Abstract
A “one bead two compound” approach to the synthesis of encoded cyclic peptoid libraries is reported.
Drug-like small molecules (<500 Daltons) generally do not bind well to the relatively shallow surfaces of proteins involved in protein-protein interactions. Thus, in order to develop effective therapeutic agents against these increasingly important targets, it is necessary to develop libraries of compounds able to cover a greater surface area and engage in multiple contacts with the target protein, as well as efficient methods to screen these libraries. With regard to their protein-binding properties, peptides are an attractive class of molecules, but linear peptides have many undesirable features. They are peptidase- and protease-sensitive, relatively cell impermeable and generally form complexes with only modest dissociation constants (KDs in the high nM to mid µM range). However, cyclic peptides can exhibit enhanced cell permeability1,2 and are much less sensitive to enzymatic degradation.3 Moreover, it is presumed that the conformational restriction imposed by cyclization may generally afford higher binding affinities, though rigorous proof for this idea is lacking.4,5 Indeed, many naturally occurring cyclic peptides and depsipeptides have been found to display potent biological activities.6–10 This interest has led to the development of facile methods for the creation of either synthetic11 or genetically encoded12,13 libraries of cyclic peptides as potential sources of drug leads.
A limitation of peptide libraries, cyclic or linear, is that only a relatively small number of building blocks are available. Moreover, although cyclic peptides, can be more cell permeable than their linear counterparts, this appears to be dependent on their ability to form intramolecular hydrogen bonds,2 a property that is likely to vary from compound to compound. Therefore, we became interested in the development of libraries of cyclic peptoids (N-substituted oligoglycines)14,15 as potential protein ligands. Large libraries of linear peptoids with a wide variety of different side chains16–18 are readily accessible using split and pool methods and efficient protocols with which to screen these libraries for protein binding have been developed.18–24 These studies have confirmed that peptoid libraries are indeed rich sources of protein ligands. We have also shown that peptoids are, in general, far more cell permeable than peptides due to their lack of polar main chain N–H bonds.25 In this report, we describe a strategy that is optimized for the creation of cyclic peptoid microarrays.
A key design issue has to do with the ability to determine the sequence of hits after screening a one bead one compound library. Since cyclic peptides or peptoids lack a free N-terminus, Edman sequencing cannot be employed. Moreover, while peptoids, like peptides, can be sequenced by tandem mass spectrometry,26 cyclic molecules will fragment at multiple positions, complicating interpretation of the MS/MS spectrum severely. This issue has limited the development of synthetic cyclic peptide libraries. Pei and co-workers addressed this problem recently by developing a “two compound, one bead” approach in which each bead contains both a linear and cyclic molecule containing the same peptide sequence.11 In other words, the linear molecule encodes the cyclic molecule. This was accomplished using the strategy of Lam27 in which different solvents were employed to segregate beads into two different domains (internal and surface-exposed) to which were attached glutamic acid residues with differentially protected carboxylate side chains. The same peptide chain was then extended from both the internal and external Glu residues. Finally, only the surface-exposed Glu side chains were deprotected, allowing them to be cyclized with the terminal amino group of the peptide. The peptides in the internal layer remained linear and thus served as the encoding strand.
