Abstract
Unc-51-like kinases (ULKs) are the most upstream kinases in the initiation of autophagy, yet the molecular mechanisms underlying their function are poorly understood. We report a new role for ULK in the induction of autophagy. ULK-mediated phosphorylation of the guanine nucleotide exchange factor DENND3 at serines 554 and 572 upregulates its GEF activity toward the small GTPase Rab12. Through binding to LC3 and associating with LC3-positive autophagosomes, active Rab12 facilitates autophagosome trafficking, thus establishing a crucial role for the ULK/DENND3/Rab12 axis in starvation-induced autophagy.
Keywords: 14-3-3, LC3, DENN domain, GEF, guanine nucleotide exchange factor
Introduction
Macroautophagy, hereafter referred to as autophagy, is an evolutionarily conserved process characterized by the sequestration of cellular structures and cytosolic proteins targeted for destruction into double membrane vesicles called autophagosomes. After autophagy is initiated, by cell starvation, for example, Atg (autophagy related) proteins are recruited to a membrane structure called the phagophore. As the phagophore expands and fuses upon itself, it creates the double membrane autophagosome. The autophagosome then fuses with lysosomes and the enclosed cellular materials are degraded, with the subsequent release of nutrients back to the cytosol for reuse 1.
Among the Atg proteins recruited to the phagophore are Unc-51-like kinases (ULKs), homologues of Atg1 in yeast and the most upstream kinases regulating the initiation of autophagy 2. The kinase activity of ULK is suppressed by mammalian target of rapamycin (mTOR), a master kinase controlling cell size and growth 3. Under starvation, the mTOR-mediated inhibition of ULK is released allowing ULK to initiate autophagy 4, 5. Though ULK activation is a critical and early step in autophagy initiation, only a few downstream targets of the kinase have been identified 6, 7, 8, 9, 10, 11 and thus the mechanisms that underlie the role of ULK in autophagy remain elusive.
Membrane trafficking events are a key aspect of the autophagy process. Rabs, which are the largest family of small GTPase, and which control numerous aspects of membrane trafficking, are emerging as important regulators of autophagy 12. For example, through regulation of membrane trafficking, Rabs contribute to autophagosome formation and maturation 12, 13, 14. Interestingly, it was also recently reported that Rab12 plays a positive role in autophagy by inhibiting mTOR, thus contributing to autophagy initiation 15.
Rabs switch between a membrane-associated GTP-bound active form and a cytosolic, GDP-bound inactive form. Guanine nucleotide exchange factors (GEFs) catalyze the dissociation of GDP from the Rab, allowing for exchange with GTP. The GTP-bound active Rab interacts with Rab effectors to regulate membrane trafficking 16. Proteins bearing a differentially expressed in normal and neoplastic cells (DENN) domain have recently emerged as the largest family of Rab GEFs. At least 26 DENN domain-containing proteins have been identified, most of which are uncharacterized 17, 18, 19. In a large-scale screen using in vitro GEF assays to match DENN domain proteins to Rab substrates, DENN domain-containing protein 3 (DENND3) was identified as a GEF for Rab12 20, and this was later confirmed using cell-based GEF assays 21.
Given that Rab12 is a positive regulator of autophagy 15, we sought to examine a role for DENND3 in this process. Here we identify a novel signaling pathway whereby starvation-induced activation of ULK leads to phosphorylation of endogenous DENND3, with subsequent activation of Rab12 and initiation of membrane trafficking events required for autophagy. These data identify DENND3 as a positive regulator of autophagy and provide a striking example of a signaling pathway impinging on membrane trafficking to control cell function.
Results and Discussion
DENND3 is required for starvation-induced autophagy
Rab12 is a positive regulator of autophagy 15, and since DENND3 is a GEF for Rab12 20, 21, it is likely that DENND3 also functions as a positive regulator of this critical cellular process. To test this directly, we used H1299 cells with stable expression of a GFP-tagged form of the autophagy marker LC3. Following initiation of autophagy, the cytosolic form of LC3 (LC3-I) is converted to a membrane-associated form (LC3-II) through covalent conjugation of phosphatidylethanolamine 22. This membrane-associated form of LC3 associates selectively with newly formed autophagosomes 1. When the H1299 cells were treated with control siRNA, starvation increased the number of GFP-LC3 puncta per cell, an indication of autophagy induction 22 (Fig1A and B). Importantly, DENND3 knockdown with two separate siRNAs significantly attenuates this change, indicating that DENND3 is a positive regulator of starvation-induced autophagy (Fig1A and B).
Figure 1.
