Abstract
Endometriosis, ectopic growth of the uterine lining (endometrium), which affects 6–11% of reproductive age women, is associated with pelvic pain and infertility. We investigated the peritoneal fluid (PF), urine and omental fat (OF) proteomes of women with endometriosis vs. individuals with no surgically visualized endometriosis. All participants were enrolled in the NICHD-funded ENDO Study. A two-step proteomic study was performed. The first, a broad survey, employed a semi-quantitative gel LC-mass spectrometry (MS) workflow: SDS PAGE fractionation, trypsin digestion and LC–MS/MS. The results showed sample integrity but failed to detect any differences between women with and without endometriosis. The second step was a quantitative analysis of OF samples. We employed another sample set (n = 30) from women ± disease and isobaric mass-tag (iTRAQ) chemistry to label peptides and 2D LC–MS/MS for protein identification and quantification. Three proteins—matrix metalloproteinase-9, neutrophil elastase, and FAM49B—were significantly lower in abundance in samples from women with endometriosis. Interestingly, neutrophil elastase and FAM49B levels were associated with higher levels of a subset of endocrine disrupting chemicals (EDCs) that were previously measured in the same samples. The results of these experiments showed the feasibility of associating endometriosis with changes in the OF protein repertoire and EDC levels.
Biological significance
Endometriosis, pathological growth of the uterine lining, is associated with significant morbidities, including pain and infertility. However, the causes of this common condition are poorly understood. This study determined whether endometriosis was associated with changes in the protein composition of peritoneal fluid, urine and/or omental fat. A protein of unknown function (FAM49B) and two proteinases (metalloproteinase-9, neutrophil elastase) were down regulated in OF samples from women with versus without endometriosis. These findings suggested proteinase imbalances at sites that were distant from the endometriotic lesions. Additionally, FAM49B and neutrophil elastase levels were associated with higher levels of a subset of environmental chemicals that were quantified in the same samples, suggesting other possible associations. Thus, this work generated hypotheses that will be tested in further studies.
Keywords: Endometriosis, Peritoneal fluid, Urine, Omental fat, Mass spectrometry, Environmental chemicals
1. Introduction
Proteomic signatures lend insights into normal and abnormal processes at cell, tissue and organismal levels. Although genetic and epigenetic profiles yield valuable information, the protein machinery they assemble gives us the best functional insights into the mechanisms underlying biological processes. However, global profiling approaches face the daunting challenge of enormous proteome complexity. To date, mass spectrometry-based techniques have been the primary tools of investigators interrogating the protein repertoire. In this regard, there has been particular interest in the proteomes of human biological fluids as they offer a window into health and disease. Those that are easily obtained—e.g., urine [1], plasma [2] and saliva [3] among others—have been intensely studied as potential sources of biomarkers. In endometriosis research, a rapid, noninvasive technique for diagnosis and follow-up using peripheral biomarkers as surrogates of disease is an active area of investigation (reviewed in [4-6]). For example, urinary enolase I, vitamin d-binding protein, and cytokeratin 19 have been proposed as potential diagnostic biomarkers [7-9], though more work is needed to validate these candidates in larger sample sizes [10]. However, there are several problems associated with proteomic analyses of body fluids, including protein abundances over several orders of magnitude. In addition, the signals of interest are often from a point source that is distal to where the sample is collected, thereby creating the additional problem of dilution [11]. These difficulties have prompted analyses of more proximal samples, such as peritoneal fluid from women with endometriosis [12-14] or the actual cell type of interest such as fat from obese individuals [15].
Endometriosis, which involves endometrial growth outside the uterus, affects 6–11% of all women of reproductive age [16,17]. A primary cause is thought to be transport and subsequent adherence of endometrial cells/tissue to the peritoneal cavity [18]. However, other theories have been proposed, including lymphatic or vascular metastases [19], immunologic deficits resulting in impaired clearance of ectopic endometrium [20] and coelomic metaplasia [21]. Estrogen, which is necessary for the proliferation and survival of endometriotic tissue, is plentiful in the peritoneal cavity with sources including the ovaries, follicular fluid, and the blood [22]. Endometrial implants elicit an inflammatory response, resulting in a host of pathologies including angiogenesis, adhesions, fibrosis, scarring, neuronal infiltration, and anatomical distortion [16,22-24]. As a result, patients with endometriosis may experience mild to severe pelvic pain and/or infertility.
Endocrine-disrupting chemicals (EDCs) are generally defined as substances in the environment, food, and consumer products that interfere with hormone biosynthesis, metabolism, or action resulting in a deviation from normal homeostatic control of reproduction [25]. Seemingly ubiquitous, endocrine disruptors are present in industrial solvents and lubricants [polychlorinated biphenyls (PCBs), polybrominated biphenyls (PBBs), dioxins], pesticides [methoxychlor, chlorpyrifos, dichlorodiphenyltrichloroethane (DDT)], fungicides (vinclozolin), pharmaceutical agents [diethylstilbestrol (DES)] and flame retardants [polybrominated diphenyl ethers (PBDEs)] [25,26]. A wealth of evidence links exposures to EDCs with causation or exacerbation of endometriosis [25,27-33]. For example, pre-natal exposure of mice to bis-phenol A (BPA) results in female offspring with an endometriosis-like condition [30,32]. Investigators have also evaluated phthalate levels in blood plasma collected from women diagnosed with endometriosis vs. age-matched women without disease, finding potential differences [27,29,31,34]. After adjusting for serum lipid levels, gravidity, and tobacco use, four PCB congeners, which are anti-estrogens, imparted a three-fold risk of developing endometriosis in a cohort of 84 women [28]. Furthermore, an evaluation of persistent organic pollutants (POPs) in omental fat (OF) and serum collected from women diagnosed with endometriosis revealed a significant positive association between α-hexachlorocyclohexane levels and endometriosis [33].
In this study, we utilized a two-stage MS-based analysis to investigate the protein repertoire of PF, urine and OF samples that were obtained from women who had been diagnosed with endometriosis and women with no surgically visualized endometriosis. The first stage, a broad survey, utilized a gel LC-MS workflow that consisted of 1D SDS PAGE protein fractionation, in-gel trypsin digestion and 1D LC–MS/MS analysis. The second stage, relative quantification focused on the OF samples, was accomplished by isobaric mass-tag (iTRAQ) labeling at the peptide level and 2D LC–MS/MS to identify and compare protein abundances. The results enabled the identification of proteins whose levels changed in endometriosis. As a final step, we investigated whether these changes were associated with EDC levels that were previously measured in the same samples [33].
2. Methods
2.1. Sample collection
For the semi-quantitative survey experiments, urine, PF and OF samples were provided by 63 women enrolled in the Endometriosis: Natural History, Diagnosis, and Outcomes (ENDO) Study. Specifically, women undergoing laparoscopy/laparotomy at one of 14 participating clinical sites in the San Francisco Bay Area were recruited for study as previously described [17]. The ENDO Study obtained Institutional Review Board approval from UCSF in addition to a formal NIH signed reliance agreement that was put in place with the UCSF Committee on Human Research. Urine samples were obtained from all study participants, aliquoted and frozen at −80 °C. Peritoneal fluid and OF samples were acquired from women who underwent laparoscopy or laparotomy procedures and frozen at −80 °C. All samples were received blinded as to the women’s disease status.
For relative quantification, a second set of 30 blinded OF samples was analyzed. They were obtained in the Salt Lake City Area of the ENDO Study and collected as described above with Institutional Review Board approval from the University of Utah Committee on Human Research.
A complete list of all the samples used in this study (including endometriosis status, donor age, race, primary reason for surgery, gravidity, and parity) is shown in Supplemental Table 1.
2.2. Reagents and chemicals
All standard reagents were obtained from Fisher Scientific unless otherwise specified. HPLC grade acetonitrile and water were obtained from Burdick & Jackson. Mammalian proteinase inhibitor cocktail P8340 was from Sigma. Sequencing grade porcine trypsin was from Promega.
2.3. Sample preparation
Peritoneal fluid samples (~0.5 mL) from 12 women with and 27 without endometriosis were defrosted at room temperature and 20 μL of proteinase inhibitor cocktail was added. Samples were centrifuged at 10,000 ×g for 25 min at 10 °C to remove particulate material. Proteins were concentrated using 0.5 mL 3000 molecular weight cutoff (MWCO) centrifugal filter units (Millipore). The retentate was washed two times with phosphate buffered saline (PBS) and then aliquoted into several fractions that were frozen at −80 °C until use.
