and 1 mM CυSO4). Finally, after washing three times with PBST, the array was scanned and the signal was quantified using the same procedure as described above. Bead-based AMPylation assays were performed using Protein G-coated Dynabeads (Life Technologies), prepared as described above. Substrate protein-coupled beads were incubated with 100 µg/mL SidM or SidMD110/112A and 250 µM N6pATP in 15 µL AMPylation solution (20 mM HEPES pH 7.4, 100 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA, 1 mM DTT) for 2 hr at 23°C. After washing three times with PBST, the detection was performed with the incubation of 20 µL click reagents (250 µM az-rho, 1 mM TCEP, 0.1 mM TBTA and 1 mM CuSO4) for 1 hr at 23°C. The reaction was stopped with the addition of 20 µl 1×SDS loading buffer containing 10% 2-mercaptoethanol. Samples were boiled 5 min and analyzed in a 4–15% Tris-Criterion™ Precast Gel (Bio-rad). The protein gel was scanned and the image was obtained using the same methods as in the bead-based pull-down assay. Subsequent to the AMPylation assay, the substrate protein on the beads was further examined using western blot, as in the bead-based pull-down assay.
Selection of host targets for L. pneumophila by physical interactions and AMPylation
Before statistical analysis, we examined the spot shape, dust and non-specific binding on the microarray images to remove any possible false-positive signals. We then normalized the raw signal intensity to decrease the background variations from slide to slide. The normalization step was executed by subtracting the background caused by the non-specific binding of probes, which was estimated by the first quartile of the printing buffer only as control. The normalized value was calculated by using the signal of each feature divided by the median background-adjusted value of all proteins on the array. Then the Z-score was calculated using the normalized value. The host target candidates were selected based on the following criteria. (1) Z-score greater than or equal to 3; (2) For physical interaction, Z-score ratio of query (LidA and SidM) to the negative control (HaloTag) higher than 1.5; for AMPylation screening, Z-score ratio of wild-type SidM to its inactive mutant (SidMD110/112A) higher than 1.5. (3) The targets have to meet the previous criteria in two independent experiments.
In addition, we also selected some potential candidates based on visual inspection of the luminous radiation (“ring”) around the microarray spot after incubation with the query proteins; LidA and SidM, in PPI, and SidM, in AMPylation assay. The display of a ring is caused by stacking of host protein to the neighboring areas of the expression spot during expression and capture, which can also bind to the query proteins in PPI or be modified by SidM through AMPylation. Previous work showed that these rings are best identified using the Array-Pro Analyzer software by adjusting the contrast of microarray image, especially when the Z-score is low (33).
Co-localization assays and fluorescence microscopy
To study protein co-localization, COS-1 cells were transiently co-transfected with pcDNA6.2/N-EmGFP and pmCherry constructs (Table 1) using Lipofectamine 2000 Transfection Reagent (Life Technologies). At 6 hr (for LidA constructs) or 16 hr (for SidM constructs) post-transfection, cells were fixed in PBS with 4% paraformaldehyde for 20 min at 23°C. Coverslips were mounted using Anti-fade reagent (Life Technologies). Cells were imaged using a 100x objective lens, AxioObserver.Z1 fluorescence microscope, and AxioVision 4.7 software (Zeiss).
To study target localization following L. pneumophila infection, CHO FcγRII cells (34) were plated in a 24-well plate and transfected the next day with GFP-tagged target plasmids as described above. The following day, Lp02 or Lp03 bacteria were opsonized by incubating for 30 min at 37°C with 1:500 dilution of anti-L. pneumophila rabbit antibodies (GenScript). Opsonized bacteria (1.5×106 CFU) were added to each well containing 1×105 cells (MOI=15), and the plates were centrifuged at 200×g for 5 min at 37°C. After 30 min in a CO2 incubator, the cells were washed and growth media was replaced with DMEM containing 0.83 mM thymidine to synchronize the infection and supplement the L. pneumophila auxotrophs. Plates were returned to the incubator for an additional 1 hr, 5.5 hr, or 9.5 hr, at which point cells were washed three times with cold PBS and fixed as above. Extracellular bacteria were stained with anti-L. pneumophila rat antibodies (1:3000) followed by anti-rat IgG complexed to AlexaFluor350 (1:1000) (Life Technologies). Cells were permeabilized by washing in ice-cold methanol for 10 sec, then washed three times in PBS. Intracellular bacteria were stained with anti-L. pneumophila rat antibodies (1:3000) followed by anti-rat IgG complexed to Texas Red fluorescent dye (1:3000) (Life Technologies). Microscopy was performed as described above.
To generate line scans, measurements of pixel intensity across the indicated line were taken using the Plot Profile tool in Image J (35).
Co-precipitation from transfected cell lysate
293T cells were transfected with plasmids encoding GFP or GFP-tagged Rab1B, Rab8B, Rab10, Rab27A, or OCEL1 using Lipofectamine 2000 Transfection Reagent (Life Technologies) according to the manufacturer’s instructions. At 24 hr post-transfection, cells were harvested and cell pellets frozen at −80°C. Cells were subsequently lysed by resuspending in Lysis Buffer (PBS, 5mM DTT, 1x protease inhibitor cocktail (Roche)) and forcing through a 20-gauge needle ten times while on ice. Lysates were spun twice at 7000 rpm for 10 min at 4°C to pellet cell debris, yielding post-nuclear supernatant (PNS). Dynabeads Epoxy M270 magnetic beads (Life Technologies) (5 µg) were prepared according to manufacturer’s instructions, and incubated with 100 µg purified LidA or commercial BSA (Sigma) overnight at 4°C with constant mixing. To perform the co-precipitation, 150 µl PNS (equivalent to one-fifth of a 10cm2 dish of confluent cells) were incubated with bait-coated beads for 2 hr at 23°C with constant mixing. Following three washes in PBS, proteins were eluted from beads by boiling 10 min at 99°C. Elutions and input samples were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti-GFP antibody (1:2000) (Life Technologies). Densitometry was performed using a quantitative scanner and QuantityOne software (both from BioRad).
RESULTS
Rationale for choosing L. pneumophila SidM and LidA
From the approximately 300 effector proteins that are translocated into human macrophages upon L. pneumophila infection, we selected two that were well characterized, SidM and LidA, as test cases in our two-pronged approach (binding and post-translational modification). Previous work had identified multiple human Rab GTPases, including Rab1, as binding partners or enzymatic targets for SidM and LidA. By characterizing these effectors with our NAPPA approach, we sought to establish proof-of-concept for our technique and reveal whether additional targets for these effectors exist. We intentionally chose one tight-binding protein (LidA) (36) and one intermediate-strength binding protein (SidM) (37) to evaluate the sensitivity of our assay and its applicability for the general study of microbial effectors with a wide variety of target affinities. Finally, the fact that SidM is an AMPylase allowed us to develop a catalysis-based assay where potential host targets are identified based on their post-translational modification with AMP rather than physical binding to the AMPylator. Notably, although the study presented here was focused on two previously characterized proteins as queries, we also succeeded in identifying novel interaction partners for less well-characterized L. pneumophila effectors (data not shown), demonstrating that the NAPPA strategy does not require any previous knowledge of the query protein or its localization or function in the cell.
Development of a high-throughput interaction assay for L. pneumophila effectors
We first evaluated the performance of two expression constructs (N-terminal and C-terminal HaloTag) and three cell-free IVTT systems (E. coli, wheat germ, and human HeLa cell extract) in the identification of known interaction partners for LidA on a partial NAPPA containing ~2,000 unique GST-tagged human proteins (Figure 1B). Whereas N-terminal HaloTag-LidA showed high background fluorescence across the array regardless of the IVTT system used, C-terminal HaloTag-LidA gave a specific fluorescent signal for one GTPase (Rab27A) using the E. coli IVTT system and four GTPases (Rab27A, Rab1A, Rab32 and Rab35) using the HeLa IVTT system (Figure 1B, C).