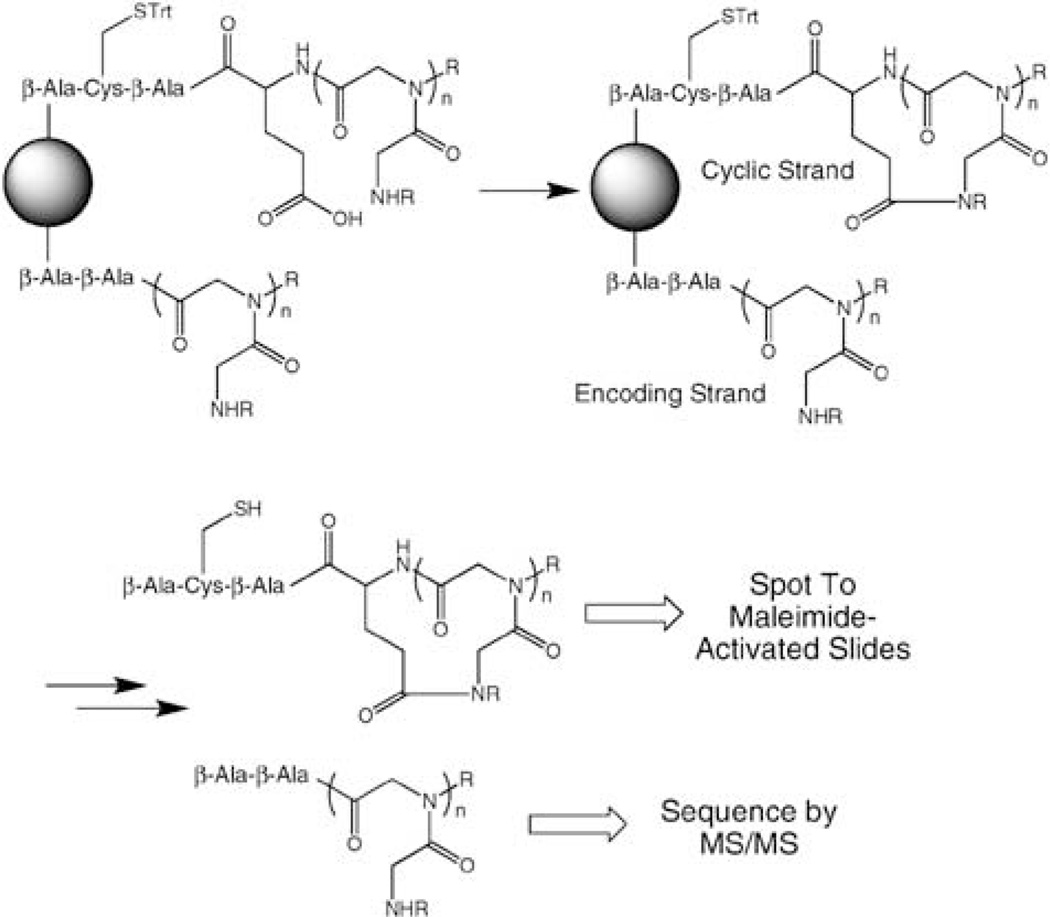
We have developed a different one bead two compound strategy that is tailored to the creation of microarrays, a useful platform for protein fingerprinting and library screening.28,29 The idea was to employ differential deprotection to create two chains, both of which contain the peptoid of interest, but only one of which contains both a glutamic acid residue to support cyclization as well as a Cys residue to allow specific conjugation of only the cyclic peptoid molecule to a maleimide-activated microscope slide30 (Fig. 1). The linear molecule would not couple to the slide, but would be present to support tandem MS-based sequencing.
Figure 1.
Schematic view of the general strategy employed to create a library in which each bead carries a cyclic peptoid and an analogous linear encoding strand. Only the cyclic molecule contains a thiol and thus will couple to a maleimide-activated glass slide.
To effect this strategy (see Fig. 2), a 7 : 1 ratio of Fmoc-Cys(Trt)-OH and ivDde-β-Ala-OH was added to β-Ala-primed Rink amide resin. This ratio was optimized empirically to provide enough linear peptoid for tandem MS sequencing from a single bead, but also produce as much cyclic peptoid as possible. After selective deprotection of Fmoc, Fmoc-β-Ala-OH was again attached to Cys followed, after a removal of this Fmoc, by addition of Fmoc-Glu(O-2-PhiPr)-OH. At this point, both the Fmoc and ivDde protetecting groups were removed and peptoid synthesis was carried out on both strands. Seven peptoid residues were incorporated using conventional sub-monomer chemistry and the nine amines shown in Fig. 2. The 2-PhiPr protecting group on the Glu side chain was then removed selectively with 1% TFA. Finally, macrocyclization was carried out using the method of Kirshenbaum and colleagues (PyBOP (3 eq), HOBt (3 eq) and DIPEA (10 eq).15 Note that the linear molecule lacks two residues present in the (presumed) cyclic molecule and thus the mass peaks derived from each can be distinguished easily, facilitating analysis and sequence determination.
Figure 2.
Synthesis of the encoded cyclic peptoid library via the one bead two compound strategy. The amines employed in the sub-monomer peptoid synthesis are shown at the bottom of the figure (one of the amines in 1,4-diaminobutane and a hydroxyl group in ethanolamine were protected).
To determine the efficacy of this procedure, individual beads were separated and treated with acid to cleave the molecules from the beads, followed by HPLC, MS and MS/MS analysis. Gratifyingly, we found that in almost every case the sequence of the peptoid on a particular bead could be determined easily by tandem MS analysis of the linear molecule. We therefore turned our attention to the determination of the efficiency of the cyclization reaction. For some of the molecules, mass spectrometry and HPLC analysis showed that cyclization of the Cys-Glu-containing molecule was clearly incomplete, as linear starting material was clearly detectable. This was not surprising, since a general problem in the creation of cyclic libraries is that not all sequences cyclize with equivalent efficiencies.31 One would presume that the nature of the N-terminal residue would have the largest effect on cyclization. Indeed, an analysis of more than 50 peptoids by MS/MS revealed that if the N-terminal residue was Nmea, the cyclization yield was almost quantitative. Therefore, this residue was made invariant in all subsequent work.