Urine samples (2–4 mL) from 17 women with and 44 without endometriosis were defrosted at room temperature and 20 μL of proteinase inhibitor cocktail was added during the process. Samples were centrifuged at 10,000 ×g for 25 min at 4 °C. Proteins were purified and concentrated using 5 mL 3000 MWCO centrifugal filter units, washed two times with PBS and frozen at −80 °C until use.
Omental fat samples (n = 17) from 3 women with and 14 women without endometriosis that were analyzed by the gel LC–MS workflow were processed on ice. Approximately 100 mg of frozen OF were excised from each sample and placed in a tube containing 4 μL of proteinase inhibitor cocktail. The sample was homogenized using a PowerGen Model 125 Homogenizer (Fisher) in 6 M urea, 250 mM Tris, pH 7.9 then centrifuged at 16,000 ×g for 30 min at 4 °C, which produced 3 discrete layers/fractions. A 150 μL aliquot of the middle (protein-containing) fraction was subjected to chloroform–methanol (1:4, v:v) extraction at room temperature. Proteins were precipitated by the addition of 400 μL methanol followed by centrifugation.
For the iTRAQ workflow, OF samples from 16 women with and 14 women without endometriosis were excised and transferred to cold lysis buffer (6 M urea, 2 M thiourea, 4% CHAPS and 0.1% SDS; 1 mg tissue:10 mg lysis buffer) in tubes on ice to which 10 μL of proteinase inhibitor cocktail was added. The samples were homogenized as described above and incubated, with shaking, at room temperature for 1 h, then centrifuged at 16,000 ×g for 20 min to remove cell debris. The supernatant (~200 μL) was transferred to a clean microfuge tube and 6 volumes of cold acetone were added. The solution was incubated at −20 °C for 4 h followed by centrifugation at 9000 ×g for 10 min to pellet the precipitated proteins. The pellet was resolubilized in 500 mM triethylammonium bicarbonate (TEAB, pH 8.5)/0.1% SDS.
Amino acid analysis was performed on aliquots of the PF and urine samples by the Texas A&M University Protein Chemistry Laboratory using a Hewlett Packard AminoQuant System (http://www.tamupcl.com/). The protein concentration of the OF samples was determined using the bicinchoninic acid (BCA) assay (Pierce).
2.4. SDS-PAGE, in gel protein digestion, and LC–MS/MS
An aliquot of each sample, equivalent to 25 μg protein, was separated by 1D SDS-PAGE using 4–12% Bis-Tris gradient gels (Invitrogen). The gels were stained with Gel Code Coomassie Colloidal G250 (Pierce). After destaining, each gel lane was rastered into 40–45 pieces, 1 mm in diameter, using a manual gel cutter. Each gel plug was transferred to one well of a 96-well microtiter plate. In-gel trypsin digestion was performed using a ProGest Protein Digestion Station (Genomic Solutions) programmed to perform SDS extraction, cysteine reduction with dithiothreitol, alkylation with iodoacetamide, trypsin digestion for 8 h at 37 °C and peptide extraction with 25 mM ammonium bicarbonate. The resulting peptides were analyzed using an LTQ XL linear ion trap mass spectrometer (ThermoFisher) equipped with an Advance CaptiveSpray ion source (Michrom Bioresources) and a nano2D LC System (Eksigent). Mobile phases A and B were 2% acetonitrile/0.1% formic acid and 80% acetonitrile/0.08% formic acid, respectively. Separation was performed on an Onyx Monolithic C18 Column (100 μm I.D. × 150 mm, Phenomenex) by using a 17-minute linear gradient from 2% to 50% B at a flow rate of 750 nL/min. Mass spectra were acquired in a data-dependent mode consisting of one survey MS scan (m/z 400–1700) and six MS/MS scans (m/z 100–1500) with dynamic exclusion set to 40 s. Normalized collision energy for CID was set to 35% with activation times of 30 ms.
2.5. In-solution trypsin digestion and iTRAQ labeling
For each sample, an aliquot containing 50 μg of OF proteins was transferred to an Eppendorf tube, concentrated to ~3 μL and then brought to a volume of 20 μL by adding an equivalent amount of 1 M TEAB/0.1% SDS. The next steps were cysteine reduction (5 mM tris[2-carboxyethyl]phosphine for 1 h at 60 °C), alkylation (200 mM iodoacetamide for 10 min at room temperature) and digestion (overnight at 37 °C with a 1:50 [w:w] trypsin to protein ratio). The peptides were labeled with iTRAQ 8-plex reagents at ambient temperature for 2 h according to the manufacturer’s (AB Sciex) protocol. The 30 samples were randomly apportioned into five sets of 8. Individual sets contained six samples, a duplicate of one of the samples to assess variability and a reference standard—an arbitrarily chosen digest included in every octet that enabled normalization across all the sample sets. Volumes were reduced to ~20 μL on a Speedvac and SDS was removed by using an SCX cartridge (AB Sciex) according to the manufacturer’s protocol.
2.6. 2D LC–MS/MS analysis of iTRAQ-labeled samples
The iTRAQ labeled peptides were separated offline by alkaline reversed phase HPLC [35,36] on a Paradigm MS4 HPLC system (Michrom Bioresources) equipped with a LEAP Technologies PAL autosampler/fraction collector with the following modifications. Mobile phases A and B were 0.1% ammonium hydroxide in water and 0.1% ammonium hydroxide acetonitrile, respectively. Separation was performed using a Zorbax Extend column (4.6 × 100 mm, Agilent) at a flow rate of 700 μL/min and the following gradient: 2–12% B over 10 min, 12–25% B over 13 min, 25–35% B over 10 min, and 35–40% B over 5 min. Fractions were collected at one minute intervals, solvents were removed by using vacuum centrifugation, and the peptide mixtures were reconstituted in 0.1% formic acid.
An LTQ Orbitrap Velos mass spectrometer (ThermoFisher) equipped with an Advance CaptiveSpray ion source (Michrom Bioresources) and a nano2D LC System (Eksigent) was used to analyze the peptide fractions. Mobile phases A and B were 2% acetonitrile/0.1% formic acid and 98% acetonitrile/0.1% formic acid, respectively. Peptides were applied to a guard column (C18 Acclaim PepMap100, 300 μm I.D. × 5 mm, 5 μm particle size, 100 Å pore size; Dionex) and washed with A, (flow rate: 20 μL/min) for 10 min prior to separation on a C18 Acclaim PepMap100 column (75 μm I.D. × 15 cm, 3 μm particle size, 100 Å pore size; Dionex) using a 60 min linear gradient from 2–40% B at a flow rate of 300 nL/min. A full MS scan (400–1600 m/z) was acquired in the Orbitrap at resolution 30,000 in profile mode, a maximum ion accumulation time of 2 s and a target value of 106. Precursor ion charge state screening was activated. The eight most intense ions were selected for fragmentation by high-energy collision-induced dissociation (HCD) with the isolation width set to 3.0 (m/z). Dynamic exclusion was activated with a duration of 20 s and 3 ppm mass tolerance. In HCD fragmentation, the following parameters were used: full scan with FTMS at resolution 7500 in profile mode, m/z 100–1500, a target value of 105, normalized collision energy of 50%, and an activation time of 100 ms.
2.7. Data processing and database searching
LTQ XL and LTQ Orbitrap Velos raw files were converted to peak lists with extract_msn using Mascot Daemon. For the gel LC–MS workflow, database searching was done using Mascot v2.1 against the human SwissProt database (v 2009_01, 20,405 entries). Search parameters were trypsin cleavage, a maximum of 2 missed cleavages, fixed modification of carbamidomethyl C, variable modifications, deamidation on N, oxidation on M, pyro-Glu on N-terminal Q, precursor mass tolerance 1.5 Da, and fragment ion mass tolerance of 0.8 Da. False discovery rate analysis was performed using a concatenated randomized version of the database. All search results were logged into the MySQL database and were reported using a program created in our laboratory for comparing protein identifications among several samples. Peptide identifications with expectation values less than 0.05 were used and only proteins with 2 or more peptides retained. Proteins were grouped using a standard “parsimony” algorithm that results in the smallest protein list that explains the peptide evidence.