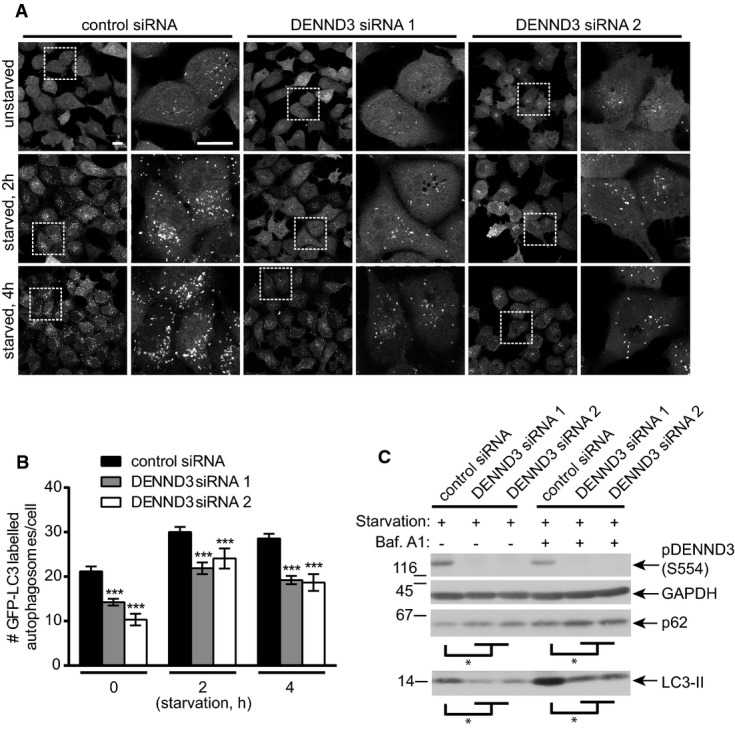
- H1299 cells stably expressing GFP-LC3 were transfected with control siRNA or two DENND3 siRNAs and were subsequently left unstarved or were starved with Earle's balanced salt solution (EBSS) for 2 or 4 h. Cells were then processed for cytochemical detection of GFP. The images on the right of each treatment are magnified views of the areas indicated by the white boxes on the image to the left. The scale bars represent 20 μm.
- The number of GFP-positive autophagosomes from images as in (A) was counted from n > 400 cells for each treatment over 3 repeats. The bars represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Dunnett's post-test. ***P < 0.001.
- Cells were transfected with control siRNA or two siRNAs targeting DENND3 and were subsequently starved with EBSS for 6 h in the presence or absence of bafilomycin A1 (Baf. A1, 100 nM). Lysates were immunoblotted with antibodies recognizing the indicated proteins (n = 3 repeats). The levels of LC3-II and p62 normalized to GAPDH level. Statistical analysis employed one-way ANOVA followed by Dunnett's post-test. *P < 0.05.
As described above, increased levels of LC3-II are an indicator of autophagy 22. We find that the level of LC3-II induced by starvation is significantly decreased following DENND3 knockdown (Fig1C). The inhibition of autophagy resulting from DENND3 knockdown is not due to a block of autophagic flux as the amount of LC3-II is still reduced by DENND3 knockdown in the presence of bafilomycin A1 (Fig1C), which blocks fusion of autophagosomes with lysosomes 22. We also monitored the levels of p62 as a measure of autophagy status. Consistent with the LC3 data, DENND3 knockdown significantly increases the levels of p62 seen following cell starvation (Fig1C). Thus, as for Rab12 15, DENND3 is a positive regulator of autophagy. We note that DENND3 knockdown decreases the number of GFP-LC3-positive puncta in both starved and unstarved cells (Fig1A and B), whereas an influence of DENND3 knockdown on the levels of endogenous LC3-II or p62 was only readily detectable following starvation. Thus, the influence of DENND3 on autophagy in the absence of starvation appears rather modest and likely requires a starvation-induced signal. It was recently reported that DENND3 knockdown does not influence autophagy in mouse embryonic fibroblasts 21. The difference between that study and the results reported here are not currently understood. However, it is notable that in the same study 21, DENND3 overexpression induced autophagy, supporting that it does have a positive role in this process.
