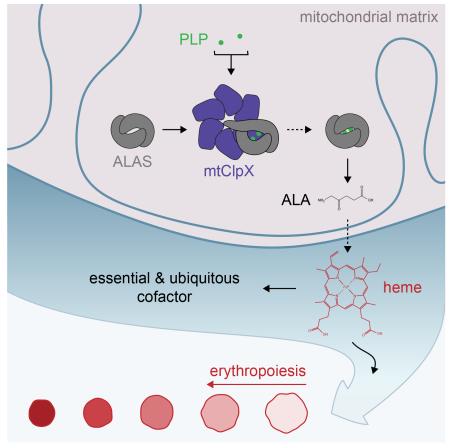
SUMMARY
The mitochondrion maintains and regulates its proteome with chaperones primarily inherited from its bacterial endosymbiont ancestor. Among these chaperones is the AAA+ unfoldase ClpX, an important regulator of prokaryotic physiology with poorly defined function in the eukaryotic mitochondrion. We observed phenotypic similarity in S. cerevisiae genetic interaction data between mitochondrial ClpX (mtClpX) and genes contributing to heme biosynthesis, an essential mitochondrial function. Metabolomic analysis revealed that 5-aminolevulinic acid (ALA), the first heme precursor, is five-fold reduced in yeast lacking mtClpX activity, and total heme is reduced by half. mtClpX directly stimulates ALA synthase in vitro by catalyzing incorporation of its cofactor, pyridoxal phosphate. This activity is conserved in mammalian homologs; additionally, mtClpX depletion impairs vertebrate erythropoiesis, which requires massive upregulation of heme biosynthesis to supply hemoglobin. mtClpX therefore is a widely conserved stimulator of an essential biosynthetic pathway, and employs a previously unrecognized mechanism for AAA+ unfoldases.
Graphical Abstract
INTRODUCTION
All organisms require AAA+ protein unfoldases to actively unfold selected proteins for protein quality control and to regulate the activity of specific substrates. The prokaryotic AAA+ unfoldase ClpX is particularly specialized for regulatory unfolding, tuning the proteome to respond to environmental stress and to orchestrate changes in cell state (Gottesman, 2003; Sauer et al., 2004). ClpX unfolds substrate proteins by ATP-driven translocation of the polypeptide chain through the central pore of its hexameric assembly. In complex with the ClpP peptidase, ClpX carries out protein degradation by translocating unfolded substrates directly into the ClpP proteolytic chamber (Sauer et al., 2004). ClpP degrades all known substrates of ClpX, although for a few substrates unfolding, and not degradation, is the biologically required event (Konieczny and Helinski, 1997; Mhammedi-Alaoui et al., 1994).
In the eukaryotic cytoplasm, the 26S proteasome, which retains the basic architecture of Clp family proteases as well as a related AAA+ unfoldase component, functionally replaces the Clp family proteases. The mitochondrion, however, maintains an autonomous machinery for proteome remodeling, including ClpX, that is largely conserved from its α-proteobacterial ancestor (Fig. S1). Mitochondrial ClpX (mtClpX) does not contribute substantially to protein quality control (Rottgers et al., 2002; van Dyck et al., 1998), suggesting that it may act primarily to control the activities of its substrates by regulatory unfolding and degradation, similarly to its prokaryotic homologs. Mitochondrial ClpP (mtClpP) is not as widely conserved as mtClpX, and mtClpX in organisms without ClpP lacks the ClpP interaction motif (Fig. S1), suggesting that mtClpX may execute a protease-independent function. The specific contributions of mtClpX to mitochondrial physiology, however, are not well understood. mtClpX is required to initiate the mitochondrial unfolded protein response (Haynes et al., 2010), and has been observed to affect mitochondrial nucleoid morphology (Bogenhagen et al., 2008; Kasashima et al., 2012), but its mechanism in these roles is unknown. The single physiological substrate identified for mtClpX, the GTPase Noa1, is degraded by mtClpXP, but how this degradation contributes to Noa1 maintenance or regulation in vivo is unclear (Al-Furoukh et al., 2014).
To uncover physiological functions and partners of mtClpX, we mined previously generated large-scale genetic and chemical interaction maps in S. cerevisiae (Costanzo et al., 2010; Hoppins et al., 2011; Lee et al., 2014). We observed strong links between the yeast mtClpX gene (MCX1) and genes involved in the first steps of heme biosynthesis (Fig. 1A), suggesting that mtClpX might act during heme biosynthesis as well.
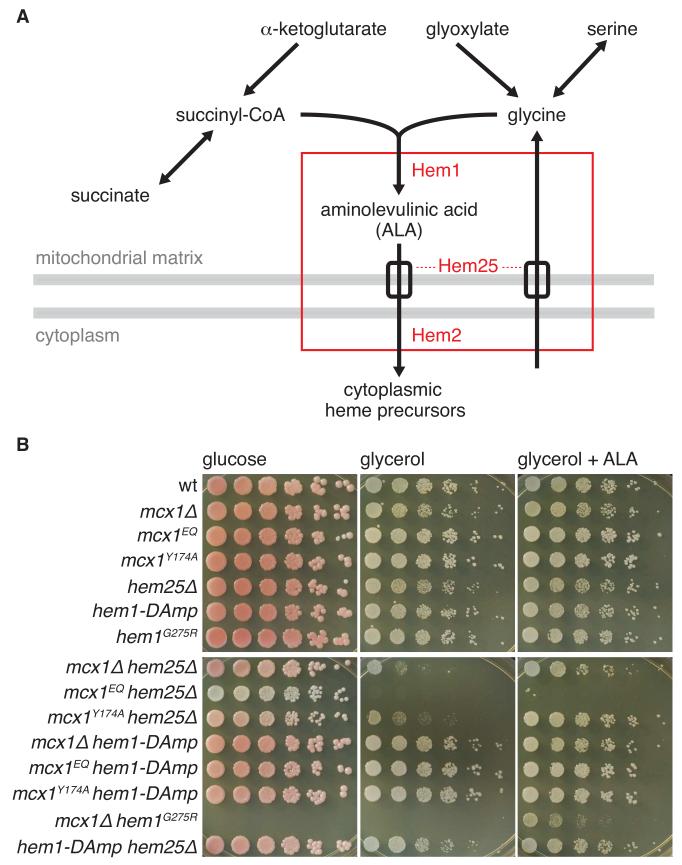
Figure 1.
MCX1 interacts chemically and genetically with the heme biosynthetic pathway. (A) The metabolic pathway for the first step of heme biosynthesis in non-plant eukaryotes. The genetic and chemical interaction profile of MCX1 is highly correlated with the profiles of yeast genes (HEM25, HEM1, and HEM2, shown in red) involved in the first steps of heme biosynthesis. Dashed lines indicate uncertainty in assigning Hem25 to glycine uptake or ALA export. Gray bars indicate mitochondrial membranes. See Fig. S1 for alignment showing conservation among ClpX homologs. (B) MCX1, HEM1, and HEM25 alleles exhibit synthetic phenotypes. Five-fold serial dilutions from cell suspensions with OD600 = 1 were pinned on YP + 2% agar, + 2% glucose or 3% glycerol. +ALA indicates 50 μg/mL ALA. Growth on glucose after 2 d and on glycerol after 3 d is shown.
