Abstract
As a critical developmental process, epithelial–mesenchymal transition (EMT) involves complex transcriptional reprogramming and has been closely linked to malignant progression. Although various epigenetic modifications, such as histone deacetylation and H3K9 methylation, have been implicated in this process, how they are coordinated remains elusive. We recently revealed that MPP8 couples H3K9 methylation and DNA methylation for E-cadherin gene silencing and promotes tumor cell migration, invasion, and EMT. Here, we show that MPP8 cooperates with the class III HDAC SIRT1 in this process through their physical interaction. SIRT1 antagonizes PCAF-catalyzed MPP8-K439 acetylation to protect MPP8 from ubiquitin-proteasome-mediated proteolysis. Conversely, MPP8 recruits SIRT1 for H4K16 deacetylation after binding to methyl-H3K9 on target promoters. Consequently, disabling either MPP8 methyl-H3K9 binding or SIRT1 interaction de-represses E-cadherin and reduces EMT phenotypes, as does knockdown of MPP8 or SIRT1 in prostate cancer cells. These results illustrate how SIRT1 and MPP8 reciprocally promote each other's function and coordinate epithelial gene silencing and EMT.
Keywords: EMT, histone deacetylation, H3K9 methylation, transcription repression
Introduction
Epithelial–mesenchymal transition (EMT) was initially defined as a developmental process which enables polarized epithelial cells reversibly lose their epithelial characteristics and acquire mesenchymal properties 1, 2. It is now evident that EMT is also an essential mechanism to endow epithelial cancer cells with migratory and invasive capabilities associated with metastatic competence 3. One hallmark of EMT is the functional loss of a key epithelial adhesion molecule E-cadherin, which is mainly achieved at the transcription level during malignant progression 4. Several transcription factors (EMT-TF), such as ZEB1/2 and SNAI1/2, directly bind to E-cadherin promoter for gene silencing, while the elevated expression of these EMT-TFs correlates with reduced E-cadherin expression and poor clinical prognosis in different epithelial cancer cells 5. It has been shown that various histone modifications are essential mediators of EMT-TFs and also play a critical role in this process 6, 7. For example, inhibition of histone deacetylation or H3K4 demethylation de-represses E-cadherin expression and prevents EMT in several tumor cell lines 8, 9, 10, 11, 12. Similar effects can also be achieved by inhibiting methylation of H3K9, H3K27, or H4K20 using different approaches 13, 14, 15, 16.
Histone deacetylation and H3K9 methylation are two intimately connected repressive modifications that have been implicated in E-cadherin silencing. For example, HDAC1/2 coexist with G9a/GLP in CtBP1 complex which coordinates the stepwise deacetylation and methylation of H3K9 on E-cadherin promoter 17. A class III HDAC SIRT1 has been implicated in E-cadherin silencing and EMT by cooperating with ZEB1 9, 12. The fact that ZEB1 was also co-purified in CtBP1 complex 17 suggests a functional link between SIRT1 and H3K9 methylation. However, the underlying mechanism is largely unknown.
Methylation of H3K9 creates binding site for a group of adaptor proteins that mediate downstream effects. We have previously established the role of MPP8, a methyl-H3K9 binding protein in E-cadherin silencing and EMT 18. Here, we demonstrate that MPP8 and SIRT1 reciprocally regulate each other's function at multiple molecular layers through their physical interaction. Disruption of MPP8-SIRT1 interaction de-represses E-cadherin expression and reduces cell motility and invasiveness, suggesting that this interplay plays critical role in MPP8- and SIRT1-mediated EMT.
Results and Discussion
MPP8 interacts with SIRT1
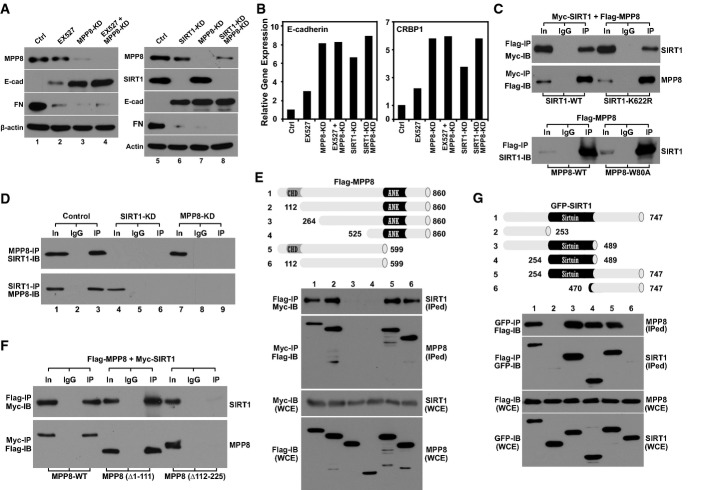
Given that MPP8 and SIRT1 displayed similar function in E-cadherin silencing and EMT 9, 12, 18, we first tested whether they have overlapping function in E-cadherin silencing. As indicated in Fig1A, stable knockdown (KD) of SIRT1 or MPP8, or treatment of EX-527, a specific SIRT1 inhibitor 19, up-regulated E-cadherin protein level in PC3 prostate cancer cells. However, EX-527 treatment of MPP8-KD cells or simultaneous knockdown of SIRT1 and MPP8 did not lead to an additive increase. Although E-cadherin mRNA level increased ∽three- to eightfold upon EX-527 treatment or knockdown of MPP8 or SIRT1, an additional increase was not detected either when SIRT1 and MPP8 were inhibited simultaneously. Similar expression changes were also observed when we analyzed another SIRT1 target gene CRBP1 12 (Fig1B). Thus, these results support the possibility that MPP8 and SIRT1 are involved in the same transcription repression pathway.