Starvation induces ULK-mediated phosphorylation of DENND3 at two unique sites
To investigate the mechanism by which DENND3 regulates autophagy, we sought to identify DENND3-binding partners. Flag-DENND3 was immunoprecipitated, and co-immunoprecipitating proteins were identified by mass spectrometry. This led to the identification of 6 of the 7 known isoforms of 14-3-3 proteins (Supplementary Table S1). The mass spectrometry data were confirmed by immunoprecipitation of Flag-DENND3 with blots for individual 14-3-3 isoforms (Supplementary Table S1) or with a pan-14-3-3 antibody (Fig2A). There are two canonical 14-3-3 binding motifs: type I (R-S-X-pS-X-P) and type II (R-X-X-X-pS-X-P) 23. The S in the motifs generally needs to be phosphorylated to enable 14-3-3 binding. The amino acid sequences around S554 and S572 in DENND3 match the 14-3-3 binding motifs with the key residues conserved across vertebrate species (Fig2B). We thus generated Flag-DENND3 S to A mutants for both S554 and S572. When Flag-DENND3 was immunoprecipitated, there was no co-immunoprecipitation of endogenous 14-3-3 proteins with the S554A mutant and greatly reduced co-immunoprecipitation with the S572A mutant when compared to wild-type DENND3 (Fig2A), thus confirming the importance of these sites for 14-3-3 binding.
Figure 2.
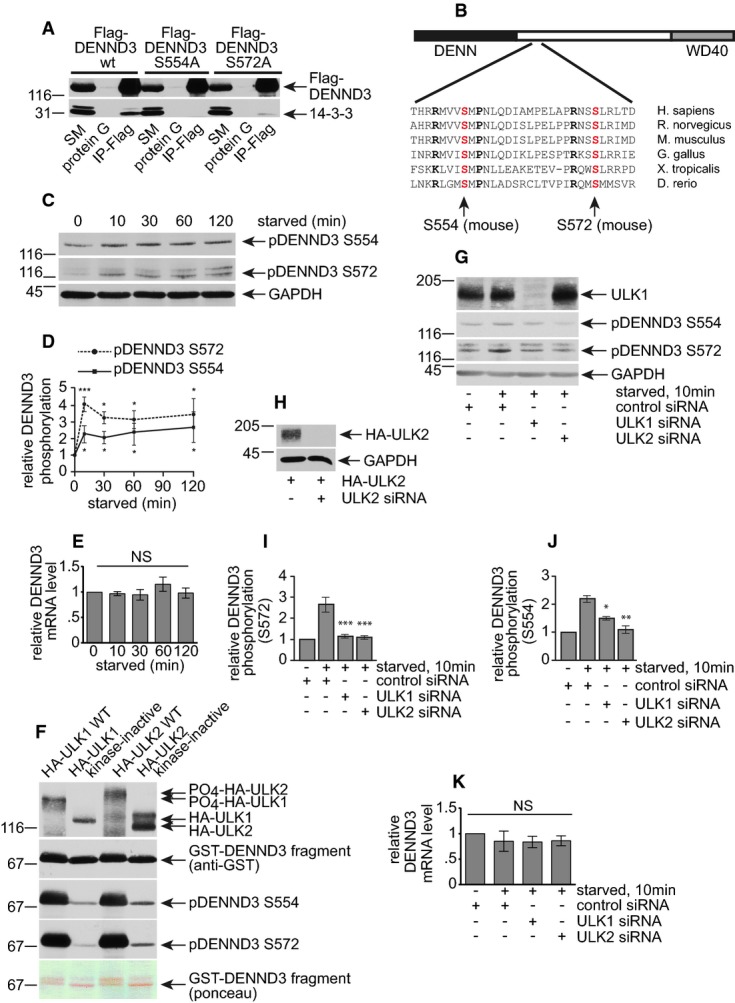
- A Lysates from HEK-293T cells transfected with Flag-DENND3 wild-type (WT), S554A or S572A mutant were incubated with protein G beads alone or protein G beads coupled to anti-Flag monoclonal antibody (IP-Flag). Proteins bound specifically to the beads were processed for Western blot with anti-Flag and anti-pan-14-3-3 antibodies. An aliquot of the cell lysate (starting material, SM) equal to 10% of that added to the beads was analyzed in parallel (n = 3 repeats).
- B Schematic diagram of DENND3 with a N-terminal DENN domain, a C-terminal WD40 domain, and a presumably weakly structured linker region (white box). The aligned sequences are from a region of the linker and indicate the high degree of conservation across various vertebrate species. S554 and S572 (position based on the mouse sequence) are indicated.
- C HeLa cells were left unstarved (0 min) or were starved with EBSS for the indicated time periods. Lysates were immunoblotted with antibodies recognizing the indicated proteins.
- D Relative DENND3 phosphorylation at serine 554 and serine 572 was determined from 3 experiments as in (C). Points represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Dunnett's post-test. ***P < 0.001, *P < 0.05.
- E HeLa cells were processed as in (C), followed by quantitative real-time PCR (n = 4 repeats). Bars represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Dunnett's post-test. NS = not significant.