The OF iTRAQ data was analyzed using ProteinPilot software (AB Sciex) with the SwissProt human database (v2012_06, 20,237 entries). Bias corrected and background subtracted relative protein abundance ratios were log transformed and compared using weighted linear regression and adjusted for multiple hypothesis using the method of Benjamini and Hochberg [37].
2.8. Clustering and gene ontology (GO)
Unsupervised hierarchical clustering of samples and proteins was performed using GenePattern [38]. GO analyses were performed using the publically available tools provided by the Lewis-Sigler Institute for Integrative Genomics at Princeton University.
2.9. Immunoblotting
The method used was published [39]. Membranes were incubated with antibodies that recognized MMP9 (Oncogene Research Products), FAM49B (Abcam), ELNE (Abcam) or β-actin (Santa Cruz Biotechnology), diluted 1:500 (MMP9 and β-actin) or 1:1000 (FAM49B and ELNE) in TBS-Tween. Antibody binding was detected via chemiluminescence.
2.10. Toxicologic analyses
Organochlorine pesticides including hexachlorocyclohexane (HCH) and its isomers gamma-hexachlorocyclohexane (γ-HCH) and beta-hexachlorocyclohexane (β-HCH), were analyzed using gas chromatography–mass spectrometry (GC–MS) as described elsewhere [40]. Briefly, 1 g of fat tissue was homogenized with anhydrous sodium sulfate and extracted in a Soxhlet apparatus using dichloromethane and hexane. The extract was concentrated and an aliquot was used for the determination of lipid content by gravimetry. The remaining extract was spiked with 13C-labeled internal standards of organochlorine compounds. Lipid was removed from the sample extracts by treatment with concentrated sulfuric acid and by passage through a series of layers of silica gel (Davisil, 100–200 mesh, Aldrich) in the following order: 2 g of silica gel, 1 g of 40% acidic-silica gel, 1 g of silica gel, 1 g of 40% acidic-silica gel, 1 g of silica gel, and a layer of dry sodium sulfate at the top. Then samples were eluted with 100 mL of 20% dichloromethane/hexane. The extract was injected into a gas chromatograph (Hewlett-Packed 6890) coupled with a mass selective detector (Hewlett-Packed, series 5973) operating in an electron impact (70 eV) selected ion monitoring (SIM) mode. Quantification was by the isotope dilution method. External calibration standards were analyzed with every set of samples and recoveries of internal standards were checked against external calibration standards. The detection limits of organochlorines were in the range of 0.01 to 0.2 ng/g, wet weight.
2.11. Statistical analysis of association between altered proteins and EDCs
Previously, we used the approach described above to quantify EDCs in biospecimens from the ENDO Study (urine, fat and peritoneal fluid) that were collected at two sites, The San Francisco Bay Area and Salt Lake City. The results showed that a subset of the analytes were significantly associated with the risk of endometriosis [33]. Here we conducted an exploratory analysis of associations between the proteins that were significantly differentially expressed in samples that were collected from women with endometriosis and EDCs that had the same relationships. These associations were based on linear regression and a permutation test was conducted to assess global associations or no associations of the EDCs considered and the mean level of each protein investigated in the current study.
3. Results
3.1. Overview of data generation
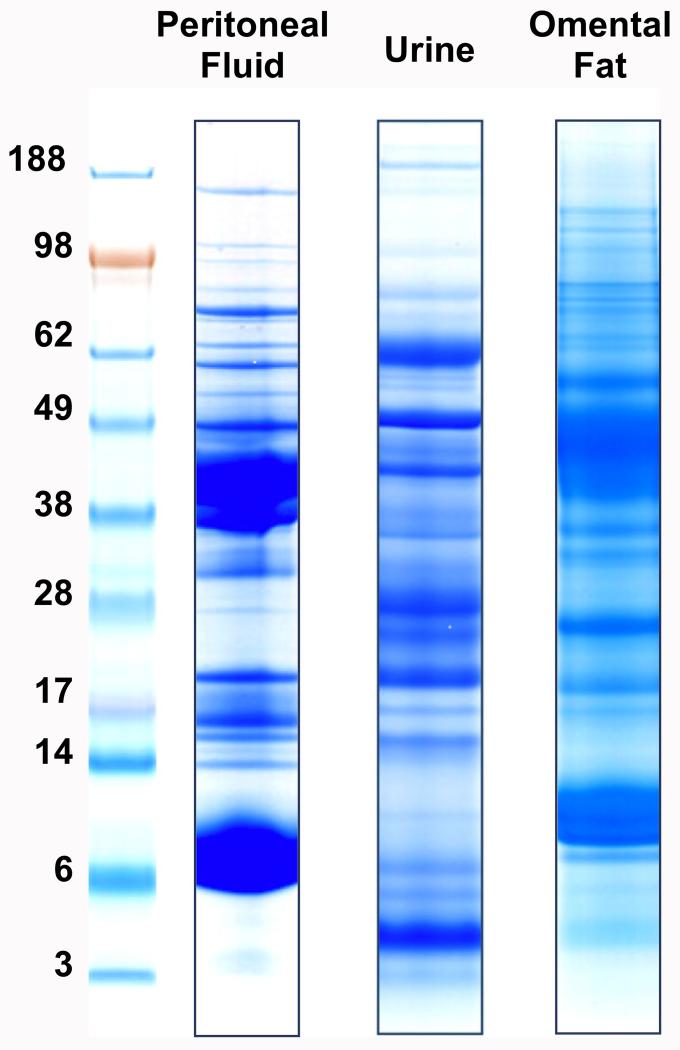
To comprehensively characterize the proteomes of PF, urine, and OF, protein preparation protocols were optimized for each sample type (see the Methods section). A workflow consisting of SDS-PAGE separation, tryptic digestion of the excised bands and LC–MS for protein identification achieved considerable depth of coverage. Fig. 1 shows representative SDS-PAGE banding patterns for the three sample types. Proteins detected at ≤5% FDR were compiled into a comprehensive inventory of the PF, urine, and OF proteomes of women with and without endometriosis.
Fig. 1. SDS-PAGE separation of proteins from peritoneal fluid, urine and omental fat samples. Each specimen type had a unique banding pattern that suggested differences in protein composition.