We synthesized N- or C-terminal HaloTag-LidA using the HeLa IVTT system and examined the protein products using in-gel fluorescence (Figure 1D). We found several lower molecular weight products in the LidA sample with N-terminal HaloTag that were not present in the sample of C-terminal HaloTag-LidA, suggesting that the N-terminal HaloTag was unstable and prone to proteolytic degradation. As the same result was observed for HaloTagged SidM (Supplementary Figure S1), all subsequent studies were performed with the C-terminally tagged effectors in order to prevent the smaller fragments from interfering with the interactions.
Finally, we compared the results obtained in two independent NAPPA assays performed on different days. We found that the same host targets were detected in each experiment (Supplementary Figure S2). Together, these results indicated that we had successfully validated the sensitivity and reproducibility of our NAPPA-based interaction assay through a combination of C-terminal HaloTag query proteins and the use of a human IVTT system.
NAPPA-based identification of host binding partners for L. pneumophila SidM and LidA
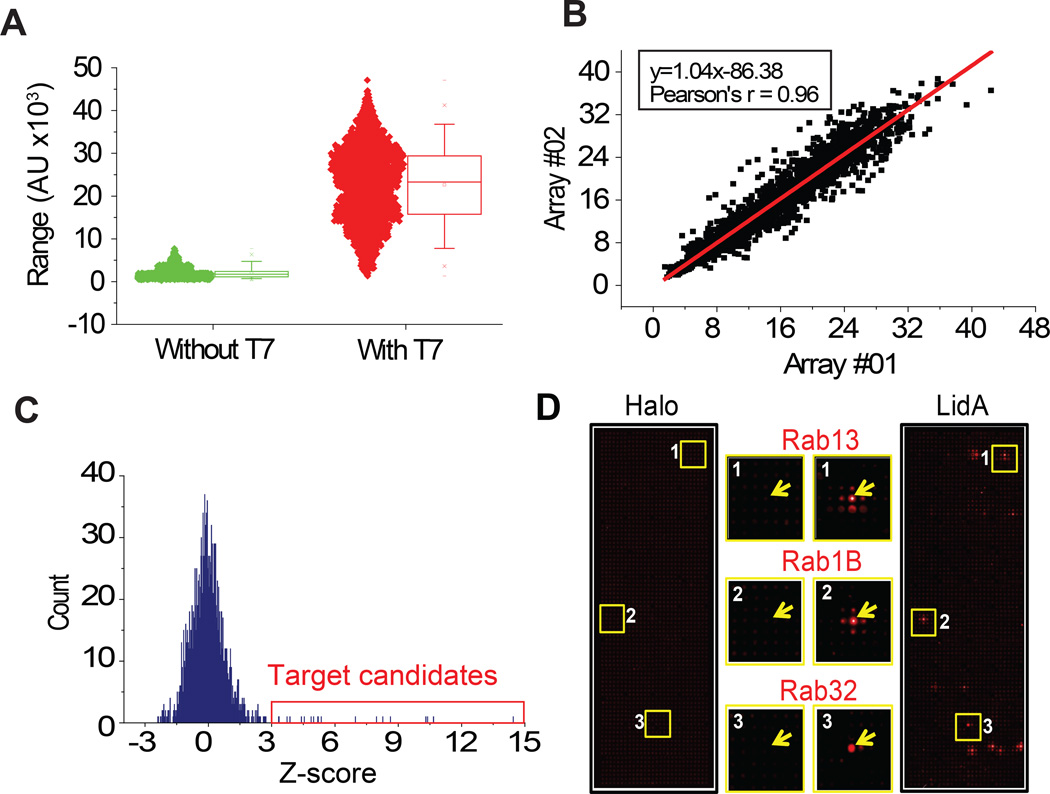
To screen for interactions between each of the two L. pneumophila effectors and the human proteome, we generated a set of NAPPA slides composed of 10,000 unique human proteins. We examined the quality of our arrays before and after IVTT (Figure 2A; Supplementary Figure S3) and confirmed the reproducibility of bait protein capture (Figure 2B). SidM and LidA were separately produced in a human HeLa cell lysate-based IVTT system as C-terminal HaloTag fusion proteins and allowed to bind to their respective targets on NAPPA during an overnight incubation. Bound effectors were detected using Alexa660-labeled Halo-ligand, and host target candidates were selected based on one of two criteria: a Z-score ≥3 (Figure 2C), or a visible luminous radiation (“ring”) surrounding a protein spot (Figure 2D).
Figure 2.
Identification of host targets for L. pneumophila effectors by physical interaction in NAPPA. (A) Protein expression and display on NAPPA with (red) and without (green) T7 polymerase. (B) Correlation of protein expression and display on two identical NAPPA arrays. (C) Selection of host target candidates for L. pneumophila effectors using a Z-score threshold of 3. (D) Representative images of NAPPA slides probed with HaloTag (control) or HaloTag-LidA. Yellow boxes are shown enlarged in center panels, and regions of interest are marked with an arrow.
We identified a total of 20 potential binding partners for LidA, and 18 potential binding partners for SidM (Table 2). Six of the 20 LidA targets (30%) had been previously reported from biochemical or structural analyses (Rab1A, Rab1B, Rab4B, Rab8A, Rab8B, Rab31) (7, 38), whereas only one (Rab1B) of the 18 potential binding partners for SidM (6%) had been previously reported (7) (Table 2). Notably, Rab GTPases comprised the largest group of targets for both LidA (14/20 or 70%) and SidM (7/18 or 39%), which is not surprising given their high sequence and structural homology to the preferred target Rab1. NAPPA successfully detected the LidA-Rab1B and SidM-Rab1B interactions, both of which were previously reported, thus demonstrating proof-of-concept for the HaloTag as a novel reporter and for L. pneumophila effectors as queries in the PPI screen. Interestingly, one of the non-Rab targets identified as a ligand for both effectors was OCEL1 (Occludin/ELL domain-containing protein 1), a protein that also emerged as a possible target from the NAPPA-based AMPylation screen (see below). Given that no specific signals were observed in the control NAPPA probed with HaloTag alone (Figure 2D), we concluded that SidM and LidA binding to their respective host targets was direct and specific.
Table 2.