Nearly all organisms (with a few known exceptions among parasites) require heme for viability (Koreny et al., 2012), and most organisms synthesize heme endogenously. Heme is an essential cofactor for many enzymes, including several members of the respiratory chain, p450 enzymes, and sterol biosynthetic enzymes, and also acts as the sensor component of multiple environmentally responsive transcription factors (Girvan and Munro, 2013; Hamza and Dailey, 2012). In non-plant eukaryotes, the first, rate-limiting step of heme biosynthesis is carried out in the mitochondrial matrix, and its product, 5-aminolevulinic acid (ALA), is exported to the cytoplasm (Fig. 1A). After several further biosynthetic steps, a heme precursor is re-imported into the mitochondrion, where synthesis is completed (Hamza and Dailey, 2012). Cells tightly control heme biosynthesis to meet demand; overstimulation of heme biosynthesis drains valuable central metabolites and can cause damage from reactive unliganded heme or accumulation of toxic heme precursors, whereas insufficient heme production limits the activity of the diverse proteins that require it as a cofactor (Girvan and Munro, 2013; Hamza and Dailey, 2012). As a consequence, causative human disease alleles of every enzyme in heme biosynthesis have been identified (Camaschella, 2009; Sassa, 2006).
In this study, we discover a stimulatory function for mtClpX in heme biosynthesis. Comparison of metabolite levels in wildtype and mcx1Δ cell extracts indicated that MCX1 acts to promote ALA synthesis, the initial step of heme biosynthesis. Mcx1 directly activates the enzyme that performs this step, ALA synthase (ALAS, Hem1 in yeast), by accelerating binding of the cofactor pyridoxal phosphate (PLP) to apoenzyme. This activation is conserved for mammalian homologs of these enzymes, and proceeds without degradation by mtClpP. mtClpX, therefore, stimulates an essential biosynthetic process through a previously unrecognized activity for AAA+ unfoldases: accelerating cofactor insertion into its protein substrate. Finally, we find that vertebrate erythropoiesis is impaired by mtClpX knockdown, commensurate with a central, conserved role for mtClpX in heme production.
RESULTS
MCX1 promotes heme biosynthesis
Using S. cerevisae genetic (Costanzo et al., 2010; Hoppins et al., 2011) and chemical (Lee et al., 2014) interaction data, we searched for genes with interaction profiles similar to MCX1, the yeast gene encoding mtClpX (van Dyck et al., 1998). Because S. cerevisiae (like several other fungi), lacks a ClpP homolog, yeast Mcx1 likely acts purely as a protein unfoldase, without coupled degradation of its substrates. MCX1 was strongly correlated (Costanzo et al., 2010; Hoppins et al., 2011; Lee et al., 2014) with several genes involved in early steps in heme biosynthesis: HEM1 (the gene encoding ALAS (EC 2.3.1.37) (Arrese et al., 1983)); HEM25 (encoding the putative mitochondrial glycine/ALA transporter SLC25A38 (Guernsey et al., 2009)); and HEM2 (encoding the cytosolic enzyme that catalyzes the second step in heme biosynthesis, ALA dehydrdatase (ALAD; EC 4.2.1.24) (Gollub et al., 1977)) (Fig. 1A).
We tested the growth of MCX1, HEM1, and HEM25 mutants singly and in combination on fermentable (glucose) or mitochondrial respiration-requiring (glycerol) carbon sources (Fig. 1B). MCX1 deletion or mutation to an ATP hydrolysis-blocked allele (mutation E206Q in the Walker B motif, mcx1EQ) did not impair fermentative or respiratory growth. A mutation in the essential gene HEM1 corresponding to a sideroblastic anemia allele of human ALA synthase (G351R in human ALAS2 (Wintrobe and Greer, 2004), G275R in yeast HEM1) also showed normal fermentative and respiratory growth, but was lethal in combination with mcx1Δ (Fig. 1B). In combination with deletion of HEM25, MCX1 deletion or mutation dramatically impaired or abrogated mitochondrial respiration (Fig. 1B). Respiratory growth of mcx1Δ hem25Δ and mcx1Δ hem1G275R was restored by supplementation with ALA, indicating that the synthetic phenotypes of Mcx1 resulted from a deficiency at the mitochondrial first step in heme biosynthesis. These strong synthetic phenotypes suggest an important role for Mcx1 in heme biosynthesis.
To directly test the contribution of MCX1 to heme biosynthesis, we measured heme levels in logarithmically growing yeast by total porphyrin fluorescence and by 55Fe incorporation. Both measurements indicated a two- and three-fold reduction of heme in mcx1Δ and mcx1EQ yeast, respectively (Fig. 2A, Fig. S2A). Mirroring the severity of its respiratory growth phenotype, mcx1Δ hem25Δ yeast (Fig. S2B) exhibited a greater heme deficiency than MCX1 mutant yeast. Supplementing the growth medium with ALA rescued the poor heme production of MCX1 mutants (Fig 2A). This rescue strongly suggests that Mcx1 promotes the first phase of heme biosynthesis, synthesis and export of ALA.
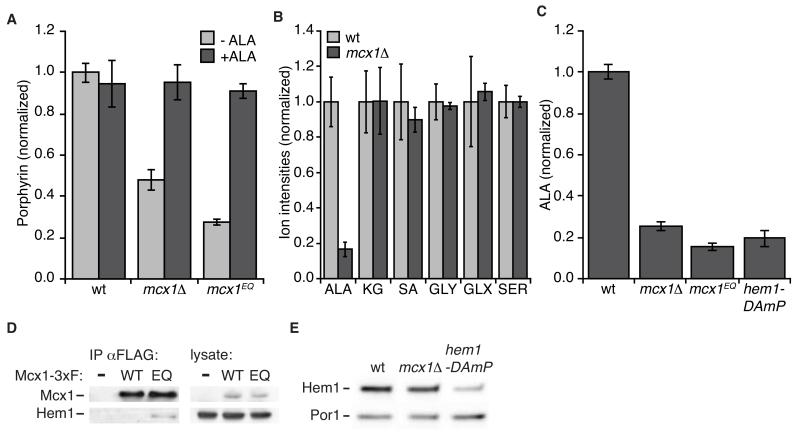
Figure 2.
Mcx1 promotes heme biosynthesis at the step of ALA synthesis. (A) Total porphyrin levels were measured by fluorescence in oxalic acid cell extracts (ex. 400 nm, em. 662 nm; p < 0.001 for difference between wt and MCX1 mutants). +ALA indicates supplementation of growth medium with 50 μg/ml ALA. See also Fig. S2. (B) Metabolites involved in the first step of heme biosynthesis (KG, α–ketoglutarate; SA, succinic acid; GLY, glycine; GLX, glyoxylate; SER, serine) were measured in extracts of the indicated yeast strains by LC-MS. P < 0.001 for ALA perturbation in mcx1Δ cells. (C) ALA levels in cell extracts were measured using modified Ehrlich’s reagent. p ≤ 10−5 for ALA reduction in MCX1 and HEM1 mutants. (D) Mcx1 was isolated with α-FLAG antibody-conjugated beads from cells harboring HEM1-3xMYC and MCX1-3xFLAG (wt or Mcx1EQ (EQ)) or untagged Mcx1 (−)) alleles at the genomic loci, and eluted with 3xFLAG peptide. The eluate was analyzed by Western blot for Mcx1 (α-FLAG) and Hem1 (α-Myc). See Extended Experimental Procedures for detailed procedure. (E) Cellular levels of Hem1-3xMyc were analyzed by Western blot, using alkaline cell extracts (von der Haar, 2007). Hem1-3xMyc intensity: in mcx1Δ = 1.1 ± 0.1 relative to wt, p = 0.35 for difference; in hem1-DAmP = 0.3 ± 0.1, p = 0.01. The mitochondrial protein Por1 was probed as a loading control. Error bars represent mean ± SD.
