Abstract
Purpose of review
To provide an update on the rapidly evolving methods for assessing prognosis and predicting response to targeted molecular therapy in uveal melanoma.
Recent findings
The techniques for assessing prognosis in uveal melanoma have evolved from simple physical features, such as tumor size, location, and cell morphology, to the slightly more sophisticated counting of chromosomal gains and losses. More recently, gene expression profiling has provided a highly accurate and biologically informative gold standard for molecular prognostication. The latest step in the evolution of molecular testing has been the recent discovery of major driver mutations that allow predictive testing of response to targeted molecular therapies. Mutations in GNAQ and GNA11 are early events that promote cell proliferation, and these mutations are sensitive to MAPK kinase, PKC, and AKT inhibitors. Mutations in BAP1, SF3B1, and EIF1AX are later events that are largely mutually exclusive. Mutations in BAP1 are strongly associated with metastasis, whereas those in SF3B1 and EIF1AX are associated with good prognosis. Uveal melanomas with BAP1 mutations demonstrate sensitivity to epigenetic modulators, such as histone deacetylase inhibitors. Clinical trials are now available to evaluate the efficacy of these targeted molecular agents in patients with uveal melanoma.
Summary
Molecular prognostic testing and enrollment of high-risk patients into clinical trials of targeted molecular therapy are rapidly becoming the standard of care in the management of uveal melanoma.
Keywords: BAP1, gene expression profiling, metastasis, prognosis, uveal melanoma
INTRODUCTION
Uveal melanoma is the most common primary intraocular malignancy, with an incidence of ~2000 new cases per year in the USA. Up to 50% of uveal melanoma patients are at risk of metastasis via hematogenous spread, most commonly to the liver [1]. Metastatic uveal melanoma is notoriously resistant to conventional chemotherapy. However, recent genetic discoveries have led to individualized patient management tools and targeted molecular therapies that are being evaluated in clinical trials. For the first time, rational molecular therapeutic interventions are possible in the treatment of high risk and metastatic uveal melanoma, which may result in improved patient survival.
CLINICOPATHOLOGIC PROGNOSTIC FACTORS
For many years, it has been known that clinicopathologic factors, such as increased patient age, increased thickness and diameter of the primary tumor, ciliary body involvement, extraocular tumor extension, and epithelioid cell type, are associated with an increased risk of metastasis [2,3]. These factors are useful for classification systems that group patients into categories based on similar prognosis, such as the American Joint Committee on Cancer Tumor-Node-Metastasis system [4]. However, such clinicopathologic classification systems are of limited value in making clinical management decisions in individual patients [5].
CHROMOSOMAL PROGNOSTIC FACTORS
For several decades, it was recognized that the copy number changes in certain chromosomes are associated with an increased risk for metastasis in uveal melanoma. The more informative changes include loss of chromosome 3 and gain of chromosome 8q [6–8]. Although cytogenetic markers are more accurate predictors of metastasis than clinical and pathological features, the methods currently available for measuring these chromosomal changes require large tissue samples that increase the risk of biopsy complications, are difficult to interpret, and are prone to sampling error [9,10].
GENE EXPRESSION PROFILING
To overcome the limitations of cytogenetic techniques and to provide clinicians with a straightforward clinical test that is practical, simple, and highly accurate for determining the prognosis of individual patients, we explored other molecular prognostic methods, such as gene expression profiling. Remarkably, we found that uveal melanomas could be divided into two basic categories based on their gene expression profile, and these categories were strongly associated with prognosis: class 1 (low risk of metastasis) or class 2 (high risk of metastasis) [11]. Gene expression profiling (GEP) was subsequently validated in multiple independent studies to provide prognostic information that is more accurate than clinical, pathologic, and cytogenetic factors [12–15]. Consequently, we developed a gene expression profile-based prognostic assay that would be simple and reliable to use in routine clinical practice. The result was a PCR-based microfluidics platform that measured the mRNA expression of 12 discriminating genes and three control genes [16▪]. The prognostic accuracy of the GEP test was validated in a prospective, multicenter study, which also verified the superiority of the GEP test over monosomy 3 and clinicopathologic features [17]. The 15-gene assay is now commercially available as the DecisionDx-UM test, which is performed by a College of American Pathologists-accredited, Clinical Laboratory Improvement Amendments-certified laboratory that maintains an unparalleled technical success rate of over 97% [18▪]. The test can be performed on samples obtained by a single fine needle aspiration biopsy, as well as tumor resection and enucleation. The test result is simple to interpret; class 1 is subdivided into class 1A and class 1B, with 2 and 21% 5-year metastatic risk, respectively, and class 2 is associated with a 72% 5-year metastatic risk. This information allows patients to be stratified promptly into risk categories, so that appropriate individualized management can be offered, as discussed later in this review.