Potential physical binding targets for LidA and SidM
| Symbol | DNASU Gene ID |
Description | NAPPA Z-score | Pull down |
Previously reported |
|
|---|---|---|---|---|---|---|
| HaloTag | Query | |||||
| For L. pneumophila LidA | ||||||
| Rab1A | 5861 | Ras-related protein Rab-1A | 0.2 | 11.4 | + | (7) |
| Rab1B | 81876 | Ras-related protein Rab-1B | 0.0 | 20.6 | + | (7) |
| Rab3B | 5865 | Ras-related protein Rab-3B | 0.1 | 12.8 | + | |
| Rab4B | 53916 | Ras-related protein Rab-4B | −0.4 | 5.4 | - | Binding (38); No binding (7) |
| Rab8A | 4218 | Ras-related protein Rab-8A | 2.1 | 16.1 | + | (7) |
| Rab8B | 51762 | Ras-related protein Rab-8B | 0.1 | 31.0 | + | (37) |
| Rab10 | 10890 | Ras-related protein Rab-10 | / | Ring | + | |
| Rab13 | 5872 | Ras-related protein Rab-13 | 0.4 | 8.9 | + | |
| Rab27A | 5873 | Ras-related protein Rab-27A | −0.2 | 5.2 | + | No binding (38) |
| Rab27B | 80718 | Ras-related protein Rab-27B | / | Ring | + | |
| Rab30 | 27314 | Ras-related protein Rab-30 | / | Ring | + | |
| Rab31 | 11031 | Ras-related protein Rab-31 (aka Rab22) |
/ | Ring | + | (38) |
| Rab32 | 10981 | Ras-related protein Rab-32 | 0.5 | 7.9 | + | |
| Rab35 | 11021 | Ras-related protein Rab-35 | −0.1 | 17.6 | + | |
| CXorf48 | 54967 | Chromosome X, open reading frame 48 |
2.2 | 3.8 | + | |
| LCN2 | 3934 | Lipocalin 2 | / | Ring | + | |
| MRPL18 | 29074 | Mitochondrial ribosomal protein L18 |
/ | Ring | + | |
| OCEL1 | 79629 | Occludin/ELL-domain- containing protein 1 |
/ | Ring | + | |
| ST3GAL3 | 6487 | ST3 beta-galactoside alpha-2,3- sialyltransferase 3 |
/ | Ring | + | |
| TOB1 | 10140 | Transducer of ErbB2, 1 | / | Ring | + | |
| For L. pneumophila SidM | ||||||
| Rab1B | 81876 | Ras-related protein Rab-1B | / | Ring | + | (7) |
| Rab8B | 23584 | Ras-related protein Rab-8B | / | Ring | + | No binding (7) |
| Rab10 | 22541 | Ras-related protein Rab-10 | / | Ring | + | |
| Rab27A | 5873 | Ras-related protein Rab-27A | / | Ring | + | |
| Rab27B | 5874 | Ras-related protein Rab-27B | / | Ring | + | |
| Rab31 | 11031 | Ras-related protein Rab-31 (aka Rab22) |
/ | Ring | + | |
| Rab35 | 11021 | Ras-related protein Rab-35 | / | Ring | + | |
| KLF15 | 28999 | Kruppel-like factor 15 | / | Ring | + | |
| GANAB | 23193 | Glucosidase, alpha; neutral AB | 1.5 | 3.8 | + | |
| MAPK4 | 5596 | Mitogen-activated protein kinase 4 |
2.5 | 5.2 | + | |
| KRTAP19- 3 |
337970 | Keratin-associated protein 19–3 | / | Ring | + | |
| PATL1 | 219988 | Protein associated with topoisomerase II homolog 1 |
/ | Ring | + | |
| HOXB6 | 3216 | Homeobox B6 | / | Ring | + | |
| OCEL1 | 79629 | Occludin/ELL-domain- containing protein 1 |
/ | Ring | + | |
| POT1 | 25913 | Protection of telomeres 1 | / | Ring | - | |
| MRPL30 | 51263 | Mitochondrial ribosomal protein L30 |
/ | Ring | - | |
| MRPL35 | 51318 | Mitochondrial ribosomal protein L35 |
/ | Ring | - | |
| PARK7 | 11315 | Parkinson protein 7 | 0.9 | 5.4 | - | |
Abbreviations: SidM*, SidM(D110/112A); /, not listed because selection based on Ring; + or -, positive or negative result in bead-based AMPylation assay.
Since many of the targets are GTPases, we wanted to know how differential nucleotide binding might affect the results of our PPI screening technique. Therefore, we generated an array comprised only of Rab GTPases, stripped them of nucleotide using EDTA, then either left them unloaded, or loaded them with GDP or GTPγS. The resulting arrays were probed with LidA as in our PPI screen. The results indicate that the Rab proteins identified as hits in our initial screen were again identified here, and that the active form of each Rab, bound by GTPγS, typically saw either no or only a modest increase in interaction with LidA (Supplementary Figure S4). Only Rab32 was a visibly better ligand for LidA when present in the active state. Rab proteins stripped of nucleotide exhibited the lowest levels of interaction with LidA, as expected for these intrinsically unstable GTPases. Due to the physiological nature of the HeLa cell-based IVTT reagent it is most likely that our standard NAPPA slides include a mixture of GDP- and GTP-bound proteins capable of capturing ligands with binding preference for either of their activation states.
Confirmation of potential binding targets by bead-based pull-down assay
To confirm the identity of putative interaction targets identified by NAPPA, we developed an independent in vitro bead-based pull-down assay (Figure 3). HaloTag-LidA and -SidM and their human interaction candidates identified by NAPPA were produced using IVTT (Supplementary Figure S5), and protein binding to LidA- or SidM-coated beads was detected with Alexa660-labeled Halo-ligand using in-gel fluorescence. We confirmed interaction of most potential targets with LidA (19 of 20) and SidM (14 of 18) (Figure 3B and Table 2). For LidA, 13 out of the 19 remaining candidates (68%) were Rab GTPases; for SidM, 7 out of the 14 (50%) were Rab proteins. Thus, the bead-based pull-down assay confirmed a total of 76% of the targets for SidM and LidA that had emerged from NAPPA.
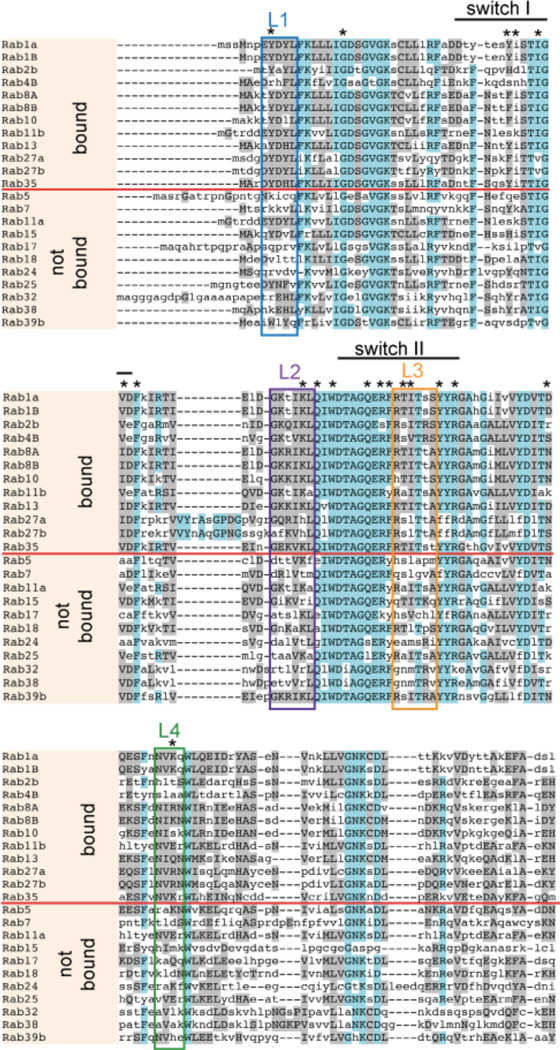
Identification of conserved regions within Rab proteins targeted by LidA
The fact that only a selected set of Rab GTPases were bound by LidA (and SidM) on the NAPPA inspired us to perform a comparative analysis of the amino acid sequences of binding vs. non-binding Rabs in order to identify potential specificity-determining regions. Rab proteins can exist in two conformations, an active GTP-bound form or an inactive GDP-loaded form. While LidA binding appears to be mostly nucleotide-independent, SidM has a strong preference for inactive Rabs (7, 8). Given that the nucleotide loading state of Rabs on the NAPPA is uncertain, we limited our in silico analysis to LidA binding vs. non-binding Rabs. We detected four clusters (L1 to L4) that were conserved among LidA-interacting Rab GTPases (namely Rab1A, 1B, 2B, 4B, 8A, 8B, 10, 13, 27A, 27b, 31, 35) but less so among Rabs that failed to bind LidA (Rab5, 7, 11, 15, 17, 18, 24, 25, 32, 38, 39B) (Figure 4). Notably, all four clusters were located in regions that in the LidA-Rab1 complex comprise the protein-protein interface (36, 38). Cluster L1 (residues 8-YDYL-11) contained Tyr8, which forms hydrogen bonds with the carbonyl oxygen of Met 414 of LidA (38), whereas cluster L2 (residues 57-GKTIKL-62) contained Lys61 which was shown to extensively interact with Ile413, Ser415, and Asn421 of LidA (Supplementary Figure S6). Both cluster L1 and L2 are located at the opposite end of the LidA-Rab1 interface relative to cluster L3 (residues 74-RTITSS-79) which contained Ile76 and Thr75, two residues whose equivalents in Rab8 were reported to be at the core of the LidA-Rab8 interaction (36). Cluster L4 (101-NVKQ-104) contained a lysine residue (Lys103) which forms a salt bridge with Asp289 of LidA. These results strongly suggest the existence of several specificity-determining regions that influence whether LidA recognizes a specific host cell Rab protein as target.