3.2. Peritoneal fluid
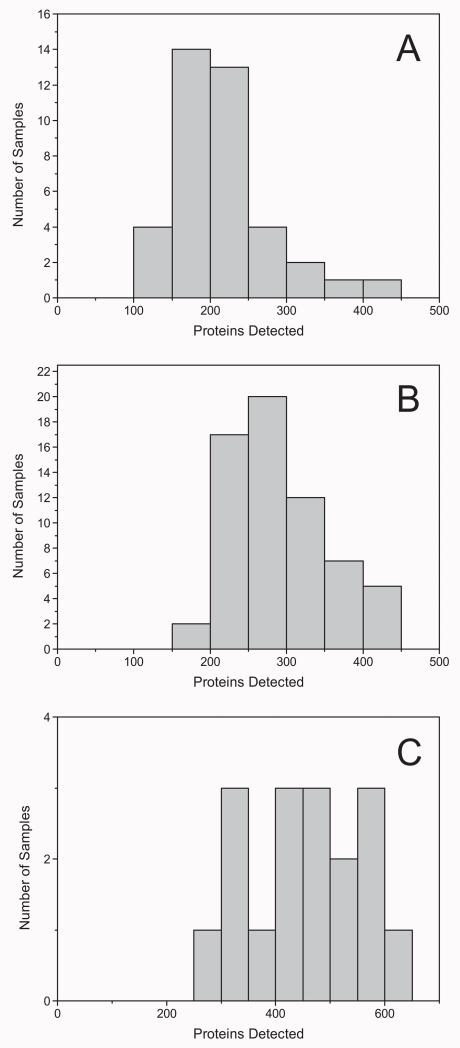
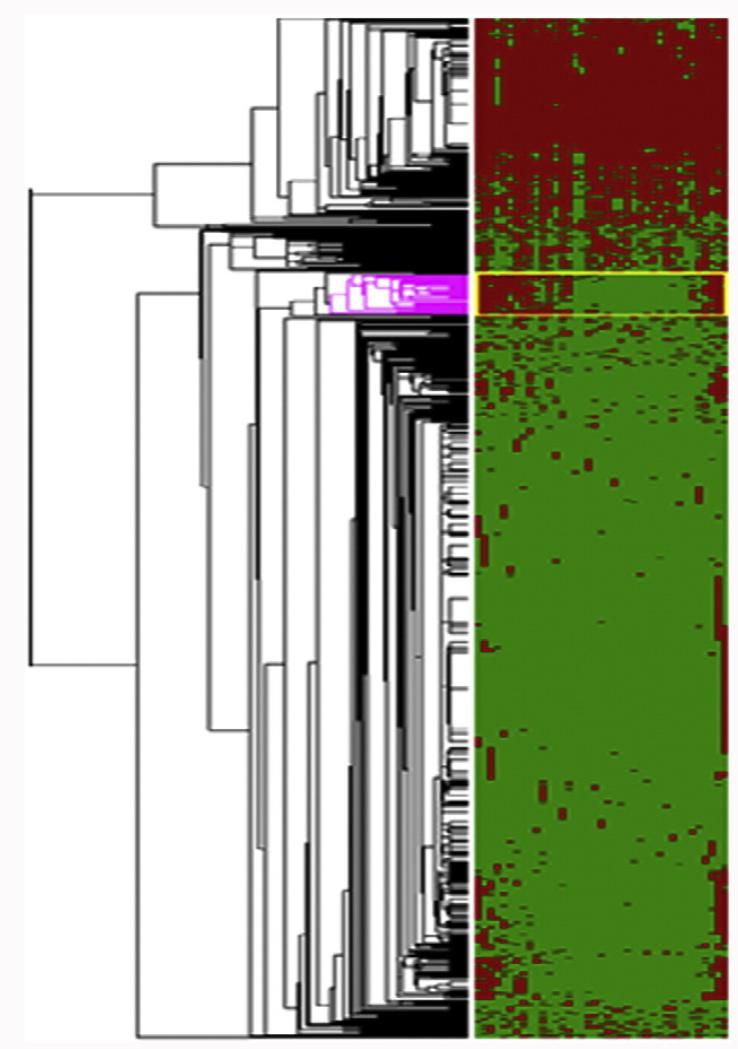
In total, 769 proteins were identified in the entire set of PF samples (Supplemental Table 2), with an average of 186 proteins detected per sample. This distribution, which is plotted in Fig. 2A, is typical of these types of shotgun proteomics analyses [41]. Likewise, the gel lanes (Fig. 1 and Supplemental Table 2) showed that protein abundances varied within and among samples, observations that were in accord with the distribution. Generally, in LC–MS/MS analyses, high-abundance proteins yield relatively high abundance peptides. Thus, the number of peptides observed and protein coverage correlate with relative protein abundances, which were approximated by the Exponentially Modified Protein Abundance Index, emPAI [42], calculated by the formula: 10Nobserved/Nobservable − 1, where Nobserved is the number of experimentally observed peptides and Nobservable is the calculated number of observable peptides for each protein (Supplemental Table 2). Hierarchical clustering and network modeling were used to visualize protein differences among the PF samples and to gain insights into their possible functions. The results are shown in Fig. 3 where the rows correspond to proteins and the columns correspond to subjects. A cluster of 35 proteins (Table 1) was identified as differentially expressed in approximately half of the samples, though this did not correlate with disease status. Their emPAI scores suggested that they were among the more abundant proteins detected. They belonged to several functional groups including kinases, antioxidants and oxidoreductases, as well as transcriptional and translational pathways.
Fig. 2. The number of peritoneal fluid (A), urine (B) or omental fat (C) proteins that were identified in individual samples.
Fig. 3. Heat map depicting unsupervised hierarchical clustering of the 39 peritoneal fluid samples (columns). Individual proteins (rows) were mapped in red (identified) or green (not identified). The colored portion of the dendrogram depicts proteins that were differentially expressed in samples collected from patients who were diagnosed with endometriosis.
Table 1. Proteins detected in peritoneal fluid that were determined to be differentially expressed in ~50% of the samples.
| Accession | Swiss-Prot ID | Molecular weight | Protein name |
|---|---|---|---|
| P63104 | 1433Z_HUMAN | 27,899 | 14-3-3 protein zeta/delta |
| P00568 | KAD1_HUMAN | 21,735 | Adenylate kinase isoenzyme 1 |
| P06733 | ENOA_HUMAN | 47,350 | Alpha-enolase |
| P13929 | ENOB_HUMAN | 47,168 | Beta-enolase |
| P53004 | BIEA_HUMAN | 33,692 | Biliverdin reductase A precursor |
| P07738 | PMGE_HUMAN | 30,027 | Bisphosphoglycerate mutase |
| P07451 | CAH3_HUMAN | 29,707 | Carbonic anhydrase 3 |
| P04040 | CATA_HUMAN | 59,816 | Catalase |
| O00299 | CLIC1_HUMAN | 27,117 | Chloride intracellular channel protein 1 |
| P23528 | COF1_HUMAN | 18,588 | Cofilin-1 |
| P13716 | HEM2_HUMAN | 36,728 | Delta-aminolevulinic acid dehydratase |
| P68104 | EF1A1_HUMAN | 50,451 | Elongation factor 1-alpha 1 |
| P30043 | BLVRB_HUMAN | 22,088 | Flavin reductase |
| P04075 | ALDOA_HUMAN | 39,720 | Fructose-bisphosphate aldolase A |
| P09211 | GSTP1_HUMAN | 23,438 | Glutathione S-transferase P |
| P78417 | GSTO1_HUMAN | 27,833 | Glutathione transferase omega-1 |
| P62826 | RAN_HUMAN | 24,448 | GTP-binding nuclear protein Ran |
| P00492 | HPRT_HUMAN | 24,661 | Hypoxanthine-guanine phosphoribosyltransferase |
| P00338 | LDHA_HUMAN | 36,819 | l-Lactate dehydrogenase A chain |
| P07195 | LDHB_HUMAN | 36,769 | l-Lactate dehydrogenase B chain |
| P14174 | MIF_HUMAN | 12,508 | Macrophage migration inhibitory factor |
| P40925 | MDHC_HUMAN | 36,500 | Malate dehydrogenase, cytoplasmic |
| P30043 | BLVRB_HUMAN | 22,088 | Flavin reductase |
| P15531 | NDKA_HUMAN | 17,309 | Nucleoside diphosphate kinase A |
| P22392 | NDKB_HUMAN | 17,401 | Nucleoside diphosphate kinase B |
| P62937 | PPIA_HUMAN | 18,098 | Peptidyl-prolyl cis-trans isomerase A |
| P30041 | PRDX6_HUMAN | 25,002 | Peroxiredoxin-6 |
| P00558 | PGK1_HUMAN | 44,854 | Phosphoglycerate kinase 1 |
| Q99497 | PARK7_HUMAN | 20,050 | Protein DJ-1 |
| P06703 | S10A6_HUMAN | 10,230 | Protein S100-A6 |
| P00491 | PNPH_HUMAN | 32,325 | Purine nucleoside phosphorylase |
| P00441 | SODC_HUMAN | 16,023 | Superoxide dismutase [Cu-Zn] |
| P37837 | TALDO_HUMAN | 37,688 | Transaldolase |
| P60174 | TPIS_HUMAN | 26,807 | Triosephosphate isomerase |
| P62988 | UBIQ_HUMAN | 8560 | Ubiquitin |
3.3. Urine
A total of 1025 proteins were identified in the entire set of urine samples (Supplemental Table 3), with an average of 257 proteins per individual specimen (Fig. 2B). Uromodulin, serum albumin, keratins, various IgGs, actin, and collagen were among the proteins detected in the majority of samples. From the composite list of proteins identified in all the urine samples, we selected those that were observed in at least 5 specimens and compared this list to a published urinary proteome with 1543 identifications [43]. We note that these investigators used much larger protein amounts than were available in our study and two separation methods at the protein level (reversed phase HPLC and SDS PAGE). Ingenuity Pathway Analysis (IPA) software was used to compare the two data sets. A total of 488 common proteins were identified. Distribution of the detected proteins across different subcellular locations and functional categories was very similar for both datasets as defined by the Gene Ontology Project (http://www.geneontology.org/). Finally, we performed hierarchical clustering of the urine protein dataset. No protein subgroups emerged (data not shown).