Mcx1 stimulates ALA synthesis, the first step of heme biosynthesis
To determine the specific perturbation leading to heme deficiency in cells lacking MCX1, we monitored total metabolites in extracts from wildtype and mcx1Δ yeast by LC-MS. Because MCX1-correlated genes function early in heme biosynthesis, we focused on metabolites directly involved in the first mitochondrial phase (Fig. 1A). Ion intensities detected for metabolites preceding ALA synthesis (α-ketoglutarate, succinate, glycine, glyoxylate, and serine) were equivalent in wildtype and mcx1Δ extracts. In contrast, the ion intensity for ALA was reduced more than 80% in mcx1Δ extracts (Fig. 2B). Chemical detection of ALA in extracts corroborated this reduction (Fig. 2C). mcx1Δ extracts exhibited 75% reduced ALA in this assay, and mcx1EQ extracts had an enhanced defect (~85% reduction) (Fig. 2C). For comparison, a reduced expression allele of Hem1 (hem1-DAmP, (Schuldiner et al., 2005)) caused an 80% reduction in cellular ALA. Thus, we conclude that Mcx1 activity promotes heme production at the first step of ALA synthesis.
Mcx1 interacts directly with the ALAS Hem1
We hypothesized that the Mcx1 unfoldase could act directly on Hem1 (the yeast ALAS) to promote ALA synthesis. ALAS is the rate limiting enzyme for heme biosynthesis in nearly all cell types (Hamza and Dailey, 2012), and as such would be a likely target for a stimulatory factor in heme biosynthesis. To test for physical interaction between Mcx1 and Hem1, we affinity purified Mcx1-3xFLAG and Mcx1EQ-3xFLAG from yeast cell extracts and probed for copurifying Hem1-3xMyc (Fig. 2D). We detected Hem1 in Mcx1EQ but not wildtype Mcx1 purified samples, consistent with the ATP dependence of ClpX-substrate interactions. These data suggest that Hem1 is a direct substrate for the Mcx1 unfoldase.
To increase ALA production by acting on Hem1, Mcx1 might increase Hem1 protein abundance, or might stimulate the activity of Hem1. Multiple mechanisms for control of ALA synthase abundance have been characterized (Hamza and Dailey, 2012; Tian et al., 2011). If Mcx1 is required to maintain Hem1 protein levels, decreased Hem1 would be expected in mcx1Δ cells. Hem1 protein abundance was equivalent in wildtype and mcx1Δ lysates (Fig. 2E). In contrast, Hem1 reduction in a hem1-DAmP strain was easily detected by this method (Fig. 2E).
To test if Mcx1 acts by increasing the enzymatic activity of Hem1, we purified these proteins and tested the effect of Mcx1 on the catalytic activity of Hem1. In contrast to the large reduction in ALA in mcx1Δ cells, Mcx1 had little effect on the rate of ALA production by Hem1 in vitro (Fig. S3), leading us to consider mechanisms by which Mcx1 might stimulate Hem1 activity other than a straightforward effect on Vmax.
Mcx1 activates Hem1 by stimulating insertion of its cofactor
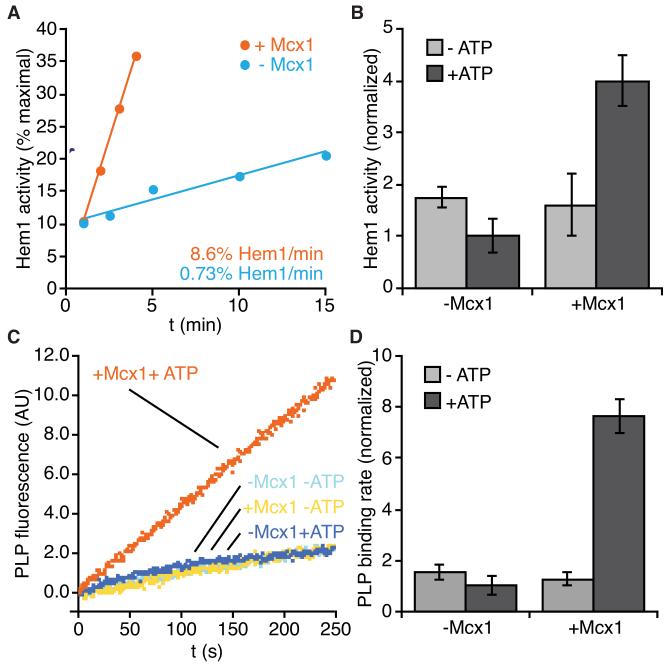
Hem1 (like all ALASs) is part of a large, evolutionarily related enzyme family that depends on the cofactor pyridoxal phosphate (PLP) for activity (the α-family of PLP-dependent enzymes (Eliot and Kirsch, 2004)). PLP binds covalently to an active site lysine, but requires other contacts buried in the interface of the homodimeric enzyme for stable binding (Astner et al., 2005; Gong et al., 1996). We hypothesized that Mcx1 might stimulate formation of the PLP-loaded holoenzyme. To test this idea, we prepared PLP-free Hem1 (apoHem1) and monitored enzyme activity after addition of PLP. As reported previously (Volland and Felix, 1984), apoHem1 regained activity slowly on its own (0.73%/min) (Fig. 3A). Inclusion of Mcx1 accelerated apoHem1 activation by a factor of ten (Fig. 3A). Acceleration by Mcx1 depended on ATP, as expected if Mcx1 acts by remodeling or unfolding Hem1 (Fig. 3B). We directly measured PLP binding to apoHem1 by monitoring formation of the fluorescent pyridoxyllysine bond. The rate of PLP binding for apoHem1 alone (1.0%/min) (Fig. 3C) closely matched the rate of activation for apoHem1 alone (Fig. 3A). Mcx1 stimulated PLP binding by a factor of eight (7.6%/min), and this stimulation was also dependent on ATP (Fig. 3C, D). Thus, Mcx1 accelerates formation of active Hem1 by stimulating cofactor binding to the apoenzyme.
Figure 3.
Mcx1 accelerates incorporation of PLP cofactor into Hem1. (A) Rate of apoHem1 activation by PLP. apoHem1 (3 μM) was incubated with PLP (50 μM) and ATP (2 μM), with (orange) or without Mcx1 (2 μM) (blue), and assayed for ALA synthase activity at indicated times using modified Ehrlich’s reagent. (B) ALAS activity resulting from 4 min incubation of apoHem1 +/− Mcx1, +/− ATP, assayed as in (A). p < 0.0001 for stimulation by Mcx1 + ATP. (C) PLP binding to apoHem1 was monitored by pyridoxyllysine fluorescence (ex. 434 nm, em. 515 nm). (D) Rates of PLP binding to apoHem1 determined by linear fits to fluorescence increase between 100-200 s in (C). p < 0.0001 for stimulation by Mcx1. Error bars represent mean ± SD. See also Fig. S3 for holoHem1 activity measurements.