Figure 4.
Conserved regions within Rab proteins targeted by LidA. Sequence alignment of Rab GTPases was performed using the MUSCLE server (http://www.bioinformatics.nl/tools/muscle.html). Rabs that interact with LidA are separated by a red line from those that do not interact. Regions of high homology are shown in magenta; regions of limited homology are shown in grey. Clusters of enhanced conservation are labeled by boxes. * indicates amino acid residues known to be involved in LidA-Rab1 binding (23).
NAPPA-based detection of AMPylated host targets for SidM
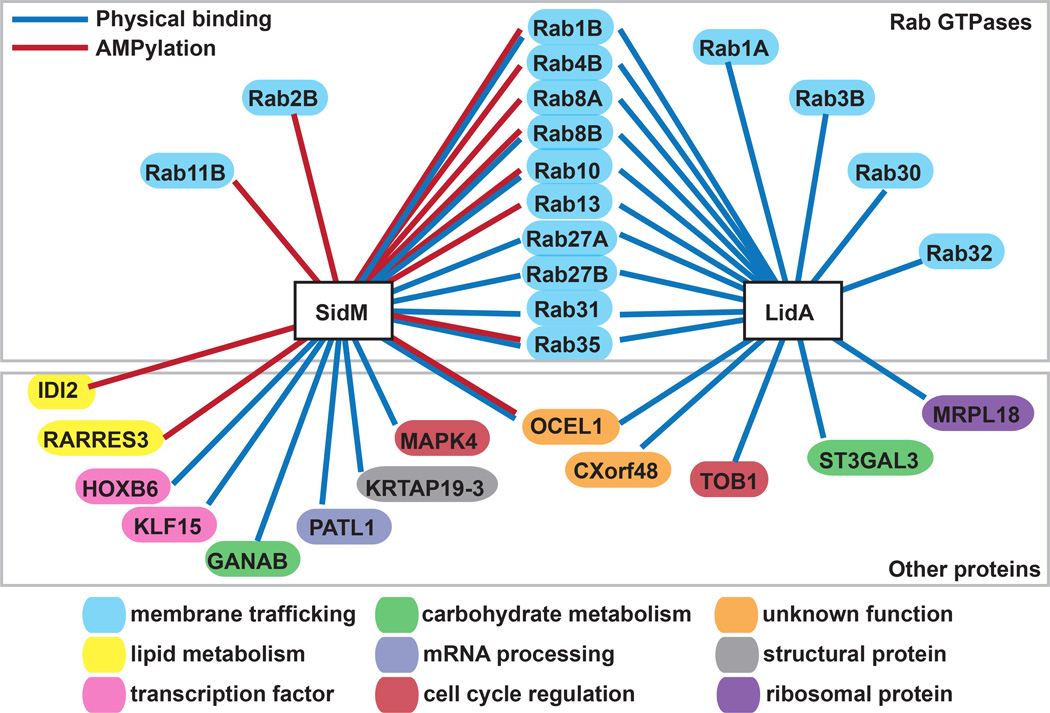
AMPylation has been recently discovered as a post-translational modification used by microbial pathogens to manipulate host protein function (9, 16, 39, 40). This activity often resides within the FIC (filamentation induced by cyclic AMP) domain, although it is important to point out that not all AMPylators possess a FIC domain (for example, SidM and GS-ATase possess a nucleotidyl transferase-like fold (9, 41)) and not all FIC domains catalyze AMPylation (some catalyze phosphocholine transfer (14)). Based on genome sequence annotation, the FIC domain is predicted to exist in more than 3,000 bacterial proteins (42), suggesting that AMPylation is widespread among bacteria. Despite their abundance, only a few FIC domain-containing proteins have been studied in detail, while the target(s) for the majority remain unknown due to the unavailability of an unbiased substrate-screening platform with high sensitivity and specificity (43–45). To address this issue, we recently developed a high-sensitivity AMPylation screening platform by combination of NAPPA and a non-radioactive click assay (also known as Copper Catalyzed Azide-Alkyne Cycloaddition (CuAAC)) in which proteins on NAPPA that are modified by an AMPylator with N6pAMP are detected using an azido-conjugated rhodamine through alkyl-azido-based click reaction (44). We recently applied this platform and successfully identified new targets for two bacterial FIC AMPylators, Vibrio parahaemolyticus VopS and Histophilus somni IbpAFic2, thus greatly expanding their repertoire of potential host substrates (21). In the present work, we extended our NAPPA platform to the study of an AMPylator (SidM) with an adenylyl transferase domain. We found that several human proteins, such as Rab8B, Rab10 and OCEL1, showed enhanced rhodamine fluorescence after incubation with SidM but not with the AMPylation-defective mutant SidMD110/112A (Supplementary Figure S6). In total, we identified 28 AMPylated targets, 12 of which (43%) were confirmed by an independent bead-based AMPylation assay (Figure 5B, Table 3). Of these 12, nine (75%) were members of the Rab GTPase family, suggesting that, indeed, Rab1 and some of its family members make up the majority of SidM targets in the host cell. We identified and confirmed AMPylation of the previously reported targets Rab1B, Rab4B, Rab8A, Rab13, and Rab35 (9, 31), and identified and confirmed eight previously unrecognized targets (Rab2B, Rab8B, Rab10, Rab11B, Rab27A, OCEL1, RARRES3, and IDI2). Notably, upon combining the targets from our two NAPPA approaches detecting physical binding or AMPylation, LidA and SidM shared a significant number of host targets, primarily Rab GTPases (Rab1B, Rab4B, Rab8A, Rab8B, Rab10, Rab13, Rab27A, Rab27B, Rab31, and Rab35) but also a non-Rab protein (OCEL1).
Figure 5.
Detection of AMPylated host targets by combining NAPPA and click chemistry. (A) Representative images of AMPylated targets (yellow boxes) on NAPPA slides after incubation with either wild-type SidM or its AMPylation-defective mutant SidMD110/112A, annotated as in Figure 2D. (B) Confirmation of target candidates using bead-based AMPylation assay with wild-type SidM (WT) or SidMD110/112A (D/A). Top panel (Az-rho) shows the detection of AMPylated targets using in-gel fluorescence with az-rho based on click reaction. Bottom panel (WB) is the detection of the host target protein (substrates) by immunoblot.
Table 3.
