Abstract
The inhibitor of DNA binding 2 (Id2) basic helix-loop-helix protein interacts genetically and physically with the pocket proteins (Rb, p107 and p130) and has been implicated as an oncogene. In other studies, however, Id2 has been shown to function as a tumor suppressor. Here, we studied the role of Id2 in a well characterized model of ocular cancer in which the three pocket proteins are inactivated by generating mice lacking one or both Id2 alleles. Id2 deficiency had no impact on tumorigenesis in the eye. Unexpectedly, however, Id2 loss significantly increased the rate of metastasis. Liver metastases in Id2 heterozygotes demonstrated significant decrease of Id2 expression and loss of the remaining Id2 allele, strongly suggesting that Id2 inactivation specifically was required for metastasis in this model. These findings provide new insights into the role of Id2 in metastasis.
Keywords: Id2, bHLH, Metastasis, Eye, Cancer, Retinoblastoma, p107, p130
Introduction
Inhibitor of DNA binding 2 (Id2) was initially identified as a dominant-negative inhibitor of basic helix-loop-helix (bHLH) proteins by interfering with their heterodimerization [1]. Id2 has also been shown to interact genetically and physically with the retinoblastoma (Rb) protein and the other pocket proteins p107 and p130 [2]. Rb-null mice die by E 14.5 of defects in neurogenesis, hematopoiesis, muscle development and other abnormalities [3]; however, Id2-Rb double knockout embryos survive to term with minimal or no defects in neurogenesis and hematopoiesis [4]. In Rb+/− mice that develop pituitary tumors, loss of Id2 resulted in a marked reduction in tumor growth and angiogenesis [5]. These findings suggested that Rb inhibits the action of Id2 on its targets, and that release of Id2 associated with Rb loss has an oncogenic effect during tumor progression. In contrast, other studies implicate Id2 as a tumor suppressor. Id2 is important for maintaining a differentiated state and noninvasive phenotype in normal breast cells and breast cancer [6], and Id2 induces differentiation and suppresses tumor formation in the intestinal epithelium [7]. Id2 is also inactivated in ocular melanoma [8].
To explore further the role of Id2 in tumor development and progression, we studied the effect of Id2 loss in a well characterized Tyr-TAg transgenic mouse eye cancer model [9]. Tyr-TAg transgenic mice express simian virus 40 (SV40) large and small tumor antigens (TAg) under the control of the mouse tyrosinase promoter and develop bilateral ocular tumors that arise from the retinal pigment epithelium (RPE). Transformation by SV40 TAg is dependent on the inactivation of all three pocket proteins by binding to TAg oncogene [9]. We found that loss of one or both Id2 alleles had no effect on the development or size of primary tumors. Unexpectedly, however, loss of Id2 significantly increased the rate of metastasis to the liver. Thus, in this animal model Id2 does not mediate the tumorigenic effects of pocket protein inactivation, but rather, suppresses metastasis. These findings provide new insights into the role of Id2 in tumorigenesis and metastasis.
Materials and methods
Animals
Animal experiments were approved by the Washington University Animal Studies Committee and conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Tyr-TAg transgenic mice on a hybrid background (C57BL/6 × BALB/c) [10] were a gift from D. M. Albert (University of Wisconsin, Madison, WI). Id2+/− mice on 129/Sv background [11] were a gift from Y. Yokota (Fukui Medical University, Fukui, Japan). Tyr-TAg mice with all three Id2 genotypes (Id2+/+-wild type, Id2+/−-heterozygote, Id2−/−-null) were generated by backcross of F1 animals from breeding Tyr-TAg mice with Id2+/− mice to Id2 heterozygotes. Animals were genotyped for SV40 TAg transgene and Id2 status by PCR analysis of tail DNA as previously described [11, 12]. Tyr-TAg positive mice with all three Id2 genotypes on the mixed genetic background (129/Sv ranged from 50 to 93.5%) were used in this study (Table 1).
Table 1.