3.4. Omental fat
In total, the gel LC–MS/MS workflow identified 1176 proteins in the entire set of OF samples (Supplemental Table 4), with an average of 425 proteins per sample, the highest number of identifications per specimen for any of the biological media that were analyzed in this study (Fig. 2C). The typical distribution of proteins detected per sample is slightly skewed due to the lower number of samples in this portion of the study (17). No clustering of the samples due to abundance differences was observed, but this was not unexpected; after unblinding on disease status it was found that only 3 of the 17 samples analyzed were from women with endometriosis. Ingenuity Pathway Analysis showed that the OF proteome functions in an interesting array of pathways including superoxide radical degradation, metabolism, and retinoic acid signaling.
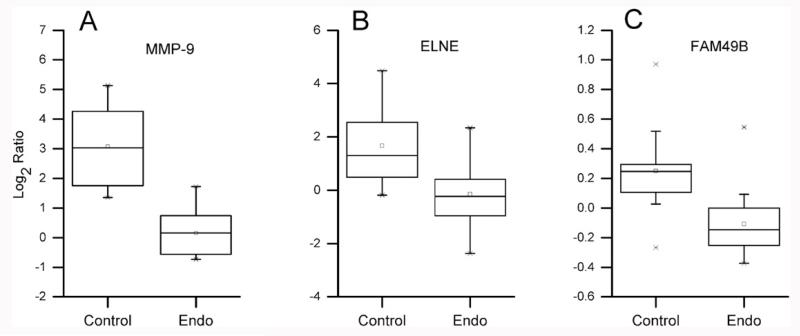
Given the depth of coverage of this proteome, the paucity of cases in our first analysis and that fat is a depot for EDCs, we went on to perform an iTRAQ analysis aimed at identifying proteins that were differentially expressed in a second set of 30 OF samples from endometriosis cases (n = 16) or control donors (n = 14). Previously we showed that concentrations of lipophilic organochlorine pollutants were higher in omental fat than serum [33] and we theorized that this could be associated with endometriosis-associated changes in protein pathways in these cells. From the OF iTRAQ analysis, 3026 proteins (Supplemental Table 5) were identified with 1605 common to all sets. The larger number of identifications was likely due to the fact that peptides were separated by high pH reversed phase chromatography prior to LC–MS/MS analyses of the individual fractions. Proteins detected in ≥5 women without and ≥5 women with endometriosis (2544) were compared by using linear regression analysis. Matrix metalloproteinase-9 (MMP-9; p = 0.0003; FDR q = 0.06), neutrophil elastase (ELNE; p = 0.0013; FDR q = 0.08) and protein FAM49B (p = 0.0015; FDR q = 0.2) were found to be in lower abundance in the OF samples obtained from endometriosis patients as compared to control individuals (Fig. 4).
Fig. 4. iTRAQ analyses of peritoneal fat samples from women with endometriosis as compared to unaffected women revealed disease-associated decreases in protein abundances. The downregulated proteins were: (A) matrix metalloproteinase-9 (MMP-9), fold change = −7.6; (B) neutrophil elastase (ELNE), fold change = −3.5; and (C) FAM49B, fold change = −1.3.
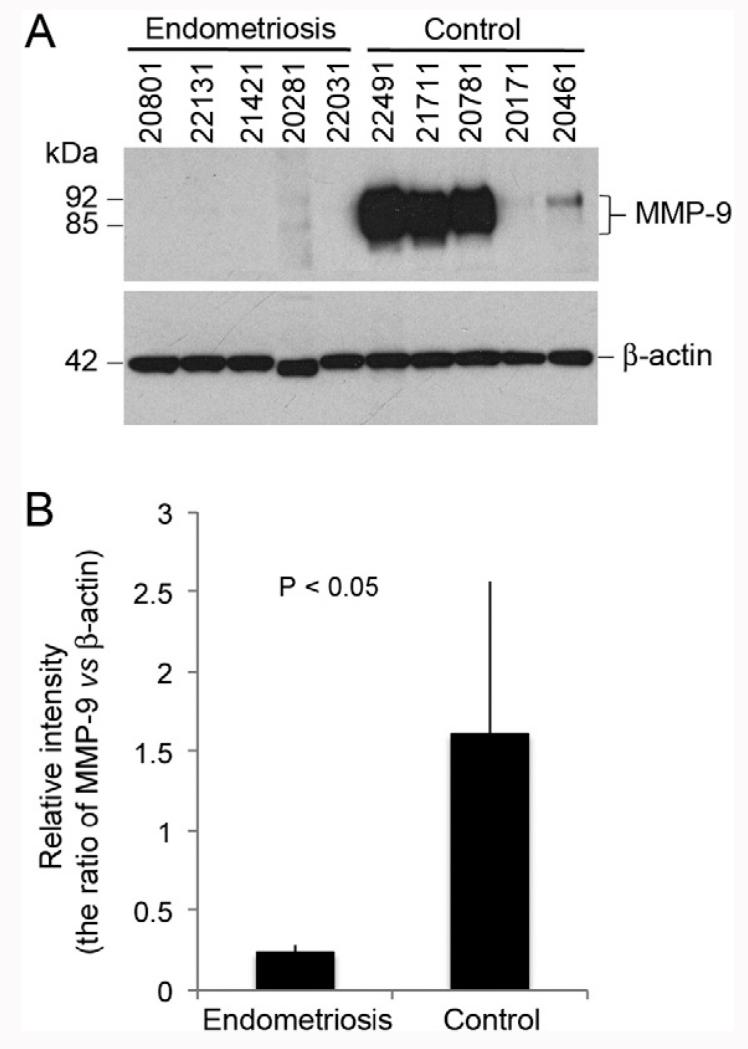
To confirm the differential abundance of these proteins, we applied an immunoblot approach to a randomly chosen subset of the samples (Fig. 5A). The results showed that MMP-9 levels, which varied among the samples from women without endometriosis, were consistently lower in the patient group. The average relative difference in MMP-9 abundance in the patient vs. control samples was 6.5-fold (Fig. 5B), in good agreement with the iTRAQ quantification data (Fig. 4A and Supplemental Table 5). Attempts to use the same approach for the relative quantitation of ELNE and FAM49B were not successful due to antibody cross-reactivity with numerous proteins in OF (data not shown).
Fig. 5.
Immunoblot verification of differential expression of matrix metalloproteinase-9 (MMP9) in OF from women with endometriosis. Sample identification numbers are shown at the top of each lane (A). Lanes 1–5 contained OF from women with endometriosis; lanes 6–10 contained OF from control patients. MMP-9 levels were normalized to the signal from the β-actin loading control (B).
3.5. Omental fat: EDC levels and proteins that are differentially expressed in endometriosis
With regard to the associations between chemicals and proteins, two important observations emerged (Table 2). First, levels of five of the chemicals (PBDE #183, PCB151, PCB156, γ-HCH, AND PCB28) measured in omental fat were significantly associated with two of the proteins found at altered levels in women with endometriosis (ELNE and FAM49B). Second, FA49B had the most chemical signals, which included γ-HCH, PBDE #183 and PCB congeners #28, 74, 151, 156, and 201. Finally, PBDE #183 was the only chemical associated with two proteins. The null hypothesis was rejected indicating that FAM49B was associated with at least one of the eight chemicals investigated.
Table 2. Associations between down-regulated proteins detected in OF from women with endometriosis with significant persistent EDCs in plasma.
| EDC | Matrix metalloproteinase-9 |
Neutrophil elastase |
Protein FAM49B |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Coeff | p value | R2 | Coeff | p value | R2 | Coeff | p value | R2 | |
| γ-HCH | 35.81 | 0.92 | 0 | −103.94 | 0.43 | 0.02 | −0.31 | 0.01 | 0.23 |
| PBDE183 | 124.16 | 0.14 | 0.13 | −55.71 | 0.03 | 0.15 | −0.05 | 0.03 | 0.16 |
| PBDE47 | −323.17 | 0.23 | 0.09 | −4.35 | 0.96 | 0 | 0.02 | 0.78 | 0 |
| PCB201 | 303.96 | 0.21 | 0.1 | −40.09 | 0.63 | 0.01 | −0.09 | 0.26 | 0.05 |
| PCB156 | −41.78 | 0.80 | 0 | 18.67 | 0.68 | 0.01 | 0.08 | 0.04 | 0.14 |
| PCB151 | 373.04 | 0.34 | 0.06 | −72.53 | 0.55 | 0.01 | −0.23 | 0.04 | 0.14 |
| PCB74 | −91.32 | 0.66 | 0.01 | 5.32 | 0.92 | 0 | 0.04 | 0.43 | 0.02 |
| PCB28 | 178.53 | 0.36 | 0.05 | 25.96 | 0.65 | 0.01 | −0.11 | 0.04 | 0.15 |
4. Discussion
The women who provided PF, urine and OF were recruited as participants in the ENDO Study funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), an institute of the National Institutes of Health (NIH). This multi-year study was implemented to learn how environmental exposure to EDCs and lifestyle factors may affect women’s reproductive health. In particular, the ENDO Study focused on how hormonally active chemicals affect the risk of developing endometriosis. This proteomics study was performed as a first step to determine whether exposure to EDCs associated with endometriosis was correlated with relative abundances of proteins identified in PF, urine or OF in women diagnosed with endometriosis relative to women without disease.