Potential host AMPylation substrates for SidM
| Symbol | DNASU Gene ID |
Description | NAPPA Z-score | AMPylation assay |
Previously reported |
|
|---|---|---|---|---|---|---|
| SidM* | SidM | |||||
| Rab1B | 391 | Ras-related protein Rab-1B | 0.5 | 10.2 | + | (9) |
| Rab2A | 5862 | Ras-related protein Rab-2A | / | Ring | - | No AMPylation (31) |
| Rab2B | 84932 | Ras-related protein Rab-2B | / | Ring | + | |
| Rab4B | 53916 | Ras-related protein Rab-4B | / | Ring | + | (9) |
| Rab8A | 4218 | Ras-related protein Rab-8A | 1.3 | 3.6 | + | AMPylation (9); No AMPylation (31) |
| Rab8B | 79165 | Ras-related protein Rab-8B | −0.8 | 21.9 | + | |
| Rab10 | 10890 | Ras-related protein Rab-10 | 0.6 | 4.8 | + | |
| Rab11A | 8766 | Ras-related protein Rab-11A | / | Ring | - | (9) |
| Rab11B | 9230 | Ras-related protein Rab-11B | / | Ring | + | |
| Rab13 | 5872 | Ras-related protein Rab-13 | / | Ring | + | (9) |
| Rab27A | 5879 | Ras-related protein Rab-27A | 0.1 | 4.3 | - | No AMPylation (9) |
| Rab35 | 11021 | Ras-related protein Rab-35 | −1.5 | 4.3 | + | (31) |
| OCEL1 | 79629 | Occludin/ELL-domain- containing protein 1 |
0.2 | 3.8 | + | |
| RARRES3 | 5920 | Retinoic acid receptor responder 3 |
/ | Ring | + | |
| IDI2 | 91734 | Isopentenyl-diphosphate delta isomerase 2 |
/ | Ring | + | |
| KIAA0652 / ATG13 |
9776 | Autophagy related 13 | / | Ring | - | |
| GGT1 | 2678 | Gamma-glutamyltransferase 1 |
1.2 | 3.4 | - | |
| DECR2 | 26063 | 2,4-Dienoyl CoA reductase 2, peroxisomal |
0.5 | 3.4 | - | |
| DERL1 | 79139 | Derlin 1 | −0.3 | 4.7 | - | |
| CREB1 | 1385 | cAMP responsive element binding protein 1 |
1.2 | 3.8 | - | |
| PAK4 | 10298 | p21 protein (Cdc42/Rac)- activated kinase 4 |
/ | Ring | - | |
| GPAM | 57678 | Glycerol-3-phosphate acyltransferase, mitochondrial |
/ | Ring | - | |
| CDCP2 | 242603 | CUB domain containing protein 2 |
/ | Ring | - | |
| HRASLS2 | 54979 | HRAS-like suppressor 2 | / | Ring | - | |
| ERMN | 57471 | Ermin, ERM-like protein | / | Ring | - | |
| TTC5 | 91875 | Tetratricopeptide repeat domain 5 |
/ | Ring | - | |
| ARHGAP11A | 9824 | Rho GTPase activating protein 11A |
/ | Ring | - | |
| C21orf2 | 755 | Chromosome 21, open reading frame 2 |
/ | Ring | - | |
Abbreviations: SidM*, SidM(D110/112A); /, not listed because selection based on Ring; + or -, positive or negative result in bead-based AMPylation assay.
Co-localization of L. pneumophila effectors and their targets within cells
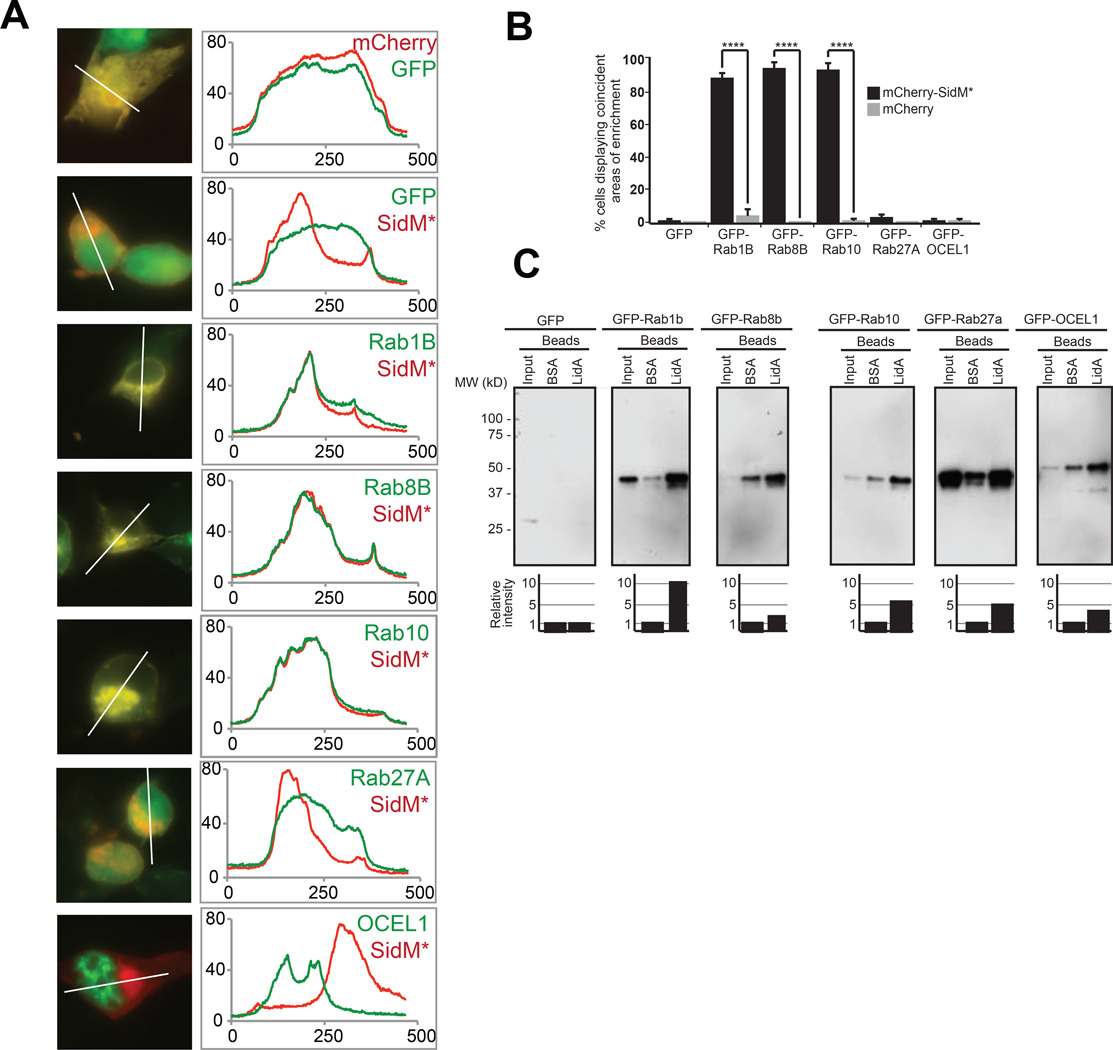
All proteins used in the aforementioned assays were synthesized using IVTT techniques. Thus, we sought to analyze a selected number of potential binding partners for their ability to interact with SidM or LidA in different cell biological contexts. Since proteins localize to different subcellular compartments, two proteins initially found to interact with one another in vitro may in fact never encounter each other during bacterial infection. Moreover, proteins may be post-translationally modified in vivo, influencing their protein interaction or stability. Thus, we performed fluorescence microscopy analysis on transiently transfected COS-1 cells producing fluorescently labeled SidM or LidA and their respective target candidates. To study co-localization with SidM, we took advantage of the AMPylation-defective mutant SidMD110/112A which exhibits strongly reduced cytotoxicity compared to AMPylation-competent SidM (9). We found that mCherry-SidMD110/112A was enriched in the perinuclear region, and that its area of enrichment matched that of GFP-tagged Rab1B, Rab8B, and Rab10 in COS-1 cells (P < 0.0001) (Figure 6A, B). Thus, it is possible that those Rab proteins may in fact be physiologically relevant targets of SidM, as has been reported for Rab1B (8, 46, 47). Rab27A exhibited partial co-localization with SidMD110/112A, whereas GFP-OCEL1 showed a distinct nuclear localization and appeared to lack any overlapping signal with the primarily extranuclear SidMD110/112A.
Figure 6.