Figure 1.
- Western blot analysis of control, MPP8, or SIRT1 single-knockdown or double-knockdown PC3 cells and EX-527 (1 μM for 24 h)-treated cells.
- RT–qPCR analysis of E-cadherin (left) and CRBP1 (right) expression in PC3 cells in which SIRT1 and MPP8 were inhibited individually or simultaneously by EX-527 (1 μM for 24 h) or shRNA knockdown. Columns represent the mean of triplicate PCRs and normalized to GAPDH.
- Myc-SIRT1 (wt or K622R) was co-expressed with Flag-MPP8 in 293T cells followed by IP-Western blot analysis using indicated antibodies (top panels). Flag-MPP8 (wt or W80A) was also expressed in 293T cells for similar IP-Western blot analysis (bottom panel).
- MPP8 interacts with SIRT1 endogenously. Cell extracts derived from control, SIRT1-KD, or MPP8-KD 293T cells were incubated with MPP8 or SIRT1 antibody for IP. Co-IPed proteins were analyzed by Western blot.
- Different Flag-tagged MPP8 deletion mutants were co-expressed with Myc-SIRT1 in 293T cells followed by IP-Western blot analysis using indicated antibodies.
- Myc-SIRT1 was co-expressed with Flag-MPP8-FL, Δ1-111, or Δ112-225 mutant in 293T cells followed by IP-Western blot analysis.
- GFP-tagged SIRT1 deletions were co-expressed with Flag-MPP8 in 293T cells. Cell lysates were IPed with GFP or Flag antibody followed by Western blot analysis.
Different histone-modifying enzymes often coexist in the same multifunctional complex and act in concert for transcription regulation 20. We thus tested whether MPP8 interacts with SIRT1 by immunoprecipitation (IP)-Western analysis. As shown in Fig1C, Flag-MPP8 and Myc-SIRT1 specifically pulled down each other when they were co-expressed in 293T cells. This interaction was not affected by either K622R mutation on SIRT1 or the methyl-lysine binding disabling mutation W80A 18, 21, 22 on MPP8, suggesting it is not mediated by the newly identified SIRT1-K622 methylation 23. Importantly, Western blot analysis detected the robust MPP8-SIRT1 interaction in 293T-wt cells but not in SIRT1-KD or MPP8-KD cells after endogenous IP using their direct antibodies (Fig1D), confirming that MPP8 specifically interacts with SIRT1 in cells.
To map MPP8-SIRT1 interaction, we generated a serial of Flag-MPP8 deletions and co-expressed them with Myc-SIRT1-wt in 293T cells. IP-Western blot analysis reveals that the deletion of first 263aa on MPP8, but not first 111aa or last 260aa or both, is sufficient to abolish its interaction with SIRT1 (Fig1E), suggesting that a small region (112–263aa) on MPP8 is critical. This result was further confirmed by a similar IP-Western blot analysis using an MPP8 internal deletion mutant (Δ112–225aa) (Fig1F). We also co-expressed Flag-MPP8 with different GFP-SIRT1 deletions for similar analysis and uncovered that the catalytic core of SIRT1 (254–489aa) is necessary and sufficient to interact with MPP8 (Fig1G).
PCAF and SIRT1 dynamically regulate MPP8-K439 acetylation
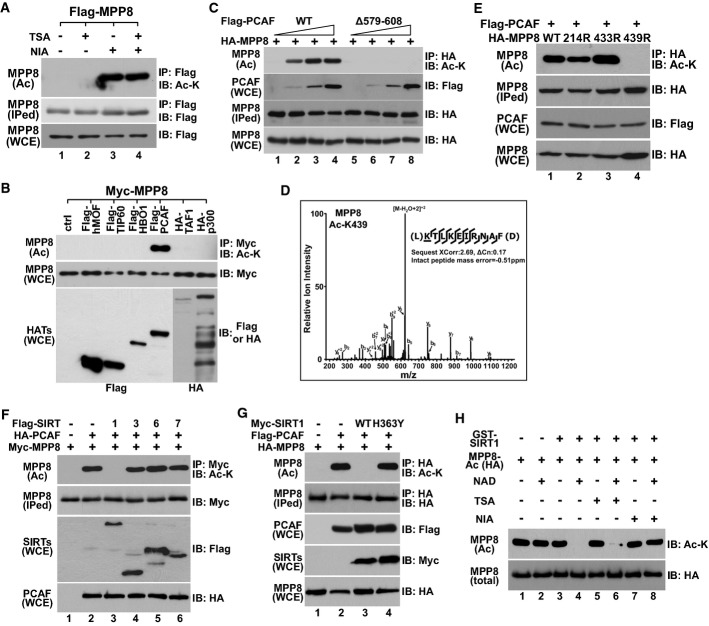
The finding that MPP8 interacts with the catalytic domain of SIRT1 prompted us to test whether MPP8 is a SIRT1 substrate. First, we expressed Flag-MPP8 in MPP8-KD 293T cells 18 followed by HDAC inhibitor treatment to test whether MPP8 is acetylated in cells. Anti-acetyl-lysine Western blot analysis of Flag-IPed MPP8 did not detect MPP8 acetylation when cells were treated with a class I/II HDAC inhibitor TSA. However, a robust acetylation signal was readily detected from cells treated with a class III HDAC inhibitor nicotinamide (NIA) and this signal did not increase additionally when cells were treated with both inhibitors. These results suggest that MPP8 is subjected to acetylation in cells and its acetylation is regulated by Sir2 family HDACs.