Summary of mice used for analysis
| Gender | Generation | Genotype | Total | ||
|---|---|---|---|---|---|
| Id2−/− | Id2+/− | Id2+/+ | |||
| M | F1 | 1 | 5 | 13 | 19 |
| N1 | 3 | 7 | 9 | 19 | |
| N2 | 2 | 11 | 9 | 22 | |
| N3 | 0 | 3 | 2 | 5 | |
| F | F1 | 0 | 1 | 16 | 17 |
| N1 | 0 | 8 | 12 | 20 | |
| N2 | 5 | 12 | 6 | 23 | |
| N3 | 0 | 3 | 4 | 7 | |
| Total | 11 | 50 | 71 | 132 | |
Analysis of primary and metastatic tumors
Animals were monitored closely for evidence of eye tumors and overall health. Mice were euthanized when they developed a large, ulcerating tumor in either eye, or at 8 months of age, whichever came first. Subsequently, a necropsy was performed, and the eyes, liver, lungs and brain were harvested, fixed in formalin and embedded in paraffin. Four micron sections were stained with hematoxylin and eosin and examined for the presence and morphology of primary and metastatic tumors. A photomicrograph of a section through the largest part of the tumor was obtained with the 4× objective, and the digital image analyzed using ImageJ software (http://rsb.info.nih.gov/ij/) to measure the area of the tumor as a percentage of the area of the entire posterior chamber of the eye. This method was chosen to minimize artifact in tumor size measurements resulting from variability in sectioning between eyes. Statistical analyses were performed as indicated using MedCalc software v. 10.4.0.0 (http:/www.medcalc.be).
DNA and RNA analysis in liver metastases
Five of 7 mice with liver metastasis were used for DNA and RNA analysis (two heterozygotes were excluded because the DNA and RNA were degraded as a result of tissue necrosis following metastasis-related death). Ten micron sections of the liver were mounted on slides covered with 2 µm PEN-membrane (Leica, Wetzlar, Germany), deparaffinized and stained with hematoxylin and eosin. Samples of normal liver and metastatic tissue were obtained by laser micro-dissection using Leica LMD 6 K (Leica). RNA and DNA were isolated from samples using Recover All Total Nucleic Acid Isolation Kit (Applied Biosystems, Forster City, CA) according to manufacturer instructions. Nucleic acid content was measured using the NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA was converted into cDNA using iScript (BioRad, Hercules, CA) and preamplified with TaqMan PreAmp Master Mix (Applied Biosystems). Genomic DNA (gDNA) and cDNA were amplified using SYBR Green JumpStart Taq ReadyMix (Sigma, St. Louis, MO) and increase in fluorescence was detected in real time using CFX96 PCR (BioRad). Primers for real-time PCR were designed for detecting the Id2 wild type gDNA allele (Forward 5′–AGT GCC AAC AAG TCC CAT GAA–3′, Reverse 5′–TGA GCG TCA TGT GAA ATC GCT A–3′) and Id2 mRNA (Forward 5′–CAC GGA CAT CAG CAT CCT GT–3′, Reverse 5′–CCC AAA TGC CAT TTA TTT AGC C–3′) with Primer Express v2.0 software (Apply Biosystems). β-actin was used as a reference gene (Forward 5′–TCCTCCTGAGCGCAAGTACTCT–3′, Reverse 5′–ACT CAT CGT ACT CCT GCT TGC TG–3′). Each primer pair was validated for efficiency using standard curve. Primer-dimer formation and nonspecific amplification were excluded by melting curve analysis. There was no amplification in “no reverse transcription” control and “no template control” with all primer sets. Relative gDNA content and mRNA expression was obtained using the standard curve method (User Bulletin #2, ABI PRISM 7700 Sequence Detection System, PE Applied Biosystems). Normal liver tissue from Id2+/+ mice served as a calibrator.