PROFILING OF DRIVER MUTATIONS
Several key mutations in uveal melanoma have been identified from sequencing of primary tissue samples. Identification and molecular characterization of these mutations has helped predict the optimal therapy for an individual patient. The role that mutational profiling can play in the treatment of uveal melanoma is discussed in this section.
GNAQ and GNA11
Mutually exclusive mutations in GNAQ and GNA11, two closely-related G-coupled protein receptor subunits, have recently been found to occur in 85–91% of uveal melanomas [19,20▪]. These mutations lead to constitutive activation of pathways involved in proliferation, differentiation, and apoptosis, including the protein kinase C (PKC) and mitogen-activated protein kinase (MAPK) pathways [19,21▪,22]. GNAQ and GNA11 mutations are thought to be early mutations or initiating events in uveal melanoma pathogenesis, as they are present in benign uveal nevi and almost all uveal melanomas regardless of cytogenetic status or GEP class, and do not correlate with patient survival [23,24,25]. Because of constitutive activation of PKC and MAPK, inhibition of these pathways has been explored as a potential treatment for uveal melanoma. In uveal melanoma cell lines and xenograft animal models with GNAQ or GNA11 mutations, combination treatment with PKC and MAPK inhibitors had a synergistic effect in reducing tumor progression, showing improved efficacy than treatment with either agent alone [21▪]. Additionally, the phosphoinositide 3-kinase (PI3K)/AKT pathway has also been shown to be activated in uveal melanoma, and may be due to GNAQ mutations or loss of phosphatase and tensin homolog (PTEN) activity [26▪,27]. Combined inhibition of MEK and AKT pathways also had a synergistic effect in decreasing cell viability and halting tumor progression in a xenograft animal model [26▪]. In clinical trials on humans with metastatic uveal melanoma, the MEK inhibitor, selumetinib, showed improved progression-free survival (15.9 compared with 7.0 weeks) and improved overall survival (10.8 compared with 9.4 months) when compared with temozolomide chemotherapy [28▪]. Further clinical studies, including combination treatment of MEK inhibitors with other agents, such as the ones mentioned above, are needed and several are currently underway.
BAP1
As loss of one copy of chromosome 3 is known to be a poor prognostic indicator and is associated with uveal melanoma metastasis [29], we conducted exome sequencing of uveal melanoma samples with only one copy of chromosome 3 (monosomy 3) to look for DNA mutations on the remaining chromosome 3 that may occur on potential tumor suppressor genes important in uveal melanoma pathogenesis. These analyses led to the discovery that BRCA1-associated protein 1 (BAP1) had mutations on 3p21.1 in 85% of class 2 uveal melanomas and almost never in class 1 tumors [30]. Thus, BAP1 may serve as a tumor suppressor in uveal melanoma, and its loss may lead to a more prometastatic uveal melanoma.
Functionally, BAP1 is an enzyme that removes ubiquitin molecules from specific proteins to regulate their function. For example, BAP1 removes ubiquitin molecules from histone H2A, which causes changes in the expression of specific genes that are regulated by this histone [31]. One set of genes that are affected by loss of BAP1 is that involved in melanocyte differentiation and function; loss of BAP1 causes uveal melanoma cells to revert to a de-differentiated, stem cell-like state that possibly contributes to their prometastatic behavior [32▪▪]. Histone deacetylase (HDAC) inhibitors, such as the readily available valproic acid, can reverse this effect of BAP1 loss and may play a role in treated patients with high-risk class 2 uveal melanomas in an adjuvant setting prior to the emergence of overt metastatic disease [32▪▪]. Consequently, several different HDAC inhibitors are now being evaluated in clinical trials [33].