Research regarding the effects of endometriosis on various proteomes has primarily relied on mass spectrometry (MS)-based methods at a global level. The emphasis has been on cataloging the endometrial, urine or PF proteomes and identifying proteins that are differentially expressed between samples collected from women diagnosed with endometriosis as compared to women without disease. To date, the majority of these analyses have utilized differential SDS-PAGE followed by either LC MS/MS using matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI). For example, the 2D SDS-PAGE patterns of eutopic endometrium from stage II to IV endometriosis cases were compared to equivalent samples from control women [44].
MS analyses of 48 spots that had unique expression patterns in the cases revealed 70 differentially expressed proteins [44]. Another study used the same experimental approach to identify 11 proteins that were differentially expressed in PF that was collected from endometriosis patients or control individuals; 8 were implicated in endometriosis pathogenesis [12]. The same group used an ELISA approach to confirm downregulation of one of the differentially expressed proteins, hemopexin, in a larger set of case and control PF samples [45]. In similarly designed studies, 12 other proteins were identified as upregulated in PF obtained from patients vs. controls [46,47]. 2D SDS-PAGE and MS/MS were also used to identify differentially expressed proteins in PF from women with endometriosis who were: 1) fertile as compared to their infertile counterparts; or 2) treated with a gonadotropin-releasing hormone (GnRH) analogue as compared to untreated control women [13,14]. The former study showed upregulation of proteins with immune functions in the infertile group and the latter study showed that a GnRH analogue downregulates the expression of proteins in this class. The urine proteome has also been studied in the context of endometriosis, a biospecimen of particular interest in the search for clinically useful biomarkers. One group used MALDI TOF MS to identify disease-specific urinary peptide profiles [48]. In addition, MS approaches identified several isoforms of cytokeratin 19 [9] and matrix metalloproteinase [49] as upregulated in urine from women with biopsy-confirmed endometriosis.
As to the experimental strategy employed in this study, we utilized two MS-based proteomic pipelines to investigate the protein repertoire of PF, urine and OF samples that were obtained from women with and without endometriosis. In total, we analyzed samples from 63 women (17 with and 46 without endometriosis) from one participating site, and another 30 samples (16 women with and 14 without endometriosis) from a second research site. One strategy was a semi-quantitative gel LC–MS workflow that consisted of SDS PAGE protein fractionation, in-gel trypsin digestion and LC–MS/MS analyses. The goal was a broad overview of these samples, which were collected by several clinics. We reasoned that the SDS PAGE step would enable identification of samples that were outliers in terms of protein degradation, a potentially important issue in studies that employ clinical specimens. As was expected, variations in banding patterns among samples of the same type were noted, but frank proteolysis was not evident, suggesting that the specimens in the ENDO bank are generally of high quality. The second workflow utilized an iTRAQ approach for relative quantification of proteins in a second set of OF samples. This enabled not only the identification of differentially expressed proteins, but also a secondary analysis in which the levels of differentially expressed proteins were associated with levels of an EDC panel that was measured in the same samples.
With regard to the results, PF was the only specimen type in which the gel LC–MS/MS approach differentially detected proteins in a subset of the samples. In this regard, both groups of women had surgical indications for laparoscopy/laparotomy. Given the often occult nature of this disease, we were interested in possible associations with this condition. The majority of differentially detected proteins have not been reported as associated with endometriosis. This category included several proteins with metabolic functions such asglucose metabolism (e.g., phosphoglycerate kinase-1, fructose-bisphosphate aldolase A, transaldolase, triosephosphate isomerase, malate dehydrogenase), possible evidence of more global disease-induced dysregulation, involving the peritoneal cavity. The detection of superoxide dismutase, catalase and glutathione transferase (omega-1) in PF samples from patient donors could be indicative of oxidative stress, an interesting observation given that reactive oxygen species have been implicated in the progression of endometriosis [50]. Another differentially detected protein, glutathione S-transferase P, which is involved in detoxification, also has functions that could be related to the presence of endometriotic lesions. Thus, the proteins that were differentially detected in patient samples had potential functional relevance to the disease process.
A surprising number of proteins have been reported as having dysregulated expression in these lesions or as playing a role in the disease process. Proteins in the category of disease-related changes in expression include purine nucleotide phosphorylase [51] and peroxiredoxin 6 [52]. Interestingly, a subgroup of women with endometriosis has autoantibodies to native and linear epitope of carbonic anhydrase [53], possible indirect evidence of upregulated expression of this protein as well. Other groups have pointed to disease-associated alterations in post-translational modifications [52], including ubiquitination [54]. In this regard, we also identified ubiquitin in this study.
Several of the differentially detected proteins have been directly implicated, at the level of the endometrium, in the disease process. Overexpression of cofilin-1 in endometrial stromal cells (ESCs) upregulates proliferation, adhesion, invasion and angiogenesis; silencing of this gene attenuates these processes [55]. Elevated levels of macrophage migration inhibitory factor (MIF) have been reported in PF samples from women with endometriosis with higher concentrations correlating with infertility [56]. In the same publication, these investigators reported that anti-MIF inhibited the ability of PF to induce proliferation of microvascular endothelial cells. Other work shows that a MIF antagonist inhibits the development of endometriosis in a nude mouse model [57]. Finally, DJ-1, which is differentially upregulated in endometriotic lesions [44], has been implicated in the pathogenesis of endometriosis by mechanisms that include modulation of the PI3/Akt pathway via PTEN [58]. The fact that these proteins were differentially detected in patient PF samples is additional evidence of a role in the pathogenesis of endometriosis.
We note that the differentially expressed proteins detected in PF did not correlate with the diagnosis of endometriosis. Perhaps menstrual cycle phase or disease severity could be among the reasons. In addition, due to the stochastic nature of MS detection, only proteins present at equally high abundance in samples from women with and without endometriosis were detected. In this regard, future experiments employing strategies that deplete higher abundance proteins prior to MS analysis would enable a deeper survey of the PF proteome.
With regard to the iTRAQ approach that was used for relative quantification of proteins in a second set of OF samples, the results showed disease-associated downregulation of MMP-9, ELNE and FAM49B. Observations concerning MMP-9 and ELNE are in contrast to the upregulation of proteinases that is usually associated with endometriotic tissue. For example, an axis involving TNF-α, MMP-9 and a new isoform of steroid receptor coactivator-1 was recently shown to be involved in the progression of endometriosis [59]. In this regard, MIF promotes MMP-9 production [58]. Little information exists about ELNE in this disease, but the secretory leukocyte inhibitor/ELNE ratio increases [60], possible evidence of a compensatory mechanism. These results suggested that endometriosis has disparate effects on proteinase production by uterine tissue as compared to the peritoneum, which is consistent with cell-type specific actions and possibly proximal vs. distal influences of the disease milieu. The function of FAM49B has yet to be elucidated, but it has been associated with diseases that are related to immune dysregulation, such as multiple sclerosis [61]. Interestingly, immune dysfunction has also been implicated in the pathogenesis of endometriosis [16]. Finally, other hypotheses emerged from this work. For example, coalescing the results of the iTRAQ workflow applied to OF and a previous quantitative analysis of EDCs in the same samples revealed a possible EDC association. Future analyses will explore this theory as well as EDC levels in the context of the proteome at the level of the endometriotic lesions.