Results
Analysis of primary ocular tumors
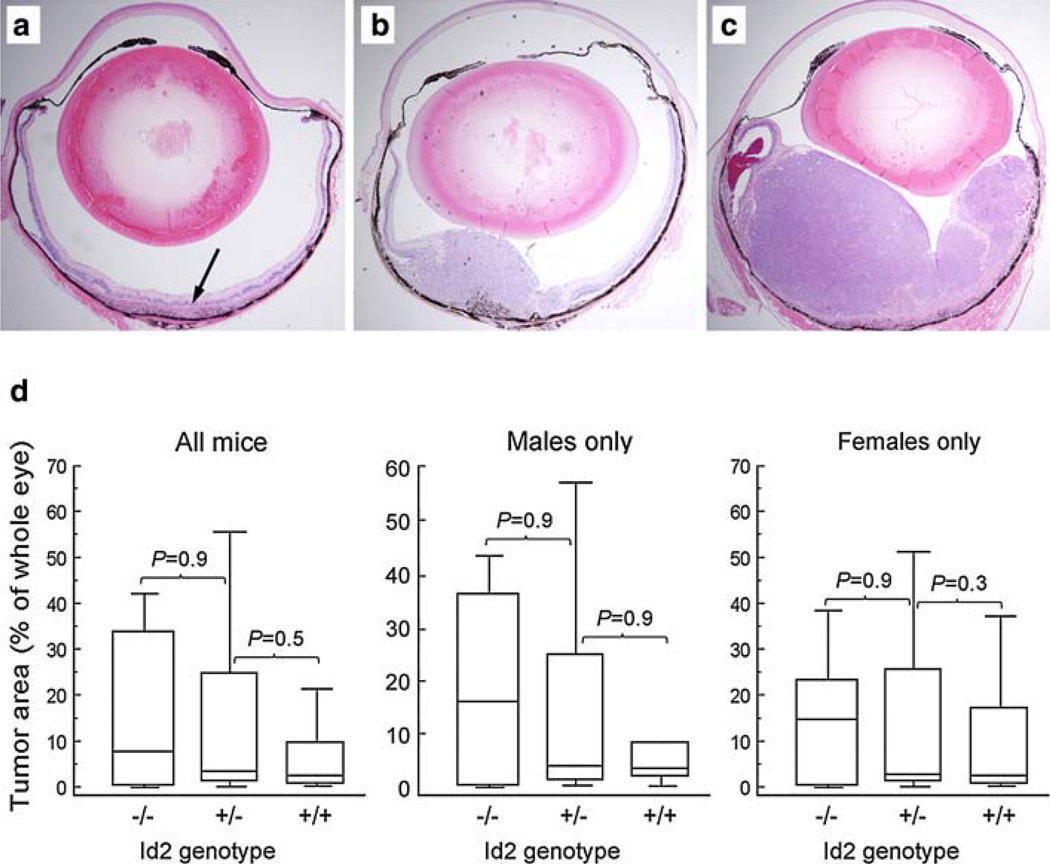
A total of 132 Tyr-TAg positive animals were analyzed from generations F1 (36 mice), N1 (39 mice), N2 (45 mice) and N3 (12 mice), including 65 males and 67 females (Table 1). Id2 genotype was Id2+/+ in 71 mice, Id2+/− in 50 mice, and Id2−/− in 11 mice. All mice developed bilateral (113 mice) or unilateral (19 mice) intraocular tumors. Primary intraocular tumors arose from the retinal pigment epithelium and exhibited malignant features as previously described [10, 13]. No difference was observed in cell morphology, nuclear pleiomorphism, intratumoral vascularity, or mitotic figures between mice with the three Id2 genotypes (data not shown). Fifty mice (25 males and 25 females; 18 Id2+/+, 26 Id2+/− and 6 Id2−/−) from the N2 and N3 generations were analyzed for tumor size. A total of 94 eyes were measured, six eyes were excluded due to inadequate ocular tissue for analysis. Tumor area as a percentage of the total area of the eye ranged from 0 to 67% (mean 12.9%, median 13.0%), which is typical of the amount of variation that we have previously observed in the parental mice (data not shown). The images of representative small (less than 5% of posterior chamber area), medium (5–50%) and large (more than 50%) tumors are presented on Fig. 1a–c. The tumor area did not differ significantly between mice with the three Id2 genotypes (Fig. 1d).
Fig. 1.