With this information in hand, we prepared ten peptoids of the form: β-Ala-Cys-Glu(Biotin)-cyclo(Glu-X-X-X-X-X-X-Nmea) (see Fig. 3), where biotin-Glu bears a side chain-conjugated biotin and X was derived from one of the amines shown in Fig. 2. The molecules were cleaved from the resin and analyzed by HPLC and tandem MS. In each case, we were able to easily sequence the linear species by tandem mass spectrometry (see Supplementary Material). Moreover, all of the detectable Cys-containing molecules were in the cyclic form (see Supplementary Material).
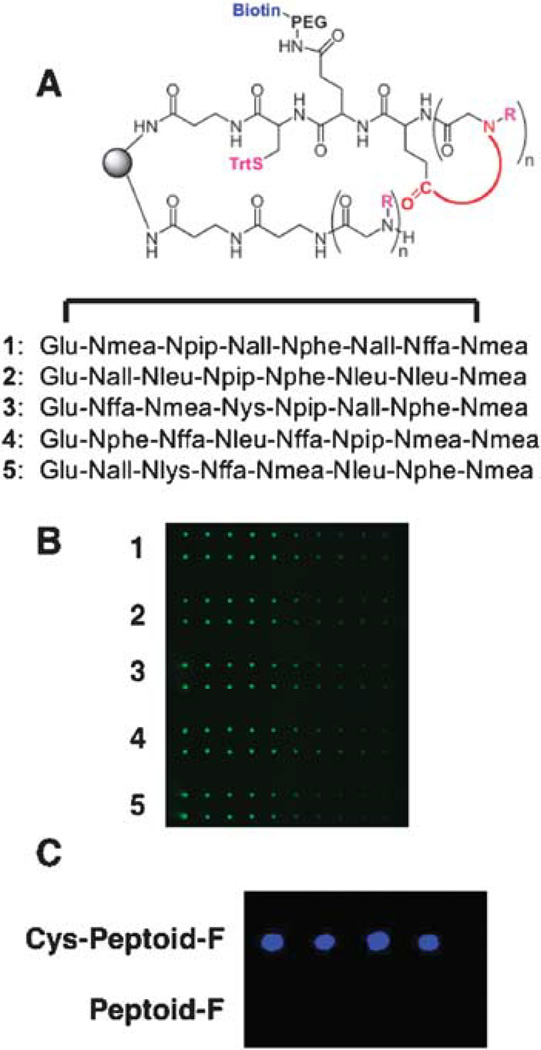
Figure 3.
Attachment of Cys-containing cyclic peptoid to a maleimide-activated glass slide. A. General structure of the cyclic and linear molecules made on each bead before cleavage and deprotection of the thiol side chain. Below: Sequences of the variable regions of five peptoids picked for the spotting experiment. B. Fluorescent image of microarrays in which each of the five peptoids have been spotted onto the activated surface. A DMSO solution of each peptoid (≈ mM) was spotted two times, the solution was diluted three-fold, spotted again, etc. After washing and drying, the arrays were hybridized with Cy3-conjugated streptavidin, washed and the slide was scanned with a fluorescence scanner (see ref. 30 for details). The spots are false-colored green. C. The Cys is essential for retention of the peptoid on the microarray. Two peptoids were synthesized. Each had the sequence Fluorescein-Nlys-Nser-Nleu-Nser-Nall-Npip-Nlys-Nlys. One peptoid also contained a C-terminal cysteine, while the other did not. The two peptoids were spotted onto a maleimide-activated glass slide. After washing, the slide was scanned using a fluorescence scanner. The fluorescence intensity is false-colored blue.
Serial dilutions of the five peptoids shown in Fig. 3 were spotted robotically onto PEGylated, maleimide-activated glass microscope slides30 and the slides were then washed rigorously. In order to demonstrate the immobilization of the cyclic peptoids, the slides were incubated with Cy3-labeled Streptavidin and scanned. As expected, the amount of protein captured decreased as the amount of peptoid spotted decreased, confirming that the fluorescence is indeed due to specific capture of the protein by the peptoid (Fig. 3B). To demonstrate that the Cys residue is critical for retention of the cyclic peptoid to the maleimide-derivatized slide, we synthesized two fluorescein-conjugated linear peptoids that were identical except for the presence and absence of Cys. These were spotted onto a slide, which was then scanned after washing. As shown in Fig. 3C, detectable fluorescence was seen only where the Cys-containing peptoid was spotted. This experiment validates an important feature of our strategy, which is that the linear encoding molecule (see Fig. 1) will not be retained on the slide when the mixture of it and the Cys-containing cyclic molecule are spotted onto the slide and thus will not interfere with screening experiments.
In summary, we have developed a new ‘one bead two compound’ strategy for the creation of encoded cyclic peptoid libraries. This particular scheme is useful for the creation of cyclic peptoid microarrays since only the cyclic peptoid, not the linear encoding molecule, contains a cysteine and thus can be spotted onto a maleimide-activated microscope slide. The use of these cyclic peptoid microarrays in screening experiments will be reported in due course.