BAP1 familial cancer syndrome
In our original report of BAP1 mutations in uveal melanoma [30], we described a patient with a germ-line BAP1 mutation, thereby providing the first evidence that BAP1 represents a hereditary cancer susceptibility gene. Since then, many groups have verified this finding, and the BAP1 familial cancer syndrome has expanded to include cutaneous melanoma, mesothelioma, renal cell carcinoma, and other cancer types [34,35]. We estimate that perhaps 2–3% of patients with uveal melanoma may have a germline BAP1 mutation. Cognizance of the BAP1 familial cancer syndrome and identification of patients with germline BAP1 mutations are now critical for physicians taking care of patients with uveal melanoma, as these patients have increased risk of developing metastasis from their uveal melanoma and are at risk for multiple other types of cancer.
SF3B1 and EIF1AX
In contrast to our findings with BAP1, we discovered that mutations in the splicing factor 3b subunit 1 (SF3B1) occurred in about 19% of uveal melanomas and were associated with a favorable prognosis [36▪▪]. SF3B1 is a component of the spliceosome and is involved in splicing pre-mRNA. Mutations in SF3B1 result in alternative splicing of a select group of mRNAs, and it is not clear how these mutations promote cancer [37▪]. Our findings have been verified by Martin et al. [38▪▪], who also identified mutations in eukaryotic translation initiation factor 1A, X-linked (EIF1AX) in 24% of uveal melanomas, which were also associated with good prognosis. EIF1AX encodes a protein involved in protein translation, and it is not clear how these mutations promote cancer. Interestingly, mutations in BAP1, SF3B1, and EIF1AX are largely mutually exclusive with one another.
CLINICAL UTILITY OF PROGNOSTIC AND PREDICTIVE TESTING IN UVEAL MELANOMA
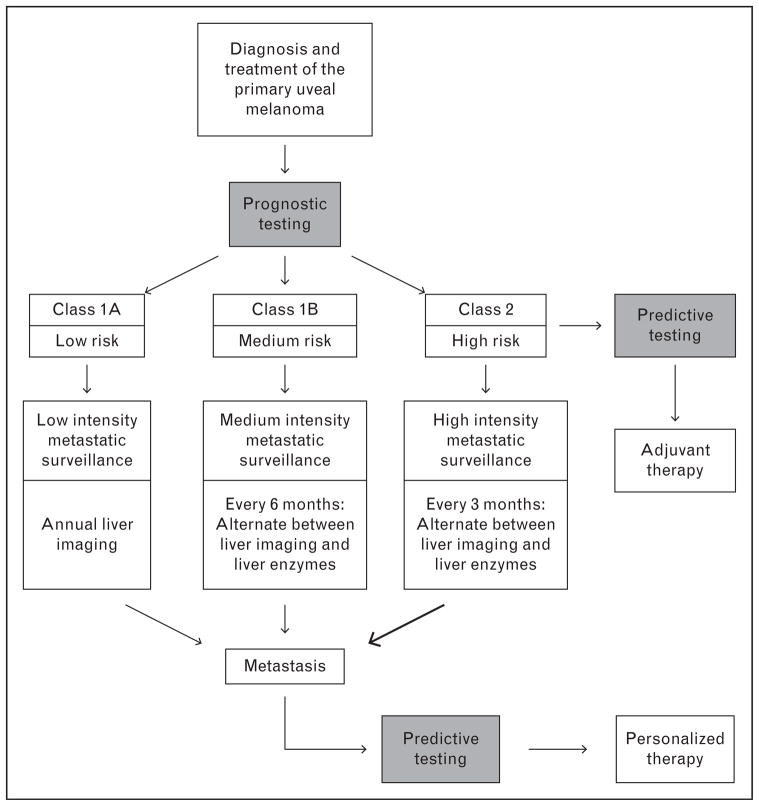
Prognostic testing allows patients to be stratified into low risk and high risk for metastasis, which can then guide an appropriate management plan (Fig. 1). Our current guidelines at the Bascom Palmer Ocular Oncology Service are as follows. For low-risk class 1A patients, we recommend annual imaging of the liver. For intermediate-risk class 1B patients, we recommend annual imaging of the liver, alternating every 6 months with liver enzymes (alkaline phosphatase, lactic dehydrogenase, and γ-glutamyl transpeptidase). For high-risk class 2 patients, we recommend liver imaging twice a year alternating every 3 months with liver enzymes. This strategy targets intensive surveillance specifically to the subset of patients who may benefit while sparing the others. We focus our surveillance on the liver as this organ is involved in over 90% of metastatic uveal melanoma and is the usual site causing patient demise [1]. Suspicious findings from this surveillance are followed up with additional imaging and biopsy as appropriate. Earlier detection of metastasis allows liver-directed therapies, such as chemoembolization, to be initiated at an earlier stage when they may prolong patient survival [39]. Even more importantly, molecular prognostic stratification allows high-risk patients to be entered into clinical trials of adjuvant therapies aimed to slow or halt the progression of micrometastatic disease [16▪].