Histopathologic photomicrographs of intraocular primary tumors demonstrating a typical small tumor (arrow) (a), medium tumor (b), and large tumor (c). Original magnification 4×. d Box-and-whiskers plots showing a primary intraocular tumor area as a percentage of the total area of the eye. Males and females are analyzed separately as indicated. The central box represents the values from the 25 to 75th percentile. The middle line represents the median. A line extends from the minimum to the maximum value. P values were calculated using the Mann–Whitney nonparametric method
Analysis of metastatic tumors
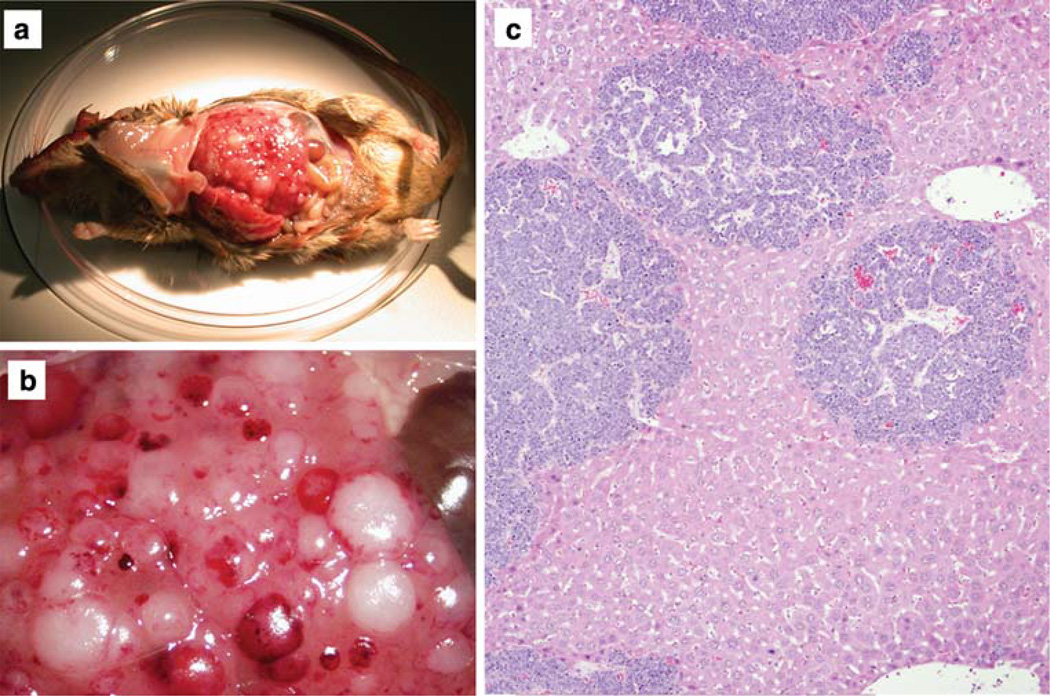
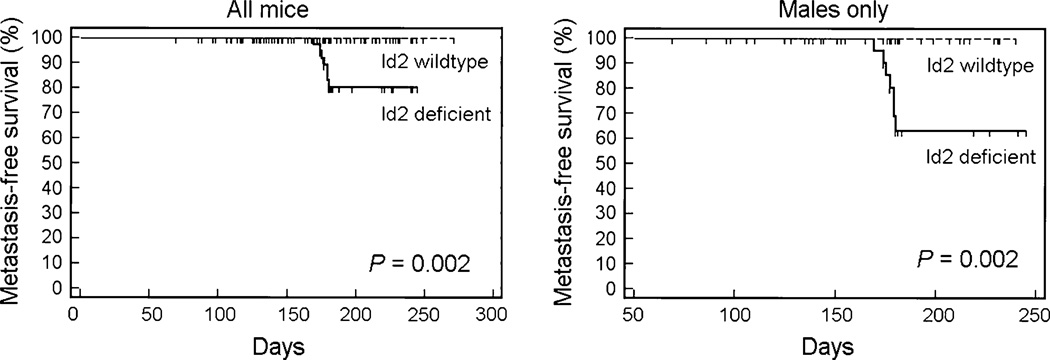
Consistent with the previous observation that metastasis is very rare in parental Tyr-TAg mice [13], metastatic disease was detected in none of the Tyr-TAg/Id2+/+ mice (Table 2). However, metastasis was observed in seven of the Id2 deficient mice (6 Tyr-TAg/Id2+/− mice and one Tyr-TAg/Id2−/− mouse) (Table 2 and Fig. 2a–c). All seven mice developed metastasis to the liver, and one also had lung metastasis. The association between metastasis and Id2 loss was highly significant (log rank test, P = 0.002) (Fig. 3). Interestingly, all seven mice with metastasis were males. The association between Id2 deficiency and metastasis among N1–N3 males was statistically significant (Fisher exact test, P = 0.04.). Only Id2 deficient males with medium or large primary tumors in at least one eye developed liver metastasis. There was a significant difference in average tumor size between Id2 deficient males with [30.3 ± 7.48 (SEM)] and without [10.8 ± 2.42 (SEM)] liver metastasis (P = 0.033 by unpaired Student’s t-Test).