Supplementary Material
Acknowledgments
This work was funded by a grant from the Welch Foundation (I-1299) and a contract from the National Heart Lung and Blood Institute for the UT Southwestern Center for Proteomics Research (NO1-HV-28185). We thank Dr Reddy Moola (University of Texas Southwestern Medical Center) for assistance with printing the molecules on the microarray.
Footnotes
Electronic supplementary information (ESI) available: Experimental details for synthesis and microarray construction, supplemental data. See DOI: 10.1039/b812735b
Notes and references
- 1.Rezai T, Bock JE, Zhou MV, Kalyanaraman C, Lokey RS, Jacobson MP. J. Am. Chem. Soc. 2006;128:14073–14080. doi: 10.1021/ja063076p. [DOI] [PubMed] [Google Scholar]
- 2.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J. Am. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 3.Satoh T, Li S, Friedman TM, Wiaderkiewicz R, Korngold R, Huang Z. Biochem. Biophys. Res. Commun. 1996;224:438–443. doi: 10.1006/bbrc.1996.1045. [DOI] [PubMed] [Google Scholar]
- 4.Udugamasooriya DG, Spaller MR. Biopolymers. 2008;89:653–667. doi: 10.1002/bip.20983. [DOI] [PubMed] [Google Scholar]
- 5.Martin SF. Pure Appl. Chem. 2007;79:193–200. [Google Scholar]
- 6.Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. Clin. Immunol. Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee S, Wang Z, Mohammad M, Sarkar FH, Mohammad RM. J. Nat. Prod. 2008;71:492–496. doi: 10.1021/np0705716. [DOI] [PubMed] [Google Scholar]
- 8.Lech-Maranda E, Robak E, Korycka A, Robak T. Mini-Rev. Med. Chem. 2007;7:1062–1069. doi: 10.2174/138955707782110178. [DOI] [PubMed] [Google Scholar]
- 9.Fouladi M. Cancer Invest. 2006;24:521–527. doi: 10.1080/07357900600814979. [DOI] [PubMed] [Google Scholar]
- 10.Hamada Y, Shioiri T. Chem. Rev. 2005;105:4441–4482. doi: 10.1021/cr0406312. [DOI] [PubMed] [Google Scholar]
- 11.Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. J. Am. Chem. Soc. 2006;128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 12.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatesh N, Im S-H, Balass M, Fuchs S, Katchalski-Katzir E. Proc. Natl. Acad. Sci. U. S. A. 2000;97:761–766. doi: 10.1073/pnas.97.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin SB, Yoo B, Todaro LJ, Kirshenbaum K. J. Am. Chem. Soc. 2007;129:3218–3225. doi: 10.1021/ja066960o. [DOI] [PubMed] [Google Scholar]
- 16.Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 17.Horn T, Lee BC, Dill KA, Zuckermann RN. Bioconjugate Chem. 2004;15:428–435. doi: 10.1021/bc0341831. [DOI] [PubMed] [Google Scholar]
- 18.Alluri PG, Reddy MM, Bacchawat-Sikder K, Olivos HJ, Kodadek T. J. Am. Chem. Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 19.Alluri PG, Liu B, P Y, Xiao X, Kodadek T. Mol. BioSyst. 2006;2:568–579. doi: 10.1039/b608924k. [DOI] [PubMed] [Google Scholar]
- 20.Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. J. Comb. Chem. 2007;9:592–600. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy MM, Bachhawatt-Sikder K, Kodadek T. Chem. Biol. 2004;11:1127–1137. doi: 10.1016/j.chembiol.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Lim H-S, Archer CT, Kodadek T. J. Am. Chem. Soc. 2007;129:7750–7751. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuckermann RN, Martin EJ, Spellmeyer DC, Stauber GB, Shoemaker KR, Kerr JM, Figliozzi GM, Goff DA, Siani MA, Simon RJ, et al. J. Med. Chem. 1994;37:2678–2685. doi: 10.1021/jm00043a007. [DOI] [PubMed] [Google Scholar]
- 24.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. J. Am. Chem. Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 25.Kwon YU, Kodadek T. J. Am. Chem. Soc. 2007;129:1508–1509. doi: 10.1021/ja0668623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. J. Comb. Chem. 2006;8:417–426. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Marik J, Lam KS. J. Am. Chem. Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 28.MacBeath G, Koehler AN, Schreiber SL. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 29.Uttamchandani M, Walsh DP, Yao SQ, Chang Y-T. Curr. Opin. Chem. Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Reddy MM, Kodadek T. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Marthandan N, Bowerman D, Garner HR, Kodadek T. Chem. Commun. 2005:581–583. doi: 10.1039/b415578e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.