Host target validation using co-localization and co-precipitation analysis. (A) COS1 cells were transiently transfected with plasmids encoding mCherry-SidMD110/112A and GFP-tagged target candidates, and protein colocalization 16 hours after transfection was determined by fluorescence microscopy. Left panels show representative fluorographs of doubly transfected cells; the right panel shows line scans denoting pixel intensity of red and green fluorescent signals along the line indicated in the image to the left. Scale bar, 1µm. (B) Quantification of (A) showing the percentage of cells with coincident areas of GFP- and mCherry-enrichment. Data are mean ± SD (error bars) for three independent experiments. ****P < 0.0001 (two-tailed t-test). (C) Pulldown assay. Beads coated with BSA (control) or purified recombinant LidA were used to precipitate GFP-tagged prey proteins from 293T cell lysate. Inputs (1%) and eluates (50%) were separated by SDS-PAGE, and prey proteins were detected by immunoblot using anti-GFP antibody. Estimated molecular weights of prey proteins: GFP (27kD), GFP-Rab1B (52kD), GFP-Rab8B (51kD), GFP-Rab10 (50kD), GFP-Rab27A (52kD), GFP-OCEL1 (56kD). The graph below each panel is a quantification of the co-precipitation data. The amount of prey protein was determined by densitometry and is shown relative to nonspecifically-bound prey protein eluted by BSA-coated beads for each group, arbitrarily set at 1. The values are representative of at least two independent experiments.
mCherry-LidA was dispersed throughout transfected COS1 cells, and the overlap of its fluorescence signal with that of GFP-tagged Rab proteins was not informative (Supplementary Figure S7). For the potential non-Rab target OCEL1, however, localization to the nucleus was distinct from the cytosolic localization pattern of mCherry-LidA. Since this assay was inconclusive in validating most potential LidA target candidates from the Rab family of proteins, we turned to an alternative assay: precipitation of GFP-tagged host proteins from transfected cell lysate. Using bead-immobilized LidA as bait, we detected robust interaction of LidA with Rab1B, Rab8B, Rab10, Rab27A, and OCEL1 as compared to BSA-coated control beads (Figure 6C). These results are consistent with our earlier report showing that Rab1B and Rab8B bind to LidA (7).
Localization of host proteins to the L. pneumophila-containing vacuole
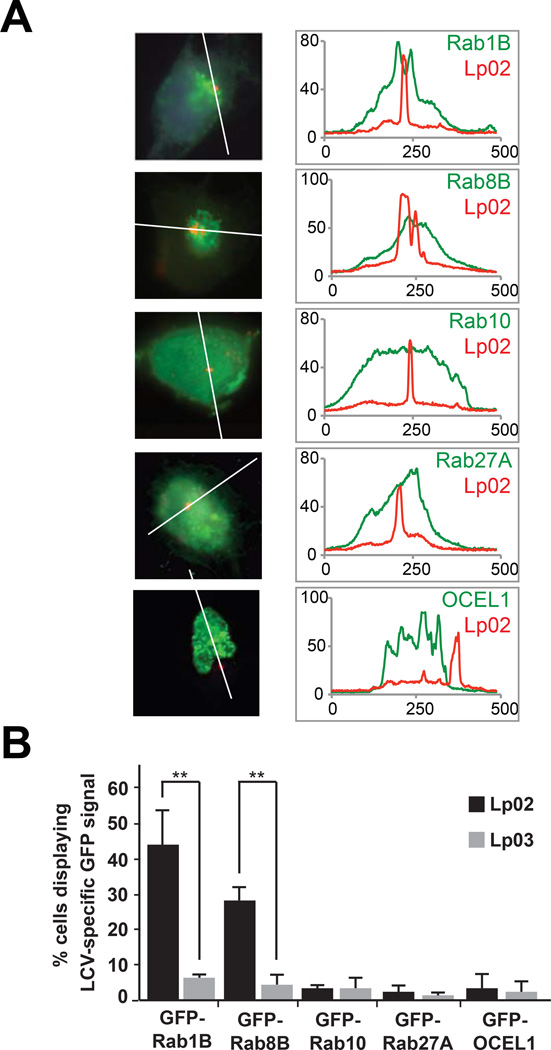
Several host targets, including Rab1, Sec22B, and Arf1, are recruited to the LCV by L. pneumophila where they contribute to the establishment of a replication compartment (reviewed in (48)). Localization to the LCV provides strong evidence that host proteins are utilized or exploited by L. pneumophila. We asked whether potential SidM or LidA target candidates identified above by NAPPA localize to the LCV during infection with virulent, but not avirulent, L. pneumophila. To that end, we transfected CHO-FcγRII cells with constructs encoding GFP-tagged host targets, challenged the cells with the parental L. pneumophila strain (Lp02) or the T4SS-defective mutant (Lp03), and looked for localization of the GFP-tagged targets to the LCV at various time points after infection (Figure 7). At 1.5 hr post-infection, GFP-Rab1B colocalized with 53% of vacuoles containing Lp02, as compared to only 5% of LCVs containing Lp03 (P < 0.01). Localization of Rab1B to the LCV was not observed at later time points (6 hr or 10 hr post-infection), in agreement with previous reports (7, 31, 46, 47). In cells transfected with GFP-Rab8B, a modest number of Lp02-containing LCVs (24%) showed co-localization with the GFP signal at 1.5 hr post-infection, as compared to 1% of Lp03-containing LCVs (P < 0.01). This was consistent with an earlier report that Rab8B localizes to LCVs in the amoebal host Dictyostelium discoideum (49). Neither GFP-Rab10, GFP-Rab27A, nor GFP-OCEL1 were detected on or around the Lp02-containing LCV at 1.5 hr, 6 hr, or 10 hr post-infection. These findings show that some but not all potential targets identified by NAPPA are recruited to the LCV as part of the L. pneumophila pathogenesis program.
Figure 7.
Localization of host targets to the LCV. (A) CHO-FcγRII cells were transfected with constructs encoding the indicated GFP-tagged Rab proteins or OCEL1, and challenged with L. pneumophila Lp02 (wild-type) or Lp03 (T4SS mutant). Intracellular bacteria (red) were detected with anti-L. pneumophila antibody. Scale bar, 1µm. Line scans (right panels) denote pixel intensity of red and green fluorescent signals along the indicated line. (B) Quantification of (A) showing the percentage of cells displaying an LCV-specific GFP signal. Data are mean ± SD (error bars) for three independent experiments. **P < 0.01 (two-tailed t-test).
DISCUSSION
The aim of the present study was to introduce an improved NAPPA screening strategy that allowed the mapping of host-pathogen interactions by detecting PPIs. Using two well-characterized L. pneumophila effectors in proof-of-concept studies, 18 and 20 candidate targets out of a total of 10,000 human proteins were identified for SidM and LidA, respectively. Of the known targets, 5 of 6 (Rab1A, Rab1B, Rab3B, Rab8A, Rab8B, Rab31) for LidA and 1 of 1 (Rab1B) for SidM were recaptured by NAPPA, yielding an accuracy of 85.7% (Table 1). 76% of all candidate targets were validated using an independent bead-based assay, demonstrating the high fidelity of NAPPA during physical protein-protein interaction screening. Moreover, three novel host interaction proteins (Rab8B, Rab10 and Rab27A) were confirmed to co-localize with SidM in COS-1 cells (Figure 6), and Rab8B was found to surround LCVs during L. pneumophila infection (Figure 7). This evidence suggests that our approach has great potential to be used in the identification of novel host targets for pathogen effectors without extensive prior knowledge of its cellular localization or function.
In addition, using a high-sensitivity and specificity nonradioactive click assay we profiled the human substrates AMPylated by SidM, an enzyme with a nucleotidyl transferase fold as opposed to the FIC domain present in a previously characterized AMPylator (21). Using the targets identified by NAPPA and validated using bead-based assays, we constructed a SidM- and LidA-host interaction map to illustrate the number and identity of these effectors’ potential host targets (Figure 8). Members of the Rab family of small GTPases comprised the majority of human targets for both LidA (14 Rabs) and SidM (7 Rabs). Mammalian cells encode more than 60 Rab proteins, which share significant sequence and structural homology. As regulators of membrane transport, Rab proteins are targeted by a number of intracellular pathogens including Brucella, Chlamydia, Mycobacteria, Salmonella, and Coxiella (reviewed in (4)). We cannot discern whether LidA or SidM specifically target each of the Rab proteins identified (Table 2), or, more likely, whether those interaction partners were identified based on their homology to Rab1 (the primary target). Within cells, Rabs localize to specific membrane compartments, and it is conceivable that LidA may never encounter, for example, Rab32, which localizes to melanosomes (50). This underscores the importance of in vivo assays to elucidate the targets of physiological relevance.
Figure 8.
Host interaction profile for L. pneumophila SidM and LidA. Potential host targets for L. pneumophila SidM and LidA are mapped according to physical binding interaction (blue lines) or AMPylation (red lines). Targets are divided into Rab GTPase family members (top panel) or non-Rab GTPases (bottom panel), and color-coded according to protein function. Only those targets confirmed by bead-based NAPPA assay are shown.