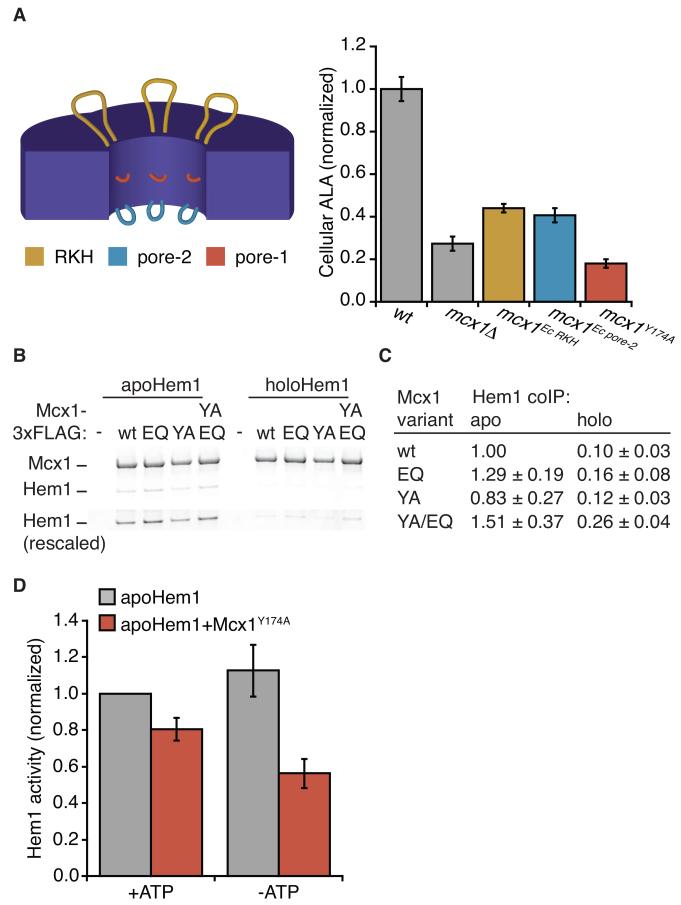
To efficiently activate apoHem1 in mitochondria, we expect that Mcx1 would interact preferentially with apoHem1 over holoHem1. Using purified proteins, we monitored the interaction of Mcx1-3xFLAG with apo- and holoHem1 by coimmunoprecipitation. We observed more efficient interaction of purified Hem1 with ATP-locked Mcx1 (Mcx1EQ) than wildtype Mcx1 (Fig. 4B,C), consistent with the interaction observed between Hem1 and ATP-locked Mcx1 in cell extracts. Importantly, we observed a ten-fold greater amount of Mcx1-bound apoHem1 than holoHem1 (Fig. 4B,C), indicating that Mcx1 has intrinsic binding specificity for the species of Hem1 that it stimulates.
Figure 4.
Mcx1 requires the ClpX translocating pore loops to activate Hem1 and promote ALA production. (A) (left) Pore loops are highlighted on a cross-section diagram of a ClpX hexamer. RKH loops are shown in yellow, pore-1 loops in dark orange, and pore-2 loops in blue. See also Fig. S1 for pore loop sequences. (right) ALA levels in cell extracts were measured by modified Ehrlich’s reagent and normalized to wildtype. Cells harbored indicated mutations at the genomic MCX1 locus. p < 0.0001 for ALA reduction in all MCX1 mutants. (B) Co-immuoprecipitation of apo- and holoHem1 with Mcx1-3xFLAG variants was tested, using purified proteins. Proteins were separated by SDS-PAGE and stained with Sypro Orange. Lower panel (“Hem1 rescaled”) shows Hem1 bands, rescaled to maximum Hem1 intensity. Mcx1 variants are indicated as follows: wt = WT; Walker B E206Q = EQ; pore-1 Y174A = YA. p < 0.001 for more apoHem1 than holoHem1 bound by each Mcx1 variant. (C) ALAS activity resulting from 4 min incubation of apoHem1 with 50 μM PLP, +/− Mcx1Y174A, +/− ATP, assayed as in Fig. 3A. p < 0.05 for suppression of Hem1 by Mcx1Y174A both with and without ATP. Error bars represent mean ± SD.
Mcx1 activates Hem1 by acting as an unfoldase
Most activities characterized for AAA+ unfoldases involve complete or large-scale unfolding of their substrates. Because the folded context of the ALAS active site is important for PLP binding, Mcx1 is unlikely to globally unfold the enzyme to activate it. We therefore sought to determine whether Mcx1 acts as an unfoldase to activate Hem1, or if it might employ an alternative mechanism. To unfold their substrates, AAA+ unfoldases translocate the substrate polyptide through the hexamer pore by the ATP-powered movement of several loops within the pore (Fig. 4A, Fig. S1) To test whether activating Hem1 requires the unfolding machinery of Mcx1, we introduced mutations into these loops at the genomic locus. The central loop (pore-1) is essential for gripping and translocating polypeptides through the pore of the hexamer; we mutated the invariant tyrosine (mcx1Y174A) within this highly conserved sequence, an alteration that abrogates the unfolding activity of many AAA+ unfoldases, including ClpX (Siddiqui et al., 2004). The RKH and pore-2 loops contribute to but are less critical for unfolding. They are more important for substrate selection, and their sequences diverge widely across evolution (Martin et al., 2008a) (Fig. S1). We replaced the RKH or pore-2 loops with the highly divergent E. coli sequences (mcx1E.c.RKH or mcx1E.c.pore-2; Fig. S1, Table S1), a substitution that was previously demonstrated to transplant E. coli ClpX substrate specificity to human mtClpX (Martin et al., 2008a). To determine the ability of these variant enzymes to activate Hem1, we monitored ALA levels within the corresponding MCX1 mutant strains. All pore loop mutants had reduced ALA, and the magnitude of reduction correlated with the importance of the pore loop to translocation (Fig. 4A). The pore-1 mutation caused a more severe ALA deficiency than MCX1 deletion, and substitution with E. coli RKH or pore-2 loops caused a milder ALA deficiency than MCX1 deletion. mcx1Y174A also exhibited genetic interactions similar to mcx1 (Fig. 1B).
To further probe the contribution of the ClpX translocation machinery to ALA synthesis, we tested the activity of purified Mcx1Y174A in vitro. As observed previously for E. coli ClpX (Martin et al., 2008b), Mcx1Y174A has mildly elevated ATPase activity (368 ± 55 ATP/hexamer/min, compared to 219 ± 23 ATP/hexamer/min for wildtype), indicating that Mcx1Y174A phenotypes do not result from loss of ATPase activity. Because Mcx1 pore loops might bind unstructured elements of Hem1, ALA reduction in Mcx1 pore loop mutants could be due to a defect in binding Hem1, rather than unfolding. To test Mcx1Y174A substrate binding, we monitored the interaction of Hem1 with Mcx1Y174A by coimmunoprecipitation using purified proteins. We observed that Hem1 interacted with equivalent strength to Mcx1Y174A and wildtype Mcx1, and also was selective for the apoenzyme by an order of magnitude (Fig. 4B, C). Therefore, mutation of the essential translocating ClpX pore loop-1 does not abolish Mcx1 stimulation of Hem1 by disrupting complex formation. Having excluded an interaction defect, we tested the effect of Mcx1Y174A on PLP activation of apoHem1 in vitro. Mcx1Y174A mildly suppressed apoHem1 activation (Fig. 4D; compare to Fig. 3B for stimulation by Mcx1), suggesting that nonproductive interactions of translocation-blocked Mcx1 with Hem1 interfere with Hem1 spontaneous activation. These data strongly indicate that Mcx1 activates Hem1 using the central polypeptide translocating activity of ClpX homologs.
mtClpX activation of ALAS is conserved in metazoans
Sequence conservation is high between fungal and metazoan ALAS and mtClpX, suggesting that activation of ALAS by mtClpX may be conserved. We measured the effect of mouse mtClpX on PLP activation of human erythroid ALAS (ALAS2) apoenzyme in vitro. mtClpX stimulated apoALAS2 activation approximately 2.5-fold (Fig. 5A); this activation required the presence of ATP (Fig. S4). mtClpX activation of ALAS therefore is broadly conserved among eukaryotes.
Figure 5.