Next, we co-expressed Myc-MPP8 with six different HATs in MPP8-KD 293T cells. Anti-myc IP-coupled Western blot analysis detected the robust MPP8 acetylation only from cells co-expressing PCAF (Fig2B), suggesting that MPP8 is acetylated by PCAF. This possibility was further confirmed by observations that MPP8 acetylation level was in PCAF dosage-dependent manner, while a catalytically inactive mutation in PCAF (Δ579–608) eliminated MPP8 acetylation (Fig2C). We next purified PCAF-acetylated MPP8 for LC–mass spectrometry and uncovered that the acetylation occurs at K439 (Fig2D). Substitution of K439 to arginine eliminated MPP8 acetylation, while K-R mutation at an acetylation consensus motif (K214KPKK) or an adjacent lysine (K433) did not (Fig2E), demonstrating that PCAF specifically acetylates MPP8 at K439.
Figure 2.
- Flag-MPP8 was expressed in MPP8-KD 293T cells followed by 4-h treatment of nicotinamide (NIA, 20 mM), trichostatin A (TSA, 3.3 μM), or both inhibitors. Flag-MPP8 was next IPed, while the protein and acetylation levels were analyzed by Western blot.
- Myc-MPP8 was co-expressed with six indicated HATs in 293T cells followed by IP-Western blot analysis using indicated antibodies.
- HA-MPP8 was co-expressed with increased amount of Flag-PCAF (wt or catalytically inactive Δ579–608 mutant) in 293T cells, while MPP8 acetylation level was analyzed by IP-Western blot analysis.
- Mass spectrometry analysis of MPP8 acetylation. Arrows are the fragment ions that confirm the location of the acetylation site as K439 (underlined).
- HA-tagged MPP8-wt or different lysine-to-arginine mutants were co-expressed with Flag-PCAF in 293T cells followed by IP-Western blot analysis using indicated antibodies.
- Myc-MPP8 was co-expressed with HA-PCAF and each of indicated Flag-Sirtuins in 293T cells followed by IP-Western blot analysis.
- HA-MPP8 and Flag-PCAF were co-expressed with or without Myc-SIRT1 (wt or H363Y catalytically inactive mutant) in 293T cells followed by HA-IP. Protein and MPP8 acetylation levels in IPed samples and whole-cell extracts were analyzed by Western blot.
- SIRT1 deacetylates MPP8-K439 in vitro. K439-acetylated HA-MPP8 was purified and incubated with purified recombinant GST-SIRT1 for in vitro deacetylation assay with or without cofactor NAD+ (1 mM), nicotinamide (NIA, 10 mM), or trichostatin A (TSA, 2 μM). MPP8 protein and acetylation levels were analyzed by Western blot.
Finally, we co-expressed MPP8 and PCAF together with four nuclear-localized Sirtuins (SIRT-1, 3, 6, 7) 24 in MPP8-KD 293T cells and examined MPP8 acetylation by IP-Western blot analysis. As shown in Fig2F, MPP8 acetylation was only abrogated in cells co-expressing SIRT1. Co-expression of a catalytically inactive mutant SIRT1-H363Y did not affect MPP8 acetylation (Fig2G), indicating that SIRT1 enzymatic activity is required. We further purified PCAF-acetylated MPP8 and incubated it with recombinant GST-SIRT1 for an in vitro deacetylation assay. As indicated in Fig2H, MPP8 acetylation was eliminated when GST-SIRT1 and cofactor NAD+ were added into the reaction and this MPP8 deacetylation was effectively inhibited by nicotinamide but not TSA. These results together demonstrate that PCAF and SIRT1 dynamically regulate MPP8 acetylation at K439.