5. Conclusions
The 1D LC–MS/MS approach failed to detect proteins in PF, urine or OF samples that were differentially associated with endometriosis. However, this approach revealed a suite of proteins in PF that were detected in only a subset of the samples, some of which have been reported as abnormally expressed in endometriosis or associated with this disease process. Further investigation is needed to determine whether any of these changes are associated with occult or early-stage disease. The iTRAQ approach that was used for relative quantification of proteins in a second set of OF samples showed the disease-associated downregulation of MMP-9, ELNE and FAM49B. Levels of the latter two proteins inversely correlated with the concentration of several chemicals, including PBDE #183, γ-HCH, and PCBs measured in the same samples. Thus, the results of these experiments show the feasibility of associating endometriosis with changes in the protein repertoire and EDC levels.
Supplementary Material
Acknowledgments
Funded by the Intramural Research Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (contracts NO1-DK-6-3428; NO1-DK-6-3427; 10001406-02). Ethicon Endo-Surgery, LLC, kindly donated the HARMONIC® ACE 36P shears and scalpel blades for use in the study through a signed Materials Transfer Agreement with the University of Utah and the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The UCSF Sandler-Moore Mass Spectrometry Core Facility acknowledges support from a Sandler New Technology Award, a gift from the Gordon and Betty Moore Foundation, and NIH/NCI Cancer Center Support Grant P30 CA082103.
Footnotes
Transparency document
The http://dx.doi.org/10.1016/j.jprot.2014.09.015Transparencydocument associated with this article can be found, in the online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2014.09.015.
REFERENCES
- [1].Albalat A, Mischak H, Mullen W. Urine proteomics in clinical applications: technologies, principal considerations and clinical implementation. Prilozi. 2011;32:13–44. [PubMed] [Google Scholar]
- [2].Prudent M, Tissot JD, Lion N. Proteomics of blood and derived products: what’s next? Expert Rev Proteomics. 2011;8:717–37. doi: 10.1586/epr.11.58. [DOI] [PubMed] [Google Scholar]
- [3].Vitorino R, Guedes S, Manadas B, Ferreira R, Amado F. Toward a standardized saliva proteome analysis methodology. J Proteomics. 2012;75:5140–65. doi: 10.1016/j.jprot.2012.05.045. [DOI] [PubMed] [Google Scholar]
- [4].May KE, Conduit-Hulbert SA, Villar J, Kirtley S, Kennedy SH, Becker CM. Peripheral biomarkers of endometriosis: a systematic review. Hum Reprod Update. 2010;16:651–74. doi: 10.1093/humupd/dmq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rizner TL. Noninvasive biomarkers of endometriosis: myth or reality? Expert Rev Mol Diagn. 2014;14:365–85. doi: 10.1586/14737159.2014.899905. [DOI] [PubMed] [Google Scholar]
- [6].Fassbender A, Vodolazkaia A, Saunders P, Lebovic D, Waelkens E, De Moor B, et al. Biomarkers of endometriosis. Fertil Steril. 2013;99:1135–45. doi: 10.1016/j.fertnstert.2013.01.097. [DOI] [PubMed] [Google Scholar]
- [7].Cho S, Choi YS, Yim SY, Yang HI, Jeon YE, Lee KE, et al. Urinary vitamin D-binding protein is elevated in patients with endometriosis. Hum Reprod. 2012;27:515–22. doi: 10.1093/humrep/der345. [DOI] [PubMed] [Google Scholar]
- [8].Yun BH, Lee YS, Chon SJ, Jung YS, Yim SY, Kim HY, et al. Evaluation of elevated urinary enolase I levels in patients with endometriosis. Biomarkers. 2014;19:16–21. doi: 10.3109/1354750X.2013.863973. [DOI] [PubMed] [Google Scholar]
- [9].Tokushige N, Markham R, Crossett B, Ahn SB, Nelaturi VL, Khan A, et al. Discovery of a novel biomarker in the urine in women with endometriosis. Fertil Steril. 2011;95:46–9. doi: 10.1016/j.fertnstert.2010.05.016. [DOI] [PubMed] [Google Scholar]
- [10].Kuessel L, Jaeger-Lansky A, Pateisky P, Rossberg N, Schulz A, Schmitz AA, et al. Cytokeratin-19 as a biomarker in urine and in serum for the diagnosis of endometriosis—a prospective study. Gynecol Endocrinol. 2014;30:38–41. doi: 10.3109/09513590.2013.856409. [DOI] [PubMed] [Google Scholar]
- [11].Wang C, Fang W, Lee CS. Recent advances in capillary electrophoresis-based proteomic techniques for biomarker discovery. In: Volpi N, Maccari F, editors. Capillary Electrophoresis of Biomolecules Methods in Molecular Biology. Humana Press; 2013. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- [12].Wolfler MM, Meinhold-Heerlein IM, Sohngen L, Rath W, Knuchel R, Neulen J, et al. Two-dimensional gel electrophoresis in peritoneal fluid samples identifies differential protein regulation in patients suffering from peritoneal or ovarian endometriosis. Fertil Steril. 2011;95:2764–8. doi: 10.1016/j.fertnstert.2011.03.061. [DOI] [PubMed] [Google Scholar]
- [13].Ferrero S, Gillott DJ, Remorgida V, Anserini P, Ragni N, Grudzinskas JG. GnRH analogue remarkably down-regulates inflammatory proteins in peritoneal fluid proteome of women with endometriosis. J Reprod Med. 2009;54:223–31. [PubMed] [Google Scholar]
- [14].Ferrero S, Gillott DJ, Remorgida V, Anserini P, Ragni N, Grudzinskas JG. Proteomic analysis of peritoneal fluid in fertile and infertile women with endometriosis. J Reprod Med. 2009;54:32–40. [PubMed] [Google Scholar]
- [15].Perez-Perez R, Garcia-Santos E, Ortega-Delgado FJ, Lopez JA, Camafeita E, Ricart W, et al. Attenuated metabolism is a hallmark of obesity as revealed by comparative proteomic analysis of human omental adipose tissue. J Proteomics. 2012;75:783–95. doi: 10.1016/j.jprot.2011.09.016. [DOI] [PubMed] [Google Scholar]
- [16].Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362:2389–98. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Buck Louis GM, Hediger ML, Peterson CM, Croughan M, Sundaram R, Stanford J, et al. Incidence of endometriosis by study population and diagnostic method: the ENDO study. Fertil Steril. 2011;96:360–5. doi: 10.1016/j.fertnstert.2011.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sampson JA. Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:422–69. [Google Scholar]
- [19].Halban J. Hysteroadenosis metastica. Wien Klin Wochenschr. 1924;37:1205–28. [Google Scholar]
- [20].Ulukus M, Arici A. Immunology of endometriosis. Minerva Ginecol. 2005;57:237–48. [PubMed] [Google Scholar]
- [21].Meyer R. Uber entzundliche neterope epithelwucherungen im weiblichen Genetalg ebiet und uber eine bis in die Wurzel des Nesocolon ausgedehnte benigne Wucherung des Dar mepithel. Virch Arch Pathol Anat. 1909;195:487. [Google Scholar]
- [22].Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–79. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- [23].Berkley KJ, Rapkin AJ, Papka RE. The pains of endometriosis. Science. 2005;308:1587–9. doi: 10.1126/science.1111445. [DOI] [PubMed] [Google Scholar]
- [24].Tokushige N, Markham R, Russell P, Fraser IS. Nerve fibres in peritoneal endometriosis. Hum Reprod. 2006;21:3001–7. doi: 10.1093/humrep/del260. [DOI] [PubMed] [Google Scholar]
- [25].Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, et al. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pellacani C, Buschini A, Galati S, Mussi F, Franzoni S, Costa LG. Evaluation of DNA damage induced by 2 polybrominated diphenyl ether flame retardants (BDE-47 and BDE-209) in SK-N-MC cells. Int J Toxicol. 2012;31:372–9. doi: 10.1177/1091581812449663. [DOI] [PubMed] [Google Scholar]
- [27].Cobellis L, Latini G, De Felice C, Razzi S, Paris I, Ruggieri F, et al. High plasma concentrations of di-(2-ethylhexyl)-phthalate in women with endometriosis. Hum Reprod. 2003;18:1512–5. doi: 10.1093/humrep/deg254. [DOI] [PubMed] [Google Scholar]