Table 2.
Summary of mice with liver metastasis
| Gender | Generation | Liver metastasis | Id2 deficient mice |
||||
|---|---|---|---|---|---|---|---|
| Id2−/− | Id2+/− | Id2+/+ | Total | Total | % With mets |
||
| M | F1 | 0 | 1 | 0 | 1 | 6 | 16.7 |
| N1 | 0 | 1 | 0 | 1 | 10 | 10.0 | |
| N2 | 1 | 1 | 0 | 2 | 13 | 15.4 | |
| N3 | 0 | 3 | 0 | 3 | 3 | 100.0 | |
| All | 1 | 6 | 0 | 7 | 32 | 21.9 | |
| F | All | 0 | 0 | 0 | 0 | 29 | 0.0 |
Fig. 2.
Liver metastasis in a Tyr-TAg/Id2+/− mouse. a Low magnification external photograph. b High magnification external photograph, showing multiple foci of metastatic lesions on the liver surface. c Histopathologic photomicrograph showing multiple dark blue foci representing intrahepatic metastases. Original magnification 40×
Fig. 3.
Kaplan-Meier survival plots comparing metastasis-free survival as a function of Id2 genotype. Id2 wild type = Tyr-TAg/Id2+/+ mice. Id2 deficient = Tyr-TAg/Id2+/− + Tyr-TAg/Id2−/− mice. A sub-analysis of males only was performed as indicated. P values were calculated using the log rank test
Loss of Id2 alleles in metastatic tumors
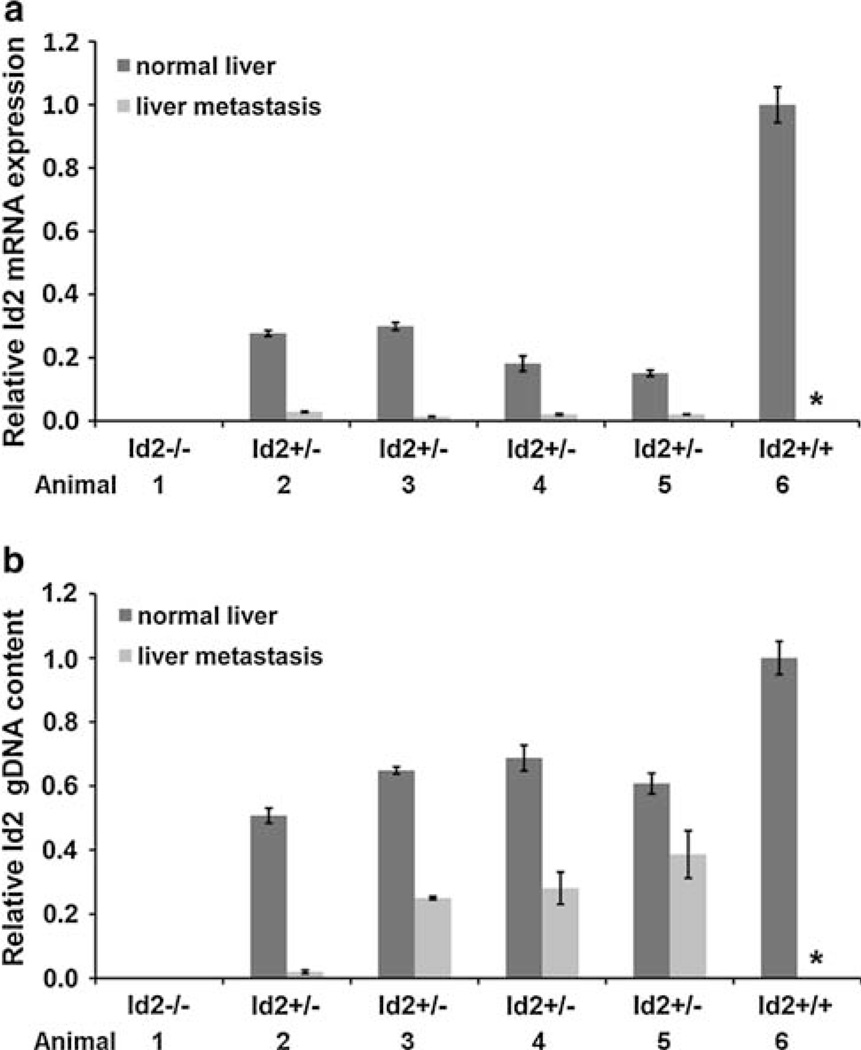
To determine whether Id2 may be specifically targeted for inactivation in Id2+/− metastatic tumors, we analyzed both Id2 mRNA expression (Fig. 4a) and Id2 gDNA content (Fig. 4b) in normal liver and liver metastasis from four Id2+/− mice. Normal liver and liver metastasis from Tyr-TAg/Id2−/− and normal liver from Tyr-TAg/Id2+/+ served as negative and positive control respectively. As expected, normal liver and liver metastasis in the Tyr-TAg/Id2−/− mouse exhibited no Id2 mRNA or wild type Id2 gDNA. Normal liver from Id2+/+ mice, which never developed liver metastasis, has higher mRNA expression and DNA content than normal liver from Id2+/− mice, consistent with one allele loss in heterozygotes. In Id2 heterozygous mice relative mRNA expression and gDNA content was significantly lower in liver metastasis than in normal liver (Fig. 4). A marked decrease of Id2 mRNA expression in liver metastasis of Tyr-TAg/Id2+/− mice was consistent with metastasis-specific inactivation of the remaining Id2 allele in the metastatic tumors. A loss of Id2 gDNA in metastatic tumors of Tyr-TAg/Id2+/− mice further supports to specifically targeted Id2 inactivation in the metastatic tumors.
Fig. 4.