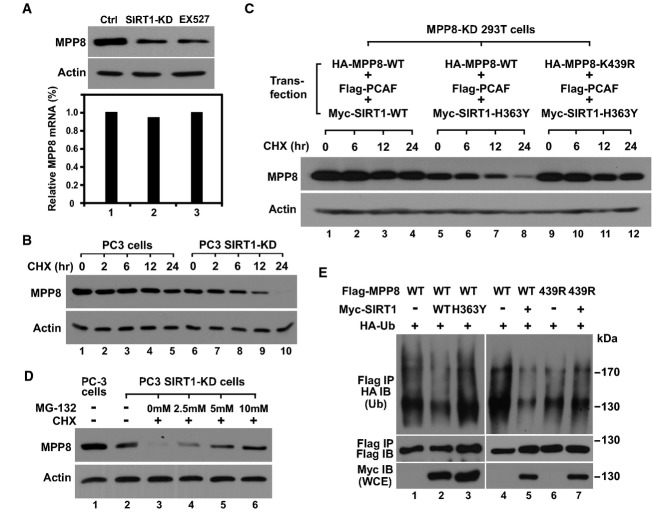
SIRT1 modulates MPP8 protein stability
The finding of MPP8-K439 acetylation prompted us to test whether it affects MPP8 methyl-H3K9 binding. This possibility was ruled out as MPP8-wt, K439Q (acetylation-mimic), and K439R (acetylation-disabled) mutant displayed the similar binding profile to H3 peptides with different methyl-K9 states (Supplementary Fig S1A). However, we noticed that MPP8 protein level, but not mRNA level, reduced in cells where SIRT1 was inhibited by EX-527 or knockdown (Figs1A and 3A), suggesting that SIRT1 affects MPP8 protein stability. We thus blocked protein synthesis in control and SIRT1-KD PC3 cells using cycloheximide (CHX) and followed endogenous MPP8 degradation by Western blot (Fig3B). In control cells, MPP8 degradation was detected only after 24-h CHX treatment. In SIRT1-KD cells, however, MPP8 protein level reduced significantly after 6-h CHX treatment and became undetectable after 24 h. We further co-expressed MPP8 (wt or K439R) and PCAF together with SIRT1 (wt or H363Y) in MPP8-KD 293T cells and carried out similar analyses. As shown in Fig3C, MPP8 protein remained stable in cells co-expressing PCAF-wt and SIRT1-wt. However, it degraded much faster when SIRT1-wt was replaced by SIRT1-H363Y mutant. On the contrary, MPP8-K439R protein remained stable when it was co-expressed with PCAF-wt and SIRT1-H363Y. Together, we conclude that SIRT1 deacetylates MPP8 at K439 to increase its protein stability.
Figure 3.
- Western blot (top) and RT–qPCR (bottom) analyses of control, EX-527-treated (1 μM for 24 h) control, and SIRT1-KD PC3 cells. Results were derived from triplicate PCRs and normalized to GAPDH.
- Control and SIRT1-KD PC3 cells were treated with 25 μg/ml CHX for the indicated time followed by Western blot analysis using MPP8 antibody. Actin served as a loading control.
- HA-MPP8 (wt or K439R) and Flag-PCAF were co-expressed with Myc-SIRT1 (wt or H363Y) in MPP8-KD 293T cells. After 40 h, transfected cells were treated with CHX (25 μg/ml) for the indicated times followed by Western blot analysis of whole-cell extract using MPP8 and actin antibodies.
- SIRT1-KD PC3 cells were treated with CHX (25 μg/ml) together with increased amount of MG132 for 24 h followed by Western blot analysis.
- Flag-MPP8 (wt or K439R) and HA-ubiquitin (HA-Ub) were co-expressed with or without Myc-SIRT1 (wt or H363Y) in MPP8-KD 293T cells. Protein and ubiquitination levels of Flag-IPed MPP8 were analyzed by Western blot. SIRT1 expression in whole-cell extracts was also examined by Western blot.
We next treated SIRT1-KD PC3 cells with CHX and MG132 together. Following Western blot analysis reveals that the degraded MPP8 protein can be restored by MG132 in a dosage-dependent manner (Fig3D), suggesting that MPP8 is degraded through proteasome pathway. As most eukaryotes proteins destined for proteasomal degradation are initially polyubiquitinated 25, we co-expressed Flag-MPP8 and HA-ubiquitin in MPP8-KD 293T cells to test whether MPP8 is polyubiquitinated. Western blot analysis of Flag-IPed MPP8 detected a strong high-molecular-mass smear signal (anti-HA) indicating polyubiquitination. MPP8 polyubiquitination reduced significantly when SIRT1-wt was co-expressed but was not affected by SIRT1-H363Y. A similar reduction in MPP8 polyubiquitination was also observed when we applied MPP8-K439R mutant and co-expression of SIRT1 did not lead to any additional reduction (Fig3E). These results not only demonstrate that MPP8-K439 deacetylation by SIRT1 protects MPP8 from ubiquitin-proteasome-mediated degradation, but also suggest that MPP8 polyubiquitination could be triggered by K439 acetylation. Although the underlying mechanism is still being investigated, we suspect that K439 acetylation could either recruit an E3 ligase or facilitate MPP8-E3 interaction, according to several recent studies 26, 27, 28. PCAF might also contribute to MPP8 ubiquitination as it possesses an E3 ligase activity 29, 30.