- [28].Louis GM, Weiner JM, Whitcomb BW, Sperrazza R, Schisterman EF, Lobdell DT, et al. Environmental PCB exposure and risk of endometriosis. Hum Reprod. 2005;20:279–85. doi: 10.1093/humrep/deh575. [DOI] [PubMed] [Google Scholar]
- [29].Reddy BS, Rozati R, Reddy BV, Raman NV. Association of phthalate esters with endometriosis in Indian women. BJOG. 2006;113:515–20. doi: 10.1111/j.1471-0528.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- [30].Signorile PG, Spugnini EP, Mita L, Mellone P, D’Avino A, Bianco M, et al. Pre-natal exposure of mice to bisphenol A elicits an endometriosis-like phenotype in female offspring. Gen Comp Endocrinol. 2010;168:318–25. doi: 10.1016/j.ygcen.2010.03.030. [DOI] [PubMed] [Google Scholar]
- [31].Kim SH, Chun S, Jang JY, Chae HD, Kim CH, Kang BM. Increased plasma levels of phthalate esters in women with advanced-stage endometriosis: a prospective case-control study. Fertil Steril. 2011;95:357–9. doi: 10.1016/j.fertnstert.2010.07.1059. [DOI] [PubMed] [Google Scholar]
- [32].Signorile PG, Spugnini EP, Citro G, Viceconte R, Vincenzi B, Baldi F, et al. Endocrine disruptors in utero cause ovarian damages linked to endometriosis. Front Biosci (Elite Ed) 2012;4:1724–30. doi: 10.2741/493. [DOI] [PubMed] [Google Scholar]
- [33].Buck Louis GM, Chen Z, Peterson CM, Hediger ML, Croughan MS, Sundaram R, et al. Persistent lipophilic environmental chemicals and endometriosis: the ENDO Study. Environ Health Perspect. 2012;120:811–6. doi: 10.1289/ehp.1104432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Buck Louis GM, Peterson CM, Chen Z, Croughan M, Sundaram R, Stanford J, et al. Bisphenol A and phthalates and endometriosis: the endometriosis: Natural History, Diagnosis and Outcomes Study. Fertil Steril. 2013;100:162–9. e1–2. doi: 10.1016/j.fertnstert.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dowell JA, Frost DC, Zhang J, Li L. Comparison of two-dimensional fractionation techniques for shotgun proteomics. Anal Chem. 2008;80:6715–23. doi: 10.1021/ac8007994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–26. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- [38].Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- [39].Zhou Y, McMaster M, Woo K, Janatpour M, Perry J, Karpanen T, et al. Vascular endothelial growth factor ligands and receptors that regulate human cytotrophoblast survival are dysregulated in severe preeclampsia and hemolysis, elevated liver enzymes, and low platelets syndrome. Am J Pathol. 2002;160:1405–23. doi: 10.1016/S0002-9440(10)62567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Johnson-Restrepo B, Kannan K, Rapaport DP, Rodan BD. Polybrominated diphenyl ethers and polychlorinated biphenyls in human adipose tissue from New York. Environ Sci Technol. 2005;39:5177–82. doi: 10.1021/es050399x. [DOI] [PubMed] [Google Scholar]
- [41].Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, et al. Performance metrics for liquid chromatography–tandem mass spectrometry systems in proteomics analyses. Mol Cell Proteomics. 2010;9:225–41. doi: 10.1074/mcp.M900223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–72. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- [43].Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rai P, Kota V, Deendayal M, Shivaji S. Differential proteome profiling of eutopic endometrium from women with endometriosis to understand etiology of endometriosis. J Proteome Res. 2010;9:4407–19. doi: 10.1021/pr100657s. [DOI] [PubMed] [Google Scholar]
- [45].Wolfler MM, Meinhold-Heerlein IM, Henkel C, Rath W, Neulen J, Maass N, et al. Reduced hemopexin levels in peritoneal fluid of patients with endometriosis. Fertil Steril. 2013;100:777–81. doi: 10.1016/j.fertnstert.2013.05.010. [DOI] [PubMed] [Google Scholar]
- [46].Ferrero S, Gillott DJ, Remorgida V, Anserini P, Leung KY, Ragni N, et al. Proteomic analysis of peritoneal fluid in women with endometriosis. J Proteome Res. 2007;6:3402–11. doi: 10.1021/pr060680q. [DOI] [PubMed] [Google Scholar]
- [47].Ferrero S, Gillott DJ, Remorgida V, Anserini P, Ragni N, Grudzinskas JG. Peritoneal fluid proteome in women with different ASRM stages of endometriosis. Gynecol Endocrinol. 2008;24:433–41. doi: 10.1080/09513590802173824. [DOI] [PubMed] [Google Scholar]
- [48].El-Kasti MM, Wright C, Fye HK, Roseman F, Kessler BM, Becker CM. Urinary peptide profiling identifies a panel of putative biomarkers for diagnosing and staging endometriosis. Fertil Steril. 2011;95:1261–6. e1–6. doi: 10.1016/j.fertnstert.2010.11.066. [DOI] [PubMed] [Google Scholar]
- [49].Becker CM, Louis G, Exarhopoulos A, Mechsner S, Ebert AD, Zurakowski D, et al. Matrix metalloproteinases are elevated in the urine of patients with endometriosis. Fertil Steril. 2010;94:2343–6. doi: 10.1016/j.fertnstert.2010.02.040. [DOI] [PubMed] [Google Scholar]
- [50].Ngo C, Chereau C, Nicco C, Weill B, Chapron C, Batteux F. Reactive oxygen species controls endometriosis progression. Am J Pathol. 2009;175:225–34. doi: 10.2353/ajpath.2009.080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, et al. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144:2870–81. doi: 10.1210/en.2003-0043. [DOI] [PubMed] [Google Scholar]
- [52].Stephens AN, Hannan NJ, Rainczuk A, Meehan KL, Chen J, Nicholls PK, et al. Post-translational modifications and protein-specific isoforms in endometriosis revealed by 2D DIGE. J Proteome Res. 2010;9:2438–49. doi: 10.1021/pr901131p. [DOI] [PubMed] [Google Scholar]
- [53].D’Cruz OJ, Wild RA, Haas GG, Jr, Reichlin M. Antibodies to carbonic anhydrase in endometriosis: prevalence, specificity, and relationship to clinical and laboratory parameters. Fertil Steril. 1996;66:547–56. doi: 10.1016/s0015-0282(16)58566-5. [DOI] [PubMed] [Google Scholar]
- [54].Hull ML, Escareno CR, Godsland JM, Doig JR, Johnson CM, Phillips SC, et al. Endometrial–peritoneal interactions during endometriotic lesion establishment. Am J Pathol. 2008;173:700–15. doi: 10.2353/ajpath.2008.071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xu YL, Wang DB, Liu QF, Chen YH, Yang Z. Silencing of cofilin-1 gene attenuates biological behaviours of stromal cells derived from eutopic endometria of women with endometriosis. Hum Reprod. 2010;25:2480–8. doi: 10.1093/humrep/deq197. [DOI] [PubMed] [Google Scholar]
- [56].Kats R, Collette T, Metz CN, Akoum A. Marked elevation of macrophage migration inhibitory factor in the peritoneal fluid of women with endometriosis. Fertil Steril. 2002;78:69–76. doi: 10.1016/s0015-0282(02)03189-8. [DOI] [PubMed] [Google Scholar]
- [57].Khoufache K, Bazin S, Girard K, Guillemette J, Roy MC, Verreault JP, et al. Macrophage migration inhibitory factor antagonist blocks the development of endometriosis in vivo. PLoS One. 2012;7:e37264. doi: 10.1371/journal.pone.0037264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rai P, Shivaji S. The role of DJ-1 in the pathogenesis of endometriosis. PLoS One. 2011;6:e18074. doi: 10.1371/journal.pone.0018074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Han SJ, Hawkins SM, Begum K, Jung SY, Kovanci E, Qin J, et al. A new isoform of steroid receptor coactivator-1 is crucial for pathogenic progression of endometriosis. Nat Med. 2012;18:1102–11. doi: 10.1038/nm.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shimoya K, Moriyama A, Ogata I, Nobunaga T, Koyama M, Azuma C, et al. Increased concentrations of secretory leukocyte protease inhibitor in peritoneal fluid of women with endometriosis. Mol Hum Reprod. 2000;6:829–34. doi: 10.1093/molehr/6.9.829. [DOI] [PubMed] [Google Scholar]
- [61].Gilli F, Navone ND, Perga S, Marnetto F, Caldano M, Capobianco M, et al. Loss of braking signals during inflammation: a factor affecting the development and disease course of multiple sclerosis. Arch Neurol. 2011;68:879–88. doi: 10.1001/archneurol.2011.32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.