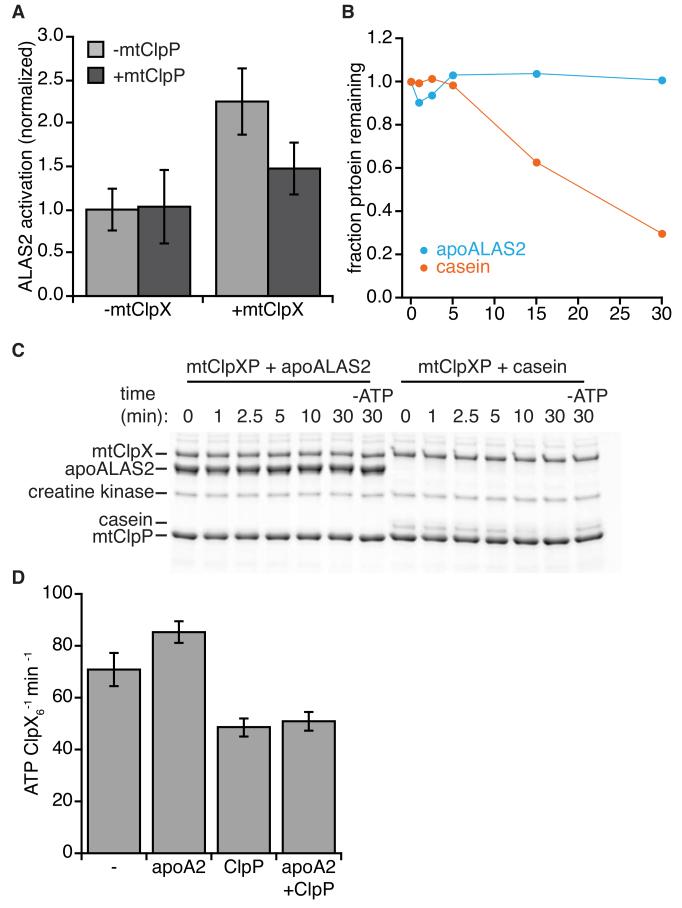
Mammalian mtClpX stimulates PLP activation of apoALAS2, and does not direct apoALAS2 for degradation by mtClpP. (A) apoALAS2 activation by PLP in vitro. Recombinant human apoALAS2 (5 μM) was incubated with PLP, with mouse mtClpX (2 μM hexamer) and human mtClpP (3 μM 14-mer) as indicated, and activation between 4 and 10 minutes was measured by an NAD-coupled assay. p < 0.01 for acceleration of apoALAS2 activation by mtClpX, and p < 0.05 for acceleration by mtClpXP. See also Fig. S4, for ATP dependence of acceleration. (B, C) mtClpXP degradation test. apoALAS2 or α-casein (5 μM each) were incubated with mouse mtClpX (0.3 μM hexamer), human mtClpP (0.8 μM 14-mer) and ATP regenerating system (including 4 mM ATP, except where noted) at 30°C, and aliquots were withdrawn and quenched with SDS at indicated timepoints. Proteins were separated by SDS-PAGE and stained with Sypro Orange. Quantitation of degradation is shown in (B), and gel is shown in (C). (D) Mouse mtClpX (0.3 μM hexamer) ATPase was monitored by NADH-coupled assay, in the presence of human mtClpP (0.8 μM 14-mer) and apoALAS2 (“apoA2”, 10 μM) as indicated. p < 0.01 for suppression of ATPase by mtClpP, and p < 0.05 for stimulation of ATPase by apoALAS2. Error bars represent mean ± SD.
Although S. cerevisisae lacks a ClpP homolog, mtClpP is present in most other eukaryotes. Because the effect of mtClpX on ALAS is activating, the presence of a mtClpP protease might oppose this action of mtClpX. Therefore, we wished to determine if mtClpP interferes with this activation by coupled degradation. In the presence of mtClpP, mtClpX still stimulated PLP activation of apoALAS2, but the magnitude of this stimulation was reduced (Fig. 5A). We monitored possible apoALAS2 degradation during incubation with mtClpXP and ATP. No degradation of apoALAS2 was observed over 30 min (Fig. 5B,C), although mtClpXP efficiently degraded casein (a model substrate for mtClpXP (Kang et al., 2002)) under the same conditions.
We considered whether mtClpP might suppress mtClpX activation of apoALAS2 by suppressing mtClpX ATPase activity. Prokaryotic ClpP partially suppresses the ATPase of prokaryotic ClpX (Kim et al., 2001). If present in the mitochondrial enzymes, ATPase suppression could explain the lower stimulation of ALAS2 by mtClpX when mtClpP is present. In the presence of mtClpP, we observed a ~30-40% suppression of mtClpX ATPase rate (Fig. 5D). This effect is commensurate with the magnitude of suppression of ALAS2 activation, and could therefore account for reduced activation by mtClpXP without invoking protein degradation. We did not observe strong stimulation of mtClpX ATPase (either basal or ClpP-suppressed) by ALAS2 (Fig. 5D). Stimulation of ClpX ATPase by its substrates is common but highly variable, and is less apparent for substrates that are more resistant to unfolding (Burton et al., 2003; Kenniston et al., 2003).
mtClpX is required for efficient erythropoiesis
In organisms with circulating blood cells, heme biosynthesis is massively upregulated during erythropoiesis to meet demand from hemoglobin production (Hamza and Dailey, 2012). Defects in heme biosynthesis cause several human anemias; congenital sideroblastic anemia is caused most commonly by mutations in ALAS2 (Camaschella, 2009). Because mtClpX activates ALAS, we reasoned it would be crucial for erythropoiesis. To facilitate increased heme production during erythropoiesis, heme biosynthetic genes are transcriptionally upregulated. We examined a previously published genome-wide transcriptional dataset for human hematopoiesis (Novershtern et al., 2011), and observed that mRNA levels of CLPX, but not CLPP, were upregulated during erythropoiesis (Fig. 6A). We also observed upregulation of CLPX during erythroid maturation in Friend mouse erythroleukemia (MEL) cells (Fig. S5A). These data suggest that mtClpX contributes to erythropoiesis. In addition, upregulation of CLPX but not CLPP suggests that in this cell type, increased mtClpX relative to mtClpP may help to avoid mtClpP suppression of ALAS activation.
Figure 6.
mtClpX is important for vertebrate heme biosynthesis and erythropoiesis. (A) Relative mRNA abundance for human CLPX, CLPP, ALAS2, and SLC25A38 (indicated as S25A38) throughout erythropoiesis as indicated in a microarray dataset described in (Novershtern et al., 2011). Erythroid development stages were defined by cell type specific markers: 1, CD34+ CD71+ GlyA−; 2, CD34− CD71+ GlyA−; 3, CD34− CD71+ GlyA+; 4, CD34− CD71low GlyA+; 5, CD34− CD71− GlyA+. See also Fig. S5A,B for expression in MEL cells and zebrafish embryos. (B) o-dianisidine staining (brown) for hemoglobinized red cells in zebrafish embyos. Embyros were grown from zygotes injected at the 1-2 cell stage with clpxa-targeting morpholinos or uninjected zygotes (control). (C) Erythrocyte development at 72 h post-fertiliztion (hpf) was quantified by flow cytometry, using dissociated cells from Tg(globin-LCR:eGFP) zebrafish. P ≤ 0.01 for erythrocyte reduction by clpxa knockdown with either morpholino. See also Fig. S5C-E for qPCR quantitation of clpxa knockdown and nontargeting morpholino injections. (D) Rescue of clpxa MOb-induced anemia by ALA supplementation. Tg(globin-LCR:eGFP) zebrafish embryos were supplemented with 2 mM ALA from 24 to 72 hpf, upon which GFP+ erythrocytes were quantified by flow cytometry. p = 0.025 for rescue of anemia in clpxa knockdown embryos by ALA supplementation (E) Heterozygous sauternes or frascati zebrafish were crossed, and progeny were grown for 72 hpf, with or without ALA supplementation as in (D). Anemia was assayed by o-dianisidine staining. p = 0.04 for rescue of anemia in sauternes+/− progeny by ALA. n=52 for sauternes −ALA; n=43 for sauternes +ALA; n=98 for frascati −ALA; n=122 for frascati +ALA. Error bars represent mean ± SD.