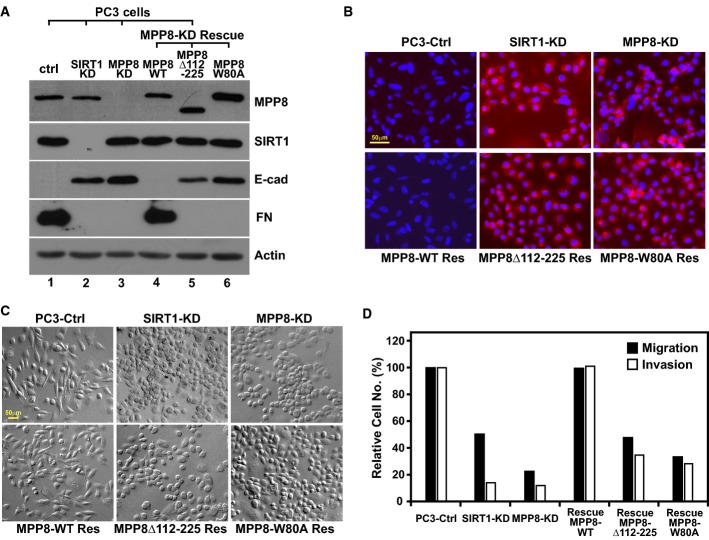
MPP8-SIRT1 interaction is important to maintain mesenchymal cell properties
To understand the role of MPP8-SIRT1 interaction in cell function, we stably rescued MPP8-KD PC3 cells with MPP8-wt, Δ112–225 (deficient in SIRT1 binding, Fig1D), or W80A mutant. Western blot analysis of EMT markers (Fig4A) reveals that rescue expression of MPP8-wt not only re-repressed E-cadherin, but also restored fibronectin expression. Because of the importance of MPP8 methyl-H3K9 binding 18, such EMT marker changes were not observed in cells rescued by MPP8-W80A mutant. However, E-cadherin expression was only slightly repressed in MPP8 Δ112–225 mutant-rescued cells, while fibronectin still remained at low level, suggesting that MPP8-SIRT1 interaction is also important for the EMT marker rearrangement.
Figure 4.
- Western blot analysis of whole-cell extract derived from control, SIRT1-KD, MPP8-KD, and different rescue PC3 cells using indicated antibodies.
- The same set of PC3 cells as in (A) were fixed with cold methanol and co-stained with E-cadherin antibody (red) and DAPI (blue).
- The same set of PC3 cells as in (A) were grown on glass bottom culture dishes while the differential interference contrast (DIC) images of live cells were taken using a Zeiss automatic fluorescent inverted microscope.
- Migration and invasion assays of control and different stable knockdown and rescue PC3 cells. Columns represent the mean of triplicate assays, while control cells were normalized as 100%.
We next examined possible morphology changes in these cells. Immunofluorescence staining results indicate that de-repressed E-cadherin in SIRT1-KD and MPP8-KD PC3 cells localized on cell membrane to re-establish cell–cell contacts. Although it completely disappeared in MPP8-wt rescued cells, the membrane-localized E-cadherin remained in MPP8 Δ112–225 or W80A mutant-rescued cells (Fig4B). Consistently, differential interference contrast microscopy images reveal that both SIRT1-KD and MPP8-KD PC3 cells changed their shape to a cuboidal form. While rescue expression of MPP8-wt impaired this morphologic change, the expression of either MPP8 Δ112–225 or W80A mutant failed to do so (Fig4C). Furthermore, we evaluated cell migration and invasion abilities using Boyden chamber assay. As shown in Fig4D, knockdown of SIRT1 or MPP8 leads to a severe reduction in migration (50–75%) and invasiveness (> 80%) of PC3 cells. Although this reduction in MPP8-KD cells was abrogated by rescue expression of MPP8-wt, MPP8 Δ112–225 or W80A mutant only displayed moderate effects. Given that MPP8 Δ112–225 mutant contains an intact chromodomain which is sufficient for methyl-H3K9 binding 21, 22 and SIRT1 binds to MPP8-wt and W80A similarly, these results demonstrate that MPP8-SIRT1 interaction as well as MPP8 methyl-H3K9 binding is necessary to maintain E-cadherin silencing and mesenchymal characteristics of PC3 cells.
MPP8 contributes to SIRT1 promoter targeting
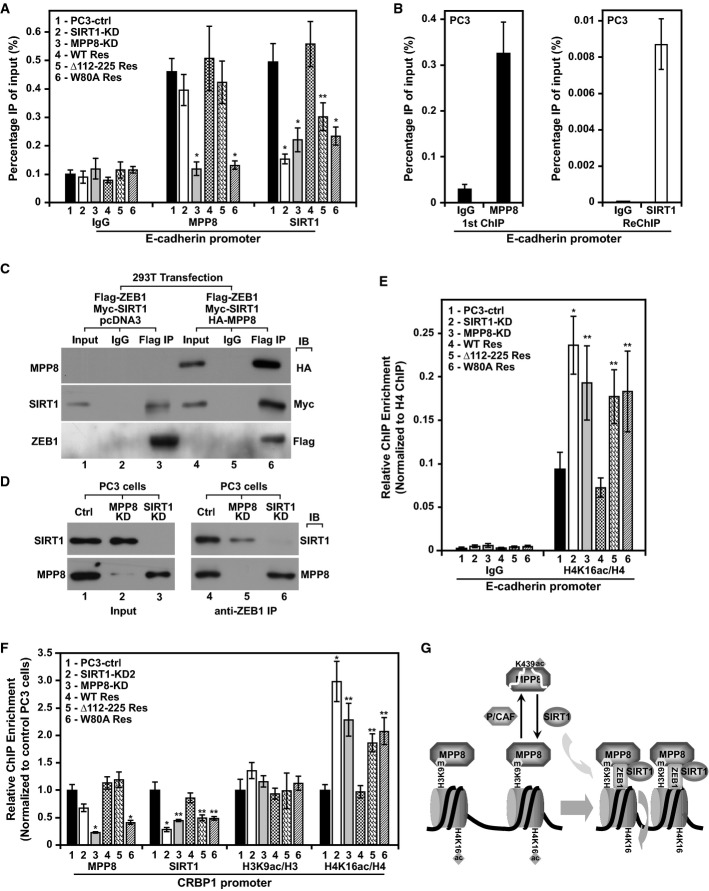
MPP8 and SIRT1 have been previously shown on E-cadherin promoter in different cancer cells 9, 18, and we thus determined whether MPP8-SIRT1 interaction plays a role in their promoter targeting. Chromatin IP-coupled qPCR (ChIP–qPCR) analyses (Fig5A) indicate that MPP8 and SIRT1 indeed target E-cadherin promoter in control PC3 cells. We also carried out ChIP-reChIP analysis, and the results indicate that MPP8-bound DNA was also occupied by SIRT1 (Fig5B), suggesting that they co-localize on E-cadherin promoter through their physical interaction. In SIRT1-KD PC3 cells, SIRT1 localization was abolished, while MPP8 