FIGURE 1.
Flow diagram demonstrating how prognostic and predictive testing in uveal melanoma can guide patient care. Following diagnosis of uveal melanoma, prognostic testing determines metastatic risk, which in turn determines the intensity of metastatic surveillance and whether the patient should be offered entry into a clinical trial of adjuvant systemic therapy.
In clinical trials of adjuvant therapy in high-risk patients and in patients with overt metastatic disease, predictive testing can be used to guide the choice of therapy and to assess response to therapy.
Mutational profiling may provide valuable predictive information for determining the optimal therapy for an individual patient. For example, pharmacologic inhibitors of MEK, AKT and/or PKC may be more effective in tumors with GNAQ/11 mutations [21▪,26▪,28▪], whereas HDAC inhibitors may play a role in tumors with BAP1 mutations [32▪▪]. This predictive information is potentially accessible by direct sampling of primary or metastatic tumor tissue, or from analysis of circulating tumor cells (CTCs) or tumor-derived nucleic acids from serum samples.
Circulating tumor DNA levels and CTC counts are being evaluated for their predictive and prognostic utility in uveal melanoma because of the less invasive nature of obtaining serum samples and the possibility that these measures could be used to monitor therapeutic effect. They have been positively correlated with the presence of hepatic metastasis, metastasis volume, and decreased survival in uveal melanoma [40▪,41▪]. In other cancers undergoing various treatment regimens, a static CTC count appeared 10–12 weeks after therapy and was shown to be a promising method to assess response to therapy [42,43]. An important aspect of clinical trial design in uveal melanoma will be to include such surrogate markers to determine which ones are of value as predictive tests and which ones may be useful for monitoring therapeutic effect.
CONCLUSION
Recent discoveries have allowed for a more targeted approach to treating uveal melanoma. Gene expression profiles have helped differentiate uveal melanoma based on the risk for metastasis. Patients with class 2 uveal melanoma can be treated more aggressively, including the usage of adjuvant therapies. In uveal melanoma patients who are assessed for DNA mutations in GNAQ, GNA11, and BAP1, the DNA mutations have been shown to turn on or off various cellular pathways. These pathways can be targeted with MEK, PKC, AKT, and HDAC inhibitors, which are now being tested in clinical trials. Knowledge of the GEP in combination with DNA mutations in each uveal melanoma tumor will likely improve treatment by determining an individualized, targeted therapeutic strategy. The effects these treatments have on morbidity and mortality of uveal melanoma are currently being assessed in clinical studies. Future studies that lead to better understanding of uveal melanoma pathogenesis and the effects of the DNA mutations will likely lead to better therapeutic strategies.
KEY POINTS.
Primary uveal melanomas can be stratified into class 1 (low metastatic risk) and class 2 (high metastatic risk) based on a 15 gene expression profile that is now available as a prospectively validated test that is used on a routine clinical basis in over 100 centers.
GNAQ or GNA11 mutations are mutually exclusive and represent early events in the development of uveal melanoma.
BAP1, SF3B1, and EIF1AX mutations appear to represent downstream mutations that are, for the most part, mutually exclusive with each other.
SF3B1 and EIF1AX mutations are associated with the class 1 gene expression profile and good prognosis, whereas BAP1 mutations are associated with the class 2 profile and poor prognosis.
An increasing number of clinical trials are becoming available for patients at high risk for metastasis (class 2) or with overt metastatic disease using targeted therapies based on GNAQ/11 and BAP1 mutations.