Irrespective of the physiological relevance of SidM- and LidA-interacting Rabs that emerged from the NAPPA, a sequence comparison between Rabs that did or did not bind to LidA revealed the existence of several clusters of conserved residues, all of which were located within the interface of the Rab-LidA protein complex (Figure 4, Supplementary Figure S6). Notably, Rabs that interacted with LidA were representatives from different evolutionary branches of the Rab subfamily, indicating that binding is not simply dictated by the overall sequence similarity among the target proteins (51). Although it is tempting to speculate that single amino acid substitutions within the specificity-determining clusters would easily convert a non-binding to a binding Rab (and vice versa), Cheng and colleagues (38) reported that they were unable to alter complex formation between LidA and Rab1 by replacing individual or even two residues within the interface. Thus, a combination of substitutions rather than individual replacements will likely be required to measurably affect binding between L. pneumophila LidA and its Rab ligands.
While this technique is not without bias (not all proteins will be soluble, folded correctly, or appropriately activated or post-translationally modified), it does reduce the bias in that among those “well-behaved’ proteins on the protein array, the conditions for query protein binding are equal. Furthermore, we speculate that the nucleotide-binding proteins (such as GTPases) on the array are bound to a heterogeneous mixture of their natural ligands thanks to the physiological nature of the HeLa cell-based IVTT system. Thus, Rab proteins are present as both GTP- and GDP-bound Rabs, increasing the chance of identifying a PPI that is dependent on one form or the other. The fact that most of the previously reported Rab targets were detected by NAPPA underscores the potential of this technique in detecting relevant PPIs.
Many of the novel NAPPA targets showed at least partial colocalization with SidM upon overproduction in transiently transfected COS1 cells, as was expected from two interacting proteins. However, not all of these Rabs colocalized with LCVs during L. pneumophila infection. While Rab1B and Rab8B selectively localized to LCVs containing virulent but not avirulent L. pneumophila (Figure 7), Rab10 and Rab27A did not localize to LCVs, suggesting that they were not recruited to the vacuole as part of the L. pneumophila virulence program. Importantly, this finding does not exclude them as bona fide targets since several other L. pneumophila effectors, including LidA, were reported to localize to subcellular regions other than the LCV (53–55). Although the targets of most L. pneumophila effectors have yet to be discovered, it is likely that some of them will manipulate host proteins at various membrane compartments, explaining why they will not be found on or close to the LCV.
One of the novel targets for LidA was Rab10, recently described as an ER-specific protein important in regulating phospholipid synthesis in and growth of ER tubules (56). Given that L. pneumophila hijacks membrane vesicles from the ER-to-Golgi trafficking route to remodel its LCV (47, 55), and that it manipulates phospholipid content of the LCV membrane (19), the newly described Rab10 target fits well within the L. pneumophila infection paradigm. Another new target of LidA was Rab27A, which is important for vesicle transport along the secretory pathway by regulating docking of exosomes at the plasma membrane (57). Considering that exosomes contribute to induction of the immune response and help protect the host cell by releasing antimicrobial peptides and reactive oxygen species into the surrounding environment, perhaps L. pneumophila disarms this process by targeting Rab27A in order to protect itself in its brief extracellular journey to a neighboring cell.
OCEL1 was an intriguing non-Rab NAPPA ligand for both LidA and SidM, and a target of AMPylation by SidM, strongly suggesting a role for this host factor during L. pneumophila infection of macrophages (Figure 3, Figure 5). Little is known about OCEL1 (occludin/ELL-domain-containing protein 1), however, its namesake, occludin, is a well-characterized eukaryotic protein implicated in microbial pathogenesis (58–60). OCEL1 primarily localizes to the cell nucleus (Figure 6A, Supplementary Figure S7) and thus seems an unlikely target for LidA or SidM, which were reported to function inside the cytosol throughout infection. Nevertheless, we cannot exclude the existence of a cytoplasmic pool of OCEL1 at some time during the course of infection, at which point LidA and/or SidM may target OCEL1 to manipulate its function and/or localization. Indeed, our co-precipitation assay suggested the possibility of an interaction between OCEL1 and LidA (Figure 6C).
Our improved NAPPA technology was also well-suited for the study of post-translational modifications such as AMPylation. We identified the previously reported AMPylation targets of SidM, namely Rab1 and Rab35 (Figure 5; Table 3), and also discovered several novel AMPylation targets whose contribution to a successful L. pneumophila infection has yet to be determined. Notably, with the exception of Rab27A, all of the newly discovered Rab proteins AMPylated by SidM contain a tyrosine residue equivalent to the tyrosine-77 (Y77) that is AMPylated in Rab1 (Supplementary Figure S8), suggesting that the modification was specific and not randomly added to other surface-exposed tyrosine residues.
The importance of post-translational modification in understanding protein mechanism, particularly in the context of bacterial pathogenesis, is increasingly appreciated. The L. pneumophila effector AnkX phosphorylcholinates host Rab1 (14), LubX mediates ubiquitination of host Clk1 (61), and Lgt1 exploits its glucosyltransferase activity to modify host elongation factor 1A (62). Other bacterial pathogens provide even more examples of post-translational modification as an important mechanism in pathogenesis (reviewed in (63, 64)), and our high-throughput approach to identify potential host targets of post-translational modification by bacterial effectors should prove valuable to the field.
Altogether, our NAPPA-based interactome profiling combines the advantages of the HeLa cell-based IVTT system for high-efficiency protein production (65) with HaloTag technology for high-specificity protein detection (19, 28). It eliminates the need for laborious protein purifications and demonstrates great potential for the initial high-throughput screen of the human proteome for targets of microbial effectors. Besides AMPylation, we anticipate that assays for other post-translational modifications will be adapted to the NAPPA platform, which could significantly accelerate the discovery of novel host-pathogen interactions and, thus, help to address important biological questions in this and other fields of research.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the Intramural Research Program of NIH (K.B.D., M.R.N., M.P.M.). We thank Jie Wang and Xiaofang Bian for the fabrication of NAPPA, and Lerys Del Moral for the technical assistance.
Footnotes
SUPPORTING INFORMATION
This material is available free of charge via the Internet at http://pubs.acs.org:
Figure S1. Expression of N-terminal and C-terminal SidM HaloTag constructs using human HeLa cell-free expression system. Figure S2. Reproducibility of target detection by LidA in two independent experiments. Figure S3. Validation of bait protein capture on NAPPA array. Figure S4. Influence of differential nucleotide binding on the interaction of GTPase targets with LidA. Figure S5. Input of HaloTag protein, HaloTag-LidA and HaloTag-SidM used in the bead-based pull-down assay. Figure S6. Interface residues conserved within LidA-binding Rabs. Figure S7. Intracellular localization of exogenous mCherry-LidA. Figure S8. Primary sequence alignment of Rab GTPases AMPylated by SidM.