To examine the contribution of mtClpX to red blood cell development, we performed morpholino-mediated knockdowns in D. rerio. Zebrafish encode two homologs of mtClpX, clpxa and clpxb. Expression of both clpxa and clpxb is ubiquitous throughout the embryo (Fig. S5B), indicating that mtClpX function is likely not restricted to developing red blood cells. clpxa mRNA was specifically reduced by two independent morpholino sequences (Table S2, Fig. S5C); both morpholinos resulted in morphologically normal embryos with reduced hemoglobin staining by o-dianisidine, indicating a specific defect in red blood cell development (Fig. 6B). Morpholinos targeting clpxb caused toxicity without specific mRNA reduction, preventing further phenotypic analysis. To quantify the anemia we observed in clpxa knockdown embyros, we performed flow cytometry of dissociated cells from embryos with GFP-marked erythroid cells, clpxa knockdown embryos from both morpholinos exhibited an ~50% reduction in GFP-positive cells at 72 h post-fertilization (hpf) (Fig 6C). We also observed a reduction in early erythroid precursors at 24 hpf (Fig. S5F), which may result from reduction in non-erythroid heme production from ALAS1 (Okano et al., 2010). ALAS1 shares very high sequence identity with the erythroid-specific ALAS2 as well as yeast ALAS, and is likely subject to the same stimulatory activity by mtClpX. Importantly, supplementation with ALA starting at 24 hpf fully rescued clpxa knockdown-induced anemia (Fig. 6D). ALA supplementation specifically rescued anemia in ALAS2 mutant (sauternes, (Brownlie et al., 1998)) embryos, but not anemia in mitochondrial iron transporter mutants (frascati, (Shaw et al., 2006)) (Fig. 6E). These results demonstrate that ALA specifically rescues defects in ALA synthesis, and not defects in later steps in heme biosynthesis. Therefore, mtClpX stimulation of ALA synthesis is conserved from S. cerevisae to vertebrates, and is important for efficient heme synthesis during erythropoiesis.
DISCUSSION
In this work, we used large-scale genetic interaction data coupled with metabolic analysis to uncover a broadly conserved stimulatory activity of mtClpX in the essential biological process of heme biosynthesis. Our biochemical studies revealed that mtClpX specifically activates the apoenzyme form of ALAS, the first enzyme in heme biosynthesis, by accelerating binding to the cofactor PLP. mtClpX activation of ALAS is not coupled to degradation by mtClpP, although the presence of mtClpP results in partial inhibition. Consistent with its function in heme biosynthesis, mtClpX is important during erythropoiesis, when heme is in extreme demand as a ligand for hemoglobin. mtClpX may represent a new factor to consider in the etiology and treatment of disorders of heme biosynthesis.
Why might ALAS need a chaperone for PLP insertion? The free PLP cofactor is highly reactive, and is maintained at a low concentration in the cell, near or below the KD for binding to many PLP-dependent enzymes (Cheung et al., 2003; Hamfelt, 1967). The very slow dissociation rate of PLP from its covalent attachment in active sites allows complex formation under these conditions, but spontaneous holoenzyme formation is likely to be inefficient. Therefore, active mechanisms for conjugation of PLP with apoenzymes have long been postulated. For example, pyridoxal kinase can interact with some PLP-dependent enzymes to shuttle newly generated PLP into the active sites of these enzymes in vitro (Cheung et al., 2003; Kim et al., 1988), but it is not known if this is a general mechanism used in vivo. There is no known pyridoxal kinase activity in the mitochondrion, which suggests that a different mechanism (such as the one described here for ALAS) might be needed to facilitate PLP conjugation within this organelle. Determining whether mtClpX acts more broadly among mitochondrial PLP-dependent enzymes to facilitate PLP binding, or exclusively on ALAS, will be important to our understanding of the maturation and function of this large and important class of enzymes.
The Hsp70/90 chaperone system actively promotes ligand binding for several other classes of proteins, the best studied of which is the glucocorticoid receptor (Kirschke et al., 2014; Pratt et al., 2008). The Hsp70/90 enzymes modulate the structure of their substrates by a very different mechanism than AAA+ unfoldases. Hsp70 and Hsp90 bind to partially unfolded intermediate structures, and the ATPase cycle does not appear coupled to an unfolding power stroke like that described for AAA+ unfoldases (Russell and Matouschek, 2014; Saibil, 2013). The requirement for a AAA+ unfoldase for apoALAS activation suggests a fundamentally different mechanism for facilitating ligand insertion, which may be dictated by the structural features of the substrate. In contrast to the structurally unstable and aggregation-prone unliganded glucocorticoid receptor (Kirschke et al., 2014), both apo- and holoALAS are well-structured dimers. This difference in the structure/folding of the substrate proteins could explain a requirement for directed unfolding by a AAA+ unfoldase, rather than trapping of partially unfolded intermediates by Hsp70/90 to accelerate PLP binding.
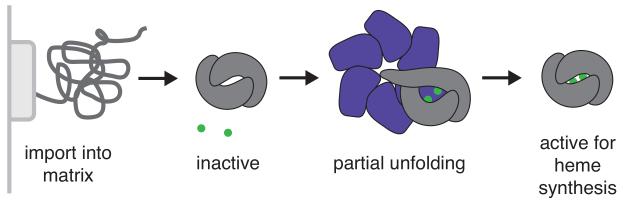
How does mtClpX accelerate PLP binding to ALAS? Multiple residues within the folded active site of ALAS form important contacts with PLP, and studies of the prototypical α-family PLP enzyme, aspartate aminotransferase, reveal that PLP is lost upon enzyme unfolding (Astner et al., 2005; Gong et al., 1996; Wu et al., 2003). Because mitochondrial proteins are unfolded by the mitochondrial translocation machinery during import, newly imported and refolded ALAS is likely in the apo state. mtClpX is unlikely to extensively re-unfold ALAS as part of its activation mechanism, as this action would return the enzyme to a non-PLP binding form, although it is possible that the directionality or rate of mtClpX-mediated complete unfolding could promote refolding through a PLP-binding intermediate. One attractive model is that mtClpX locally unfolds or distorts the structure of ALAS to expose the buried active site and promote efficient PLP binding (Fig. 7). The strong preference mtClpX exhibits for binding apoHem1 indicates that it must specifically recognize a structural feature or exposed motif that is specific to the apoenzyme to initiate this activity. Interestingly, two distantly related AAA+ ATPases that function as dedicated activators for red- or green-type Rubisco stimulate a reverse event, release of an inhibitory ligand (Mueller-Cajar et al., 2011; Wang and Portis, 1992). The better-characterized red-type activase has been proposed to trigger ligand release by partial unfolding of Rubisco (Mueller-Cajar et al., 2011; Wang and Portis, 1992). Although stimulating ligand release and stimulating cofactor association are reverse biological processes, they may be mechanistically related. It will be interesting to compare the characteristics of the unfoldase-induced structural alterations in these distantly related systems.
Figure 7.
Model for mtClpX activation of ALAS. ALAS is unfolded by the mitochondrial import machinery, and refolds in the mitochondrial matrix. Newly folded ALAS binds PLP slowly on its own; partial unfolding by mtClpX renders the active site of ALAS more accessible to PLP, thus accelerating holoenzyme formation. Mitochondrial import machinery (light gray), ALAS (dark gray), mtClpX (purple), and PLP (green) are diagrammed.