localization only decreased slightly. However, MPP8 knockdown severely reduced the occupancy of SIRT1 in addition to MPP8, suggesting that the promoter localization of MPP8 is necessary for SIRT1 targeting. This possibility is also supported by results that rescue-expressed MPP8-wt restored promoter localization of both MPP8 and SIRT1. Although MPP8 Δ112–225 mutant displayed an intact promoter targeting, it failed to restore SIRT1 localization, whereas MPP8-W80A mutant did not restore either MPP8 or SIRT1 localization. We further generated the same set of stable knockdown and rescue MDA-MB-231 cells, while Western blot and ChIP–qPCR analyses detected similar changes on E-cadherin expression and promoter localization of MPP8 and SIRT1 (Supplementary Fig S2). Therefore, these results illustrate a scenario that MPP8 occupies E-cadherin promoter by binding to methyl-H3K9 and in turn recruits SIRT1 through their physical interaction. Although fibronectin expression was also effected in these knockdown and rescue cells, it is likely an indirect effect as MPP8 does not target FN1 promoter 18.
Figure 5.
- ChIP–qPCR analysis of chromatin derived from control, SIRT1-KD, MPP8-KD, and different rescue PC3 cells. MPP8 and SIRT1 antibodies were used for ChIP, while qPCR was conducted using primers for E-cadherin promoter.
- ChIP-ReChIP analysis of chromatin derived from PC3 cells using MPP8 (left) and SIRT1 (right) antibodies sequentially.
- Flag-ZEB1 was co-expressed with myc-SIRT1 with or without HA-MPP8 in 293T cells. Cell lysates were next analyzed by IP-Western blot analysis using indicated antibodies.
- Cell lysates derived from control, MPP8-KD, or SIRT1-KD PC3 cells were incubated with ZEB1 antibody for IP followed by Western blot analysis using indicated antibodies.
- ChIP–qPCR analysis of chromatin derived from same set of PC3 cells as in (D) using antibodies for H4 and H4K16ac and primers for E-cadherin promoter. ChIP enrichment of H4K16ac in each analyzed cells was normalized to H4.
- ChIP–qPCR analysis of same set of PC3 cells as in (D) using antibodies for MPP8, SIRT1, H3, H3K9ac, H4, and H4K16ac. qPCR was conducted using primers for CRBP1 promoter. ChIP enrichment of each antibody in control PC3 cells was normalized to 1 in each set of cells. H3K9ac/H3 and H4K16/H4 represent normalized H3K9 and H4K16 acetylation (to H3 and H4, respectively).
- A cartoon illustration of the proposed model in which MPP8 and SIRT1 cooperate for E-cadherin silencing during EMT.
It has been shown that SIRT1 is recruited to E-cadherin promoter by ZEB1 through their interaction 9. We next wondered whether MPP8 contributes to SIRT1 recruitment by ZEB1 by regulating SIRT1-ZEB1 interaction. To test this possibility, we co-expressed Flag-ZEB1 and myc-SIRT1 in 293T cells with or without HA-MPP8. IP-Western blot analysis not only confirmed ZEB1-SIRT1 interaction, but also detected a robust ZEB1-MPP8 interaction when they were co-expressed. Intriguingly, ZEB1-SIRT1 interaction increased significantly in the presence of MPP8 (Fig5C). We next carried out endogenous IP using ZEB1 antibody and cell extracts derived from control, MPP8-KD, and SIRT1-KD PC3 cells. Western blot analysis reveals that endogenous MPP8 and SIRT1 also interacted with ZEB1 specifically. While knockdown of SIRT1 did not affect ZEB1-MPP8 interaction, knockdown of MPP8 severely reduced endogenous ZEB1-SIRT1 interaction (Fig5D). These results suggest that ZEB1 could recruit both SIRT1 and MPP8 to target promoters through their physical interactions; importantly, MPP8 also bridges or stabilizes ZEB1-SIRT1 interaction to facilitate SIRT1 recruitment by ZEB1.