Analysis of Id2 mRNA expression and Id2 gDNA content in normal liver and liver metastases by real-time PCR. a Id2 mRNA expression in normal liver and liver metastasis from Id2+/− mice normalized to β-actin relative to Id2+/+ normal liver. b Id2 gDNA content normal liver and liver metastasis from Id2+/− mice normalized to β-actin relative to Id2+/+ normal liver. Each bar graph represents mean and SEM for samples performed in triplicate. Specimens from Id2−/− and Id2+/+ mice served as negative and positive controls, respectively. The asterisk indicates that there were no metastatic tumors in Id2+/+ mice
Discussion
Based on previous work showing that Id2 loss could mitigate the effects of Rb inactivation in tumorigenesis, we predicted that loss of Id2 would lead to fewer and/or smaller primary tumors in this cancer model. Surprisingly, Id2 loss had no effect on the frequency of occurrence or the size of primary tumors. Unexpectedly, however, Id2 loss significantly increased the rate of metastasis. The finding that the Id2 gene was deleted in some metastatic tumors suggests strongly that Id2 was specifically targeted for silencing. Since loss of Id2 did not alter the primary tumor phenotype, our findings do not support a role for Id2 as a major effector of the tumorigenic consequences of pocket protein deficiency in this model. Rather, our findings suggest that Id2 functions downstream in tumor progression, at a stage where the primary tumor gains metastatic capacity.
Since Id2 plays a critical role in the development of peripheral lymphoid organs and natural killer cells [11], one might speculate that Id2 deficient mice are simply immunodeficient and unable to prevent the spread of metastatic tumor cells. However, Id2 heterozygotes in which most of the metastatic events were observed have normal immune function [14]. Rather than such an indirect effect, the silencing and deletion of the remaining Id2 allele in metastatic tumors in Id2 heterozygotes strongly implies that Id2 loss plays a direct role in metastasis. A curious finding was that metastasis occurred only in males. One potential explanation is that this was a chance occurrence and that a larger number of animals would have included females. Another possibility is that there is a genetic interaction between Id2 and factors on the X chromosome. Interestingly, the prognosis for survival is worse in males than females in human ocular melanoma [15]. This association needs to be explored in future studies.