A limited-unfolding model for ALAS activation by ClpX is consistent with the lack of ALAS degradation by mtClpXP. If unfolding by mtClpX is sufficiently limited, then the substrate protein would never be translocated far enough through the mtClpXP complex to reach the proteolytic active sites within the chamber of mtClpP. Because substrates of protease-coupled AAA+ unfoldases are often identified or validated by their degradation, such unfolding without coupled degradation may be a much more widespread activity than indicated by the repertoire of known substrates.
How does ClpX discriminate between activation and degradation? ClpX is best understood as part of the ClpXP proteolytic machine, and all previously identified substrates of ClpX are subject to degradation by ClpP. For two substrates, the tetrameric MuA transposase and the dimeric plasmid replication factor TrfA, unfolding rather than degradation is the biologically crucial event, although both substrates can be degraded by ClpXP (Konieczny and Helinski, 1997; Mhammedi-Alaoui et al., 1994). The function of ClpX in these cases is to extract one subunit from the complex by complete unfolding, altering the conformation of the remaining protein(s); the subsequent degradation by ClpP of the unfolded subunit therefore is not deleterious. In the case of ALAS, where the dimer must remain intact to function, ClpX must instead activate the specific molecule it exerts force upon. Therefore, proteolysis by ClpP must not occur; this ClpP-independent action of ClpX is what we observe.
Within the context of the cell, AAA+ unfoldases might be specified for nonproteolytic functions by sublocalization or interaction with other binding partners that is mutually exclusive with binding to their cognate protease. For example, yeast mtClpX associates tightly with the mitochondrial inner membrane (van Dyck et al., 1998); this association might sterically block mtClpP association in eukaryotes which encode it, as well as facilitate efficient activation of ALAS newly imported across the membrane.
Such complete biochemical uncoupling of substrate unfolding from proteolysis has not been previously observed for any protease-coupled AAA+ unfoldase, but two recently described activities of AAA+ proteases are informative for how this may be accomplished. The C. crescentus DNA polymerase clamp loader subunit (DnaX) is partially degraded by ClpXP to produce a functional isoform (Vass and Chien, 2013). To trigger N-end rule substrate delivery to ClpAP from the E. coli adaptor protein ClpS, the ClpA translocation pore engages ClpS itself without causing ClpS degradation (Rivera-Rivera et al., 2014; Roman-Hernandez et al., 2011). In these cases, local structure that is highly resistant to unfolding (in the case of DnaX and ClpS) and/or local peptide sequence that is poorly gripped by the unfoldase (DnaX) appear to release the protein from the grasp of the unfoldase, thereby attenuating or preventing degradation. Both of these types of elements also delimit the degraded region in two transcription factors that are processed the 26S proteasome (Tian et al., 2005). Limited unfolding and translocation of ALAS by mtClpX such as we propose may be dictated by similar sequence and/or structural elements within ALAS. Defining this signal will help to delineate an emerging set of rules by which the fates of AAA+ unfoldase and protease substrates are determined.
EXPERIMENTAL PROCEDURES
Statistics
Error bars indicate standard deviation, calculated from at least three biological replicates. p values were calculated using Student’s t-test unless otherwise indicated.
Yeast strain construction and culture
Strains used in this study are listed in Supplemental Table 1. Yeast genes were modified at chromosomal loci using standard homologous recombination techniques. Yeast strains were grown for experimental purposes in synthetic defined medium (YNB + CSM, Sunrise Sciences) with 2% dextrose at 30°C with shaking.
Heme and ALA measurement
Cellular heme levels were monitored by porphyrin fluorescence and 55Fe labeling. ALA levels in cell extracts were quantified using modified Ehrlich’s reagent. ALA production by purified proteins was quantified using modified Ehrlich’s reagent or by an NAD-coupled assay as indicated. See Extended Experimental Procedures for details.
Metabolic profiling
Metabolite extracts were made by rapid vacuum filtration of yeast liquid culture, followed by incubation of the cell-laden filter in extraction solvent (40% acetonitrile, 40% methanol, 20% water). Extracts were analyzed for relative metabolite levels by LC-MS. See Extended Experimental Procedures for further details.
Protein purification and biochemical assays
mtClpX, mtClpP, and ALAS proteins were recombinantly produced in their mature forms (lacking the mitochondrial presequence). Biochemical assays for ALAS activity and PLP reconstitution were performed at 30°C in 25 mM Hepes pH 7.6, 5 mM MgCl2, and 10% glycerol with ATP regenerating system (5 mM creatine phosphate and 50 mg/mL creatine kinase, with 2 mM ATP when indicated), supplemented with 100 mM KCl (Hem1) or 130 mM KCl and 0.75 mg/ml BSA (ALAS2). 50 μM PLP was included in reconstitution experiments. Protein concentrations during PLP reconstitution were: 3 μM apoHem1, 2 μM Mcx1 (hexamer), 5 μM apoALAS2, 2 μM mouse ClpX (hexamer), 3 μM human ClpP-His6 (14-mer). For detailed procedures, see Extended Experimental Procedures.
Zebrafish maintenance and studies
Wild-type (AB), dino (dintt250) (Hammerschmidt et al., 1996) and Tg(globin-LCR:eGFP) (Ganis et al., 2012) zebrafish (Danio rerio) were maintained, bred and staged according to standard methods (Lawrence et al., 2003). Zebrafish studies were conducted with the IACUC approval at Boston Children’s Hospital. Embryos were stained for hemoglobinized cells with o-dianisidine as previously described (Amigo et al., 2009). Flow cytometry was used to quantify erythrocytes from embryos (Cooney et al., 2013).
Morpholino-mediated knockdown in zebrafish
Splice site-blocking antisense morpholino oligomers (Table S2) were injected into 1–2 cell-stage embryos. Knockdown in morphant embryos was confirmed with qRT-PCR using Taqman probes (Applied Biosystems). For details of ALA complementation, see Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
The mitochondrial ClpX unfoldase is required for efficient heme biosynthesis.
mtClpX activates a key enzyme in heme biosynthesis by catalyzing cofactor binding.
mtClpX activates ALAS without committing it to degradation by mtClpP.
mtClpX is important for erythropoiesis, when demand for heme is high.