MPP8 recruits SIRT1 to preferentially deacetylate H4K16
As histone deacetylation is one of the major functions of SIRT1 31, we examined E-cadherin promoter acetylation at two SIRT1 direct target sites, H3K9 and H4K16 12, 31, 32, 33, 34, 35, in different knockdown and rescue PC3 cells. As indicated in ChIP–qPCR results (Fig5E and Supplementary Fig S3), knockdown of SIRT1 significantly increased H4K16 acetylation on E-cadherin promoter but only slightly affected H3K9 acetylation. As expected, we observed similar changes in MPP8-KD PC3 cells. Although rescue expression of MPP8-wt completely abrogated the increased H4K16 acetylation, MPP8 Δ112–225 or W80A mutant only showed modest effects. We thus conclude that MPP8 recruits SIRT1 for H4K16 deacetylation and E-cadherin silencing. In addition, ChIP–qPCR analysis detected similar changes on H3K9 and H4K16 acetylation as well as the occupancy of MPP8 and SIRT1 on CRBP1 promoter (Fig5F and Supplementary S4A–F), but not on the control GAPDH promoter (Supplementary Fig S4G), suggesting that the recruitment of SIRT1 and promoter H4K16 deacetylation could be a general mechanism for MPP8-mediated transcription repression.
While MPP8 and SIRT1 preferentially affect H4K16 acetylation, promoter H3K9 acetylation has also been associated with E-cadherin expression 12, 13, 14, 17. We thus treated control and SIRT1-KD PC3 cells with TSA, a class I/II HDAC inhibitor that has being widely used to restore H3K9 acetylation to test whether they are independent mechanisms in E-cadherin silencing. Although E-cadherin expression is de-repressed in SIRT1-KD cells, TSA treatment increased E-cadherin protein and mRNA levels in both control and SIRT1-KD cells in a dosage-dependent manner (Supplementary Fig S5A and B). When normalized to control treatment, E-cadherin mRNA displayed similar up-regulation pattern in control and SIRT1-KD PC3 cells (Supplementary Fig S5C), indicating that TSA and SIRT1 knockdown induce E-cadherin expression in a non-overlapping manner. Upon TSA treatment, ChIP–qPCR analysis detected a significant increase in H3K9 acetylation on E-cadherin promoter in both control and SIRT1-KD PC3 cells (Supplementary Fig S5D). However, we only observed modest increases on H4K16 acetylation (Supplementary Fig S5E). These results suggest that H3K9 deacetylation and SIRT-mediated H4K16 deacetylation both play important roles in E-cadherin silencing but contribute to different repression pathways.
Collectively, our results uncovered a novel molecular mechanism by which two repressive chromatin modifiers MPP8 and SIRT1 positively regulate each other's function through protein interaction. While SIRT1 counteracts MPP8-K439 acetylation catalyzed by PCAF to increase MPP8 protein stability, MPP8 recruits SIRT1 to deacetylate H4K16 on target promoters for gene silencing. Disruption of MPP8-SIRT1 interaction reduces cellular motility, invasiveness, and other mesenchymal properties, indicating that this interaction plays an important role in MPP8- and SIRT1-mediated EMT. The finding that MPP8 interacts with ZEB1 and greatly facilitates SIRT1-ZEB1 interaction (Fig5C and D) suggests that MPP8 not only is targeted by ZEB1 to promoters, but also bridges SIRT1 recruitment by this key EMT-TF (Fig5G). Since ZEB1 was co-purified with G9a/GLP in CtBP1 complex 17, we suspect it also facilitates the retention of MPP8 on target promoters by recruiting G9a/GLP for H3K9 methylation. Disabling MPP8 methyl-H3K9 binding does not affect MPP8-SIRT1 interaction but impairs SIRT1 promoter targeting and E-cadherin silencing (Figs1 and 5, Supplementary Fig S2), suggesting that the stable binding of MPP8 is critical for assembly of ZEB1-MPP8-SIRT1 complex on target promoters.
Consistent with the fact that SIRT1 preferentially deacetylates H4K16 at the physiological concentration 35, our results did not detect significant changes on promoter H3K9 acetylation in SIRT1-KD or MPP8-KD cells. However, we demonstrate that H3K9 deacetylation plays a pivotal role in E-cadherin silencing in a SIRT1-independent manner (Supplementary Fig S5). H3K9 could be deacetylated by HDAC1/2-containing Sin3A and NuRD complexes as they are recruited to E-cadherin promoter by Snail and Twist, respectively 11, 36. HDAC1/2 also exist in ZEB1-CtBP1 complex 17 while knockdown of ZEB1 decreases HDAC1/2 binding 8 and increases H3K9 acetylation on E-cadherin promoter 9, suggesting that ZEB1 recruit HDAC1/2 to deacetylate H3K9 as well. As H3K9 deacetylation is a prerequisite for the stepwise H3K9 methylation, we predict it occurs prior to promoter targeting of MPP8 and SIRT1. In addition, SIRT1 might also indirectly facilitate H3K9 methylation and MPP8 binding as a positive feedback as SIRT1 deacetylates SUV39H1 at K266 to increase its H3K9 MTase activity 37.