This study provides further evidence that Id2 can function not only as an oncogene [4], and a tumor suppressor [6, 7], but also as a metastasis suppressor. In hepatocellular carcinoma, loss of Id2 promotes metastasis apparently by altering VEGF expression and cell motility [16]. In primary ocular melanoma, Id2 is frequently silenced during tumor progression and leads to a switch from non-metastasizing to highly metastatic tumors, at least in part by altering E-cadherin expression [8]. Thus, the role of Id2 appears to be tissue-specific and context-dependent [17]. These findings provide important new insights into the role of Id2 in cancer biology in general and metastasis in particular.
Acknowledgments
This research was supported by R01 EY1316905 from the National Eye Institute, Knights Templar Foundation, Research to Prevent Blindness, Horncrest Foundation (JWH), and departmental grants from Research to Prevent Blindness and National Eye Institute Vision Core Grant P30 EY 02687. The authors thank the Immunomorphology Core Lab for preparation of histopathologic sections.
Abbreviations
- Id2
Inhibitor of DNA binding 2
- Rb
Retinoblastoma
- bHLH
Basic helix-loop-helix
- SV40
Simian virus 40
- TAg
Large and small tumor antigens
- RPE
Retinal pigment epithelium
- gDNA
Genomic DNA
- VEGF
Vascular endothelial growth factor
References
- 1.Sun XH, Copeland NG, Jenkins NA, et al. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Molec Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Molec Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee EY, Chang CY, Hu N, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359(6393):288–894. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 4.Lasorella A, Noseda M, Beyna M, et al. Id2 is a retinoblastoma protein target and mediates signaling by Myc oncoproteins. Nature. 2000;407(6804):592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 5.Lasorella A, Rothschild G, Yokota Y, et al. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005;25(9):3563–3574. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itahana Y, Singh J, Sumida T, et al. Role of Id-2 in the maintenance of a differentiated and noninvasive phenotype in breast cancer cells. Cancer Res. 2003;63(21):7098–7105. [PubMed] [Google Scholar]
- 7.Russell RG, Lasorella A, Dettin LE, et al. Id2 drives differentiation and suppresses tumor formation in the intestinal epithelium. Cancer Res. 2004;64(20):7220–7225. doi: 10.1158/0008-5472.CAN-04-2095. [DOI] [PubMed] [Google Scholar]
- 8.Onken MD, Ehlers JP, Worley LA, et al. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006;66(9):4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalvide J, Stubdal H, DeCaprio JA. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18(3):1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syed NA, Windle JJ, Darjatmoko SR, et al. Transgenic mice with pigmented intraocular tumors: tissue of origin and treatment. Invest Ophthalmol Vis Sci. 1998;39(13):2800–2805. [PubMed] [Google Scholar]
- 11.Yokota Y, Mansouri A, Mori S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 12.Windle JJ, Albert DM, O’Brien JM, et al. Retinoblastoma in transgenic mice. Nature. 1990;343(6259):665–669. doi: 10.1038/343665a0. [DOI] [PubMed] [Google Scholar]
- 13.Bradl M, Klein-Szanto A, Porter S, et al. Malignant melanoma in transgenic mice. Proc Natl Acad Sci USA. 1991;88(1):164–168. doi: 10.1073/pnas.88.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikawa T, Fujimoto S, Kawamoto H, et al. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98(9):5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virgili G, Gatta G, Ciccolallo L, et al. Survival in patients with uveal melanoma in Europe. Arch Ophthalmol. 2008;126(10):1413–1418. doi: 10.1001/archopht.126.10.1413. [DOI] [PubMed] [Google Scholar]
- 16.Tsunedomi R, Iizuka N, Tamesa T, et al. Decreased ID2 promotes metastatic potentials of hepatocellular carcinoma by altering secretion of vascular endothelial growth factor. Clin Cancer Res. 2008;14(4):1025–1031. doi: 10.1158/1078-0432.CCR-07-1116. [DOI] [PubMed] [Google Scholar]
- 17.Kowanetz M, Valcourt U, Bergstrom R, et al. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol. 2004;24(10):4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]