Acknowledgments
Financial support: This work was supported by grants to J.W.H. from the National Cancer Institute (R01 CA125970), Melanoma Research Alliance, Melanoma Research Foundation and Tumori Foundation, as well as NIH Core Grant P30EY014801, Research to Prevent Blindness Unrestricted Grant, and Department of Defense Grant #W81XWH-09-1-0675 to the Bascom Palmer Eye Institute. The funding organizations had no role in the design or conduct of this research.
Footnotes
Conflicts of interest
J.W.H. is the inventor of intellectual property described in this article and receives royalties from its commercialization. He is a paid consultant for Castle Biosciences, licensee of intellectual property presented in this article. M.G.F. has no disclosures.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Collaborative Ocular Melanoma Study Group. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch Ophthalmol. 2001;199:670–676. doi: 10.1001/archopht.119.5.670. [DOI] [PubMed] [Google Scholar]
- 2.Augsburger JJ, Gamel JW. Clinical prognostic factors in patients with posterior uveal malignant melanoma. Cancer. 1990;66:1596–1600. doi: 10.1002/1097-0142(19901001)66:7<1596::aid-cncr2820660726>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Seddon JM, Albert DM, Lavin PT, Robinson N. A prognostic factor study of disease-free interval and survival following enucleation for uveal melanoma. Arch Ophthalmol. 1983;101:1894–1899. doi: 10.1001/archopht.1983.01040020896012. [DOI] [PubMed] [Google Scholar]
- 4.Finger PT 7th Edition, AJCC-UICC Ophthalmic Oncology Task Force. The 7th edition AJCC staging system for eye cancer: an international language for ophthalmic oncology. Arch Pathol Lab Med. 2009;133:1197–1198. doi: 10.5858/133.8.1197. [DOI] [PubMed] [Google Scholar]
- 5.Harbour JW, Augsburger JJ, Char DH. Gene expression profiling versus TNM classification. Ophthalmology. 2013;120:e52–e53. doi: 10.1016/j.ophtha.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Horsman DE, Sroka H, Rootman J, White VA. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet Cytogenet. 1990;45:249–253. doi: 10.1016/0165-4608(90)90090-w. [DOI] [PubMed] [Google Scholar]
- 7.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J Natl Cancer Inst. 1990;82:1765–1769. doi: 10.1093/jnci/82.22.1765. [DOI] [PubMed] [Google Scholar]
- 8.Sisley K, Rennie IG, Cottam DW, et al. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes Cancer. 1990;2:205–209. doi: 10.1002/gcc.2870020307. [DOI] [PubMed] [Google Scholar]
- 9.Dopierala J, Damato BE, Lake SL, et al. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Invest Ophthalmol Vis Sci. 2010;51:4898–4905. doi: 10.1167/iovs.09-5004. [DOI] [PubMed] [Google Scholar]
- 10.Maat W, Jordanova ES, van Zelderen-Bhola SL, et al. The heterogenous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007;131:91–96. doi: 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- 11.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill HS, Char DH. Uveal melanoma prognostication: from lesion size and cell type to molecular class. Can J Ophthalmol. 2012;47:246–253. doi: 10.1016/j.jcjo.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 13.Petrausch U, Martus P, Tonnies H, et al. Significance of gene expression analysis in comparison to standard risk factors for risk assessment of subsequent metastases. Eye. 2007;22:997–1007. doi: 10.1038/sj.eye.6702779. [DOI] [PubMed] [Google Scholar]
- 14.Van Gils W, Lodder EM, Mensink HW, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008;49:4254–4262. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- 15.Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res. 2007;13:1466–1471. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 16▪.Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Bio. 2014;1102:427–440. doi: 10.1007/978-1-62703-727-3_22. Describes the technical aspects of the 15-gene expression profile uveal melanoma prognostic test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onken M, Worley L, Char D, et al. Collaborative Ocular Oncology Group. Report No. 1. Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪.Harbour JW, Chen R. The DecisionDx-UM gene expression profile test provides risk stratification and individualized patient care in uveal melanoma. PLoS Curr. 2013;5:ii. doi: 10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. Describes the development and clinical validation of the 15-gene expression profile uveal melanoma prognostic test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20▪.Daniels AB, Lee JE, MacConaill LE, et al. High throughput mass spectrometrybased mutation profiling of primary uveal melanoma. Invest Ophthalmol Vis Sci. 2012;53:6991–6996. doi: 10.1167/iovs.12-10427. Over 90% of uveal melanomas were found to have a mutation in either GNAQ or GNA11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Chen X, Wu Q, Porter D, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2013 doi: 10.1038/onc.2013.555. Epub ahead of print. PKC and MEK inhibitors were found to decrease cell growth in uveal melanoma cell lines with GNAQ or GNA11 mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onken MD, Worley LA, Long MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:5230–5234. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauer J, Kilic E, Vaarwater J, et al. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br J Cancer. 2009;101:813–815. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koopmans AE, Vaarwater J, Paridaens D, et al. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br J Cancer. 2013;109:493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪.Ambrosini G, Musi E, Ho AL, et al. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther. 2013;12:768–776. doi: 10.1158/1535-7163.MCT-12-1020. Inhibitors of the adenosine monophosphate-activated protein kinase pathway were shown to induce cell death in uveal melanoma cell lines with GNAQ mutations. [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Rahman MH, Yang Y, Zhou XP, et al. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol. 2006;24:288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 28▪.Selumetinib shows promise in metastatic uveal, melanoma. Cancer Discov. 2013;3:OF8. doi: 10.1158/2159-8290.CD-NB2013-086. The MEK inhibitor selumetinib was shown to improve patient survival in individuals with metastatic uveal melanoma. [DOI] [PubMed] [Google Scholar]
- 29.Prescher G, Bornfeld N, Hirche H, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 30.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32▪▪.Landreville S, Agapova OA, Matatall KA, et al. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012;18:408–416. doi: 10.1158/1078-0432.CCR-11-0946. HDAC inhibitors reverse the biochemical effects of BAP1 mutations, resulting in a shift of class 2 uveal melanomas to a class 1 phenotype and growth arrest. This finding suggested a potential role for HDAC inhibitors for adjuvant therapy in patients with high-risk class 2 uveal melanomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbour JW, Chao DL. A molecular revolution in uveal melanoma: implications for patient care and targeted therapy. Ophthalmology. 2014 doi: 10.1016/j.ophtha.2013.12.014. pii:S0161-6420(13)01189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013;45:116–126. doi: 10.1097/PAT.0b013e32835d0efb. [DOI] [PubMed] [Google Scholar]
- 35.Carbone M, Yang H, Pass HI. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36▪▪.Harbour JW, Roberson ED, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. This paper was the first report of mutations in SF3B1 in uveal melanoma, and the first to show that these mutations are associated with a favorable prognosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37▪.Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. This study showed that SF3B1 mutations are associated with alterations in the mRNA splicing of certain genes, which may provide insights into the role of these mutations in the pathogenesis of uveal melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38▪▪.Martin M, Maβhöfer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. This paper was the first report of EIF1AX mutations in uveal melanoma, and the first to show that these mutations are associated with a favorable prognosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dayani PN, Gould JE, Brown DB, et al. Hepatic metastasis from uveal melanoma: angiographic pattern predictive of survival after hepatic arterial chemoembolization. Arch Ophthalmol. 2009;127:628–632. doi: 10.1001/archophthalmol.2009.45. [DOI] [PubMed] [Google Scholar]
- 40▪.Bidard FC, Madic J, Mariani P, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2013:1207–1213. doi: 10.1002/ijc.28436. Circulating uveal melanoma tumor cells and circulating tumor DNA may be useful prognostic indicators for metastatic disease and could be used to monitor improvements in disease status with therapy. [DOI] [PubMed] [Google Scholar]
- 41▪.Madic J, Piperno-Neumann S, Servois V, et al. Pyrophosphorolysis-activated polymerization detects circulating tumor DNA in metastatic uveal melanoma. Clin Cancer Res. 2012;18:3934–3941. doi: 10.1158/1078-0432.CCR-12-0309. Circulating tumor DNA in metastatic uveal melanoma can be detected and quantified based on GNAQ or GNA11 mutations found in the primary tumor. [DOI] [PubMed] [Google Scholar]
- 42.Gorges TM, Pantel K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol Immunother. 2013;62:931–939. doi: 10.1007/s00262-012-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coumans FA, Ligthart ST, Terstappen LW. Interpretation of changes in circulating tumor cell counts. Transl Oncol. 2012;5:486–491. doi: 10.1593/tlo.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]