AUTHOR CONTRIBUTIONS
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Current opinion in microbiology. 2009;12(1):67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annual review of cell and developmental biology. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 3.Vogel JP, Isberg RR. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 1999;2(1):30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 4.Sherwood RK, Roy CR. A rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell host & microbe. 2013;14(3):256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machner MP, Chen Y. Catch and release: Rab1 exploitation by Legionella pneumophila. Cell Logist. 2011;1(4):133–138. doi: 10.4161/cl.1.4.18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neunuebel MR, Machner MP. The taming of a Rab GTPase by Legionella pneumophila. Small GTPases. 2012;3(1):28–33. doi: 10.4161/sgtp.18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Developmental cell. 2006;11(1):47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nature cell biology. 2006;8(9):971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 9.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 10.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338(6110):1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1316376111. Published online before print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, Buchrieser C. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12(9):1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 13.Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: a Legionella pneumophila activator of NF-kappaB. Cellular microbiology. 2010;12(8):1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyi Y, Tartakovskaya D, Tais A, Fitzke E, Tzivelekidis T, Jank T, Rospert S, Aktories K. Elongation factor 1A is the target of growth inhibition in yeast caused by Legionella pneumophila glucosyltransferase Lgt1. J Biol Chem. 2012;287(31):26029–26037. doi: 10.1074/jbc.M112.372672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woolery AR, Yu X, LaBaer J, Orth K. AMPylation of Rho GTPases subverts multiple host signaling processes. J Biol Chem. 2014 doi: 10.1074/jbc.M114.601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305(5680):86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 18.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 2012;30(5):1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 20.Yu B, Xiao B, Throop A, Song L, Moral L, Park J, Seiler CY, Fiacco M, Steel J, Hunter P, Saul J, Wang J, Qiu J, Pipas J, LaBaer J. Exploration of Panviral Proteome: High-Throughput Cloning and Functional Implications in Virus-Host Interactions. Theranostics. 2014;4(8):808–822. doi: 10.7150/thno.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Woolery AR, Luong P, Hao YH, Grammel M, Westcott N, Park J, Wang J, Bian X, Demirkan G, Hang HC, Orth K, LaBaer J. Click chemistry-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M114.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabay JE, Horwitz MA. Isolation and characterization of the cytoplasmic and outer membranes of the Legionnaires' disease bacterium (Legionella pneumophila) The Journal of experimental medicine. 1985;161(2):409–422. doi: 10.1084/jem.161.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. Journal of clinical microbiology. 1979;10(4):437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Molecular microbiology. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 25.Berger KH, Isberg RR. Intracellular survival by Legionella. Methods in cell biology. 1994;45:247–259. doi: 10.1016/s0091-679x(08)61855-5. [DOI] [PubMed] [Google Scholar]
- 26.Hutchison CA, 3rd, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M. Mutagenesis at a specific position in a DNA sequence. The Journal of biological chemistry. 1978;253(18):6551–6560. [PubMed] [Google Scholar]
- 27.Berger KH, Merriam JJ, Isberg RR. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Molecular microbiology. 1994;14(4):809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Barker K, Steel J, Park J, Saul J, Festa F, Wallstrom G, Yu X, Bian X, Anderson KS, Figueroa JD, LaBaer J, Qiu J. A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin Appl. 2013;7(5–6):378–383. doi: 10.1002/prca.201200062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Wallstrom G, Magee DM, Qiu J, Mendoza DE, Wang J, Bian X, Graves M, LaBaer J. Quantifying antibody binding on protein microarrays using microarray nonlinear calibration. Biotechniques. 2013;54(5):257–264. doi: 10.2144/000114028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DS, Qiu J, Mendoza EA, Barker K, Rooney ME, LaBaer J. Circulating and synovial antibody profiling of juvenile arthritis patients by nucleic acid programmable protein arrays. Arthritis research & therapy. 2012;14(2):R77. doi: 10.1186/ar3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333(6041):453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5(6):535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montor WR, Huang J, Hu Y, Hainsworth E, Lynch S, Kronish JW, Ordonez CL, Logvinenko T, Lory S, LaBaer J. Genome-wide study of Pseudomonas aeruginosa outer membrane protein immunogenicity using self-assembling protein microarrays. Infect Immun. 2009;77(11):4877–4886. doi: 10.1128/IAI.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joiner KA, Fuhrman SA, Miettinen HM, Kasper LH, Mellman I. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science. 1990;249(4969):641–646. doi: 10.1126/science.2200126. [DOI] [PubMed] [Google Scholar]
- 35.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 36.Schoebel S, Cichy AL, Goody RS, Itzen A. Protein LidA from Legionella is a Rab GTPase supereffector. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(44):17945–17950. doi: 10.1073/pnas.1113133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh HY, Lee DW, Lee KH, Ku B, Choi SJ, Woo JS, Kim YG, Oh BH. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. The EMBO journal. 2010;29(2):496–504. doi: 10.1038/emboj.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng W, Yin K, Lu D, Li B, Zhu D, Chen Y, Zhang H, Xu S, Chai J, Gu L. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS pathogens. 2012;8(3):e1002528. doi: 10.1371/journal.ppat.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323(5911):269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 40.Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE. The fic domain: regulation of cell signaling by adenylylation. Mol Cell. 2009;34(1):93–103. doi: 10.1016/j.molcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Carr PD, Vasudevan SG, Ollis DL. Structure of the adenylylation domain of E. coli glutamine synthetase adenylyl transferase: evidence for gene duplication and evolution of a new active site. J Mol Biol. 2010;396(3):773–784. doi: 10.1016/j.jmb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One. 2009;4(6):e5818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, Johannessen CM, Silver SJ, Nguyen C, Murray RR, Hieronymus H, Balcha D, Fan C, Lin C, Ghamsari L, Vidal M, Hahn WC, Hill DE, Root DE. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8(8):659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grammel M, Luong P, Orth K, Hang HC. A chemical reporter for protein AMPylation. J Am Chem Soc. 2011;133(43):17103–17105. doi: 10.1021/ja205137d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broncel M, Serwa RA, Tate EW. A new chemical handle for protein AMPylation at the host-pathogen interface. Chembiochem. 2012;13(2):183–185. doi: 10.1002/cbic.201100743. [DOI] [PubMed] [Google Scholar]
- 46.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450(7168):365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 47.Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. The Journal of experimental medicine. 2004;199(9):1201–1211. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nature reviews. Microbiology. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic. 2009;10(1):76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- 50.Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, Seabra MC. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. The Journal of cell biology. 2006;175(2):271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004(250):RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Machner MP. Targeting of the small GTPase Rab6A' by the Legionella pneumophila effector LidA. Infection and immunity. 2013;81(6):2226–2235. doi: 10.1128/IAI.00157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degtyar E, Zusman T, Ehrlich M, Segal G. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cellular microbiology. 2009;11(8):1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 54.Li T, Lu Q, Wang G, Xu H, Huang H, Cai T, Kan B, Ge J, Shao F. SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO reports. 2013;14(8):733–740. doi: 10.1038/embor.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derre I, Isberg RR. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infection and immunity. 2005;73(7):4370–4380. doi: 10.1128/IAI.73.7.4370-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nature cell biology. 2013;15(2):169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature cell biology. 2010;12(1):19–30. doi: 10.1038/ncb2000. suppp 1–13. [DOI] [PubMed] [Google Scholar]
- 58.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. The Journal of cell biology. 1993;123(6 Pt 2):1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Z, Nybom P, Magnusson KE. Distinct effects of Vibrio cholerae haemagglutinin/protease on the structure and localization of the tight junction-associated proteins occludin and ZO-1. Cellular microbiology. 2000;2(1):11–17. doi: 10.1046/j.1462-5822.2000.00025.x. [DOI] [PubMed] [Google Scholar]
- 60.Simonovic I, Rosenberg J, Koutsouris A, Hecht G. Enteropathogenic Escherichia coli dephosphorylates and dissociates occludin from intestinal epithelial tight junctions. Cellular microbiology. 2000;2(4):305–315. doi: 10.1046/j.1462-5822.2000.00055.x. [DOI] [PubMed] [Google Scholar]
- 61.Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Molecular microbiology. 2008;67(6):1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 62.Lu W, Du J, Stahl M, Tzivelekidis T, Belyi Y, Gerhardt S, Aktories K, Einsle O. Structural basis of the action of glucosyltransferase Lgt1 from Legionella pneumophila. Journal of molecular biology. 2010;396(2):321–331. doi: 10.1016/j.jmb.2009.11.044. [DOI] [PubMed] [Google Scholar]
- 63.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143(5):694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salomon D, Orth K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe. 2013;14(3):269–279. doi: 10.1016/j.chom.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saul J, Petritis B, Sau S, Rauf F, Gaskin M, Ober-Reynolds B, Mineyev I, Magee M, Chaput J, Qiu J, LaBaer J. Development of a full-length human protein production pipeline. Protein Sci. 2014 doi: 10.1002/pro.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.