ACKNOWLEDGEMENTS
We thank S. Bell, J. Weissman, R. Vale, J. Nunnari, and N. Bradshaw for critical reading of the manuscript, L. Zon for the Tg(globin-LCR:eGFP) transgenic line, K. Hoffmeister for FACS machine use, and C. Lawrence and his team for zebrafish husbandry. This work was supported by grants from the National Institutes of Health (RO1 GM049224, T.A.B.; F32DK095726, J.R.K.; T32HL007574 and F32DK098866, Y.Y.Y.; R01DK070838 and P01HL032262, B.H.P.), March of Dimes Foundation (6-FY09-289, B.H.P.), and Brazilian CAPES and FAPESP Foundations (D.S.B.). K.Y.R. is the recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences and of support from the William Randolph Hearst Foundation. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Al-Furoukh N, Kardon JR, Kruger M, Szibor M, Baker TA, Braun T. NOA1, a Novel ClpXP Substrate, Takes an Unexpected Nuclear Detour Prior to Mitochondrial Import. PLoS One. 2014;9:e103141. doi: 10.1371/journal.pone.0103141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amigo JD, Ackermann GE, Cope JJ, Yu M, Cooney JD, Ma D, Langer NB, Shafizadeh E, Shaw GC, Horsely W, et al. The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood. 2009;114:4654–4663. doi: 10.1182/blood-2008-12-189910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese M, Carvajal E, Robison S, Sambunaris A, Panek A, Mattoon J. Cloning of the δ-aminolevulinic acid synthase structural gene in yeast. Curr Genet. 1983;7:175–183. doi: 10.1007/BF00434887. [DOI] [PubMed] [Google Scholar]
- Astner I, Schulze J.o.r.O., Heuvel J.v.d., Jahn D, Schubert W-D, Heinz DW. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J. 2005;24:3166. doi: 10.1038/sj.emboj.7600792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- Brownlie A, Donovan A, Pratt SJ, Paw BH, Oates AC, Brugnara C, Witkowska HE, Sassa S, Zon LI. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat Genet. 1998;20:244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: Linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Science: A Publication of the Protein Society. 2003;12:893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaschella C. Hereditary sideroblastic anemias: pathophysiology, diagnosis, and treatment. Seminars in hematology. 2009;46:371–377. doi: 10.1053/j.seminhematol.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Cheung PY, Fong CC, Ng KT, Lam WC, Leung YC, Tsang CW, Yang M, Wong MS. Interaction between pyridoxal kinase and pyridoxal-5-phosphate-dependent enzymes. J Biochem. 2003;134:731–738. doi: 10.1093/jb/mvg201. [DOI] [PubMed] [Google Scholar]
- Cooney JD, Hildick-Smith GJ, Shafizadeh E, McBride PF, Carroll KJ, Anderson H, Shaw GC, Tamplin OJ, Branco DS, Dalton AJ, et al. Teleost growth factor independence (gfi) genes differentially regulate successive waves of hematopoiesis. Dev Biol. 2013;373:431–441. doi: 10.1016/j.ydbio.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear E, Sevier C, Ding H, Koh J, Toufighi K, Mostafavi S, et al. The Genetic Landscape of a Cell. Science. 2010;327:425. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, Jia Z, Sabo PJ, Weaver M, Sandstrom R, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366:185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan HM, Munro AW. Heme sensor proteins. J Biol Chem. 2013;288:13194–13203. doi: 10.1074/jbc.R112.422642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub EG, Liu KP, Dayan J, Adlersberg M, Sprinson DB. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- Gong J, Kay CJ, Barber MJ, Ferreira GC. Mutations at a glycine loop in aminolevulinate synthase affect pyridoxal phosphate cofactor binding and catalysis. Biochemistry. 1996;35:14109–14117. doi: 10.1021/bi961296h. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Campagna DR, Evans SC, Ferguson M, Kellogg MD, Lachance M, Matsuoka M, Nightingale M, Rideout A, et al. Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat Genet. 2009;41:651–653. doi: 10.1038/ng.359. [DOI] [PubMed] [Google Scholar]
- Hamfelt A. Pyridoxal phosphate concentration and aminotransferase activity in human blood cells. Clinica chimica acta; international journal of clinical chemistry. 1967;16:19–28. doi: 10.1016/0009-8981(67)90264-1. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, et al. dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development. 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- Hamza I, Dailey HA. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta. 2012;1823:1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The Matrix Peptide Exporter HAF-1 Signals a Mitochondrial UPR by Activating the Transcription Factor ZC376.7 in C. elegans. Molecular Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang N-N, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem. 2002;277:21095–21102. doi: 10.1074/jbc.M201642200. [DOI] [PubMed] [Google Scholar]
- Kasashima K, Sumitani M, Endo H. Maintenance of mitochondrial genome distribution by mitochondrial AAA+ protein ClpX. Exp Cell Res. 2012;318:2335–2343. doi: 10.1016/j.yexcr.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Kim YT, Kwok F, Churchich JE. Interactions of pyridoxal kinase and aspartate aminotransferase emission anisotropy and compartmentation studies. J Biol Chem. 1988;263:13712–13717. [PubMed] [Google Scholar]
- Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny I, Helinski DR. The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc Natl Acad Sci U S A. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koreny L, Sobotka R, Kovárová J, Gnipová A, Flegontov P, Horváth A, Oborník M, Ayala FJ, Lukes J. Aerobic kinetoplastid flagellate Phytomonas does not require heme for viability. Proc Natl Acad Sci USA. 2012;109:3808–3813. doi: 10.1073/pnas.1201089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JM, Plank LR, Lawrence AL. The effect of feeding frequency on consumption of food, absorption efficiency, and gonad production in the sea urchin Lytechinus variegatus. Comp Biochem Physiol A Mol Integr Physiol. 2003;134:69–75. doi: 10.1016/s1095-6433(02)00222-2. [DOI] [PubMed] [Google Scholar]
- Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, et al. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 2014;344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Molecular Cell. 2008a;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008b;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhammedi-Alaoui A, Pato M, Gama MJ, Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature. 2011;479:194–198. doi: 10.1038/nature10568. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Zhou L, Kusaka T, Shibata K, Shimizu K, Gao X, Kikuchi Y, Togashi Y, Hosoya T, Takahashi S, et al. Indispensable function for embryogenesis, expression and regulation of the nonspecific form of the 5-aminolevulinate synthase gene in mouse. Genes to cells: devoted to molecular & cellular mechanisms. 2010;15:77–89. doi: 10.1111/j.1365-2443.2009.01366.x. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Rivera I, Román-Hernández G, Sauer RT, Baker TA. Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1414933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Hernandez G, Hou JY, Grant RA, Sauer RT, Baker TA. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell. 2011;43:217–228. doi: 10.1016/j.molcel.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottgers K, Zufall N, Guiard B, Voos W. The ClpB homolog Hsp78 is required for the efficient degradation of proteins in the mitochondrial matrix. J Biol Chem. 2002;277:45829–45837. doi: 10.1074/jbc.M207152200. [DOI] [PubMed] [Google Scholar]
- Russell R, Matouschek A. Chance, Destiny, and the Inner Workings of ClpXP. Cell. 2014;158:479–480. doi: 10.1016/j.cell.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa S. Modern diagnosis and management of the porphyrias. British journal of haematology. 2006;135:281–292. doi: 10.1111/j.1365-2141.2006.06289.x. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- Tian Q, Li T, Hou W, Zheng J, Schrum LW, Bonkovsky HL. LONP1-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M110.215772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyck L, Dembowski M, Neupert W, Langer T. Mcx1p, a ClpX homologue in mitochondria of Saccharomyces cerevisiae. FEBS Lett. 1998;438:250–254. doi: 10.1016/s0014-5793(98)01310-6. [DOI] [PubMed] [Google Scholar]
- Vass RH, Chien P. Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2013;110:18138–18143. doi: 10.1073/pnas.1311302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland C, Felix F. Isolation and properties of 5-aminolevulinate synthase from the yeast Saccharomyces cerevisiae. Eur J Biochem. 1984;142:551–557. doi: 10.1111/j.1432-1033.1984.tb08321.x. [DOI] [PubMed] [Google Scholar]
- von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE. 2007;2:e1078. doi: 10.1371/journal.pone.0001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Portis AR. Dissociation of ribulose-1,5-bisphosphate bound to ribulose-1,5-bisphosphate carboxylase/oxygenase and its enhancement by ribulose-1,5-bisphosphate carboxylase/oxygenase activase-mediated hydrolysis of ATP. Plant physiology. 1992;99:1348–1353. doi: 10.1104/pp.99.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintrobe MM, Greer JP. Wintrobe’s clinical hematology. 11th edn Lippincott Williams & Wilkins; Philadelphia: 2004. [Google Scholar]
- Wu T-H, Oses-Prieto JA, Iriarte A, Martinez-Carrion M. Release of pyridoxal 5′-phosphate upon unfolding of mitochondrial aspartate aminotransferase. Biochim Biophys Acta. 2003;1647:315–320. doi: 10.1016/s1570-9639(03)00081-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.