MPP8-wt and Δ112–225 mutant displayed similar promoter enrichment (Fig5A and F) suggesting that MPP8 interaction with SIRT1 does not affect its binding to methyl-H3K9 or DNMT3A-K47me 22. We thus postulate that MPP8 recruits DNMT3A and SIRT1 independently. It has been shown that CpG methylation prevents promoter binding by RNA Pol II, while H4K16 acetylation promotes the release of Pol II from promoter-proximal pausing and the transition into active elongation 38, MPP8-directed promoter DNA methylation and H4K16 deacetylation may introduce multiple barriers for transcription. SIRT1 interacts with DNMT1 39 and positively regulates its enzymatic activity 40, indicating that SIRT1 may also recruit DNMT1 to facilitate DNA methylation. Moreover, the facts that H4K16 acetylation directly de-compacts chromatin in vitro 41, 42, 43, 44 and SIRT1 promotes facultative heterochromatin formation by deacetylating H1 35 suggest that MPP8-SIRT1 pathway might also compact local chromatin directly for transcription repression.
Materials and Methods
Knockdown and rescue
Knockdown and shRNA sequence targeting MPP8 was described previously 18, while shRNA targeting SIRT1 are “GTATTGCTGAACAGATGGA” and “GGAAATATATCCTGGACAA.” For rescue, lentiviral vector expressing shRNA-resistant (1102-AACCAG-1107 of NM_017520) Flag-MPP8-wt, Δ112-225, and W80A was transfected into 293T cells with helper vectors for virus production. MPP8-KD cells were then infected with lentiviruses and selected by both blasticidin (10 μg/ml) and puromycin (1 μg/ml).
In vitro deacetylation assay
To generate substrate for in vitro deacetylation assays, HA-MPP8 (FL) and Flag-PCAF were co-expressed in MPP8-KD 293T cells. K439-acetylated MPP8-FL protein was next purified by HA-IP. To generate enzyme, GST-SIRT1 45 was expressed and purified from E. coli BL21 (DE3) Codon-plus (Stratagene) using standard protocol. To set up the deacetylation reaction, purified HA-MPP8-K439ac on HA-7 agarose was incubated with purified 1 μg GST-SIRT1 at 30°C for 10 min with rotation in a 40 μl reaction containing 50 mM Tris–Cl (pH 8.0), 0.8 mM MgCl2, 1 mM NAD+. Control, nicotinamide (NIA, 10 mM), or trichostatin A (TSA, 2 μM) was added in different reactions. The reactions were then stopped and analyzed by Western blot.
ChIP and ReChIP assays
ChIP assays were carried out as previously described 18 with some modifications. Briefly, formaldehyde-cross-linked cells were sonicated using a Bioruptor (Diagenode), while sheared chromatin was incubated with different antibodies. ChIPed complexes were captured on G-protein-conjugated magnetic beads (Cell Signaling) and then reverse-cross-linked. For ReChIP assays, chromatin bound to MPP8 antibody was eluted with reChIP elution buffer (50 mM Tris–Cl, 10 mM DTT, 1% SDS, pH 8.0) and diluted tenfold with reChIP dilution buffer (50 mM Tris–Cl, 167 mM NaCl, 2.2 mM EDTA, 1.1% Triton X-100, pH 8.0). SIRT1 antibody was next added for 2nd IP followed by the same process. Elution was purified using PCR Cleanup kit (Invitrogen) and assayed by real-time qPCR using primer pairs listed in the Supplementary Experimental Procedures.
Other assays
All other assays including pull-down, IP-Western blot analysis, immunostaining, microscopy, LC-MS/MS analysis, migration, and invasion assays were carried out as we described previously 18, 22.
Acknowledgments
We thank Drs. Srikumar Chellappan and John Cleveland for their criticisms, suggestions, and discussions. This work was supported by NIH grant (R01CA172774) and Florida Department of Health grant (3KN02) to J.F., NIH grant (R01CA169210) to E.S., and NIH grants (CA141244 and CA109636) and Florida Department of Health grant (4BB14) to J.C.
Author contributions
LS designed and performed the experiments and analyzed the data; KK participated in MPP8 protein stability analysis; VI and JMK performed peptide acetylation analysis using the LC-MS/MS approach; ES and JC provided important intellectual input and reagents; JF conceived the study, designed the experiments, analyzed the data, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Experimental Procedures and Figures
Review Process File
References
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shang Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res. 2013;319:160–169. doi: 10.1016/j.yexcr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Büchler M, Evert M, Lerch MM, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, Dai Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG, Chi YI, Evers BM, Zhou BP. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2013;32:1351–1362. doi: 10.1038/onc.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol. 2011;408:807–814. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Sun L, Kokura K, Horton JR, Fukuda M, Espejo A, Izumi V, Koomen JM, Bedford MT, Zhang X, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun. 2011;2:533. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, Wolf-Yadlin A, Gozani O. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50:444–456. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Poyurovsky MV, Zhu Y, Prives C. Mdm2 and PCAF increase Chk2 ubiquitination and degradation independently of their intrinsic E3 ligase activities. Cell Cycle. 2009;8:430–437. doi: 10.4161/cc.8.3.7624. [DOI] [PubMed] [Google Scholar]
- Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, Lacroix M, Le Cam L, Coux O, Benkirane M. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- Martinez-Redondo P, Vaquero A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer. 2013;4:148–163. doi: 10.1177/1947601913483767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Németh A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, Nordenskiold L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lu C, Yang Y, Fan Y, Yang R, Liu CF, Korolev N, Nordenskiold L. Influence of histone tails and H4 tail acetylations on nucleosome-nucleosome interactions. J Mol Biol. 2011;414:749–764. doi: 10.1016/j.jmb.2011.10.031. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Experimental Procedures and Figures
Review Process File