- F Purified GST-DENND3 (538–973) was subjected to in vitro phosphorylation by purified HA-ULKs (n = 3 repeats). Note that the WT ULK1 and ULK2 run higher on SDS–PAGE than the kinase-inactive forms, due to autophosphorylation, and a shift of the phosphorylated GST-DENND3 can also be seen on a Ponceau-stained transfer.
- G HeLa cells were transfected with control siRNA or siRNA targeting ULK1 or ULK2 and were subsequently left unstarved or starved with EBSS for 10 min. Lysates were immunoblotted with the indicated antibodies (n = 3 repeats).
- H For validating the ULK2 siRNA knockdown efficiency without workable antibody, HeLa cells were transfected with HA-tagged ULK2 with or without siRNA for ULK2 and lysates were immunoblotted with the indicated antibodies (n = 3 repeats).
- I, J Relative DENND3 phosphorylation at S572 (H) and S554 (I) was determined from three repeats as in (F). Bars represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Tukey's post-test. ***P < 0.001, **P < 0.01. *P < 0.05.
- K HeLa cells were processed as in (G), followed by qPCR (n = 4 repeats). Bars represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Tukey's post-test. NS = not significant.
14-3-3 binding predicts that S554 and S572 are phosphorylated and we thus generated anti-phosphopeptide antibodies targeting these sites. To test whether the antibodies are specific for the phosphorylated sites, Flag-DENND3 wild-type, S554A or S572A was expressed in HEK-293T cells and lysates were blotted with anti-Flag, anti-pS554 or anti-pS572 antibody. Anti-pS554 and anti-pS572 fail to detect the S554A and S572A mutants, respectively, despite even loading of the proteins as revealed by Flag blot (Supplementary Fig S1A). Moreover, treatment of immunoprecipitated Flag-DENND3 with calf intestinal alkaline phosphatase (CIP) eliminates the reaction of anti-pS572 and strongly reduces reactivity of anti-pS554 (Supplementary Fig S1B). The remaining signal with the anti-pS554 antibody after CIP treatment suggests that there may be some phosphorylation-independent DENND3 antibodies in the anti-pS554 sample. Finally, siRNA-mediated knockdown of DENND3 leads to a reduced signal detected with anti-pS554 and anti-pS572 antibodies (Supplementary Fig S1C), indicating that the antibodies detect endogenous phosphorylated protein. Together, these data validate that the antibodies recognize DENND3 phosphorylated at S554 and S572 and demonstrate that these sites are at least partially phosphorylated at steady state.
Since DENND3 functions in starvation-induced autophagy, we asked whether starvation regulates DENND3 phosphorylation. Cells were left unstarved or were starved for times ranging from 10 to 120 min and then blotted with the anti-pDENND3 antibodies. Intriguingly, starvation enhances phosphorylation of both S554 and S572 of the endogenous DENND3 protein in HeLa cells, and phosphorylation of both sites peaks by 10 min and stays elevated for 120 min (Fig2C and D) and even as long as 6 h (data not shown). Due to the lack of a pan-DENND3 antibody, we were unable to directly determine whether starvation had any influence on the total levels of DENND3 protein. However, quantitative real-time PCR revealed no changes in DENND3 mRNA levels resulting from starvation (Fig2E). Thus, the enhanced signal detected for the anti-pDENND3 antibodies (Fig2C and D) upon starvation is due to enhanced phosphorylation, indicating that DENND3 phosphorylation is a downstream event of cell starvation.
We next sought to identify the starvation-activated kinase responsible for DENND3 phosphorylation. ULK is an upstream kinase of starvation-induced autophagy and is thus a potential candidate for this activity. There are two isoforms of ULK in mammals, ULK1 and ULK2, and interestingly, overexpression of wild-type forms of either kinase, but not overexpression of kinase-inactive mutants, significantly increases the phosphorylation of S554 and S572 (Supplementary Fig S2A–C), while the mRNA levels of DENND3 stay the same among these overexpression conditions (Supplementary Fig S2D). To determine whether ULKs phosphorylate DENND3 directly, we performed in vitro kinase assays using HA-tagged ULK1/2 immunoprecipitated from HEK-293T cells and purified bacterial GST-DENND3 encoding amino acids 538–973. ULK1 and ULK2 wild-type kinases phosphorylate DENND3 at both S554 and S572 when compared to the kinase-inactive mutants (Fig2F). Moreover, the starvation-induced phosphorylation of DENND3 at both S554 and S572 is suppressed by ULK1/2 knockdown (Fig2G–J), while the mRNA levels of DENND3 do not change between these treatments (Fig2K). Thus, it appears that ULK directly phosphorylates DENND3 although it remains formally possible that ULK influences DENND3 indirectly through another kinase. Since ULKs can bind their kinase substrates 11, we investigated whether ULK binds to DENND3. Because we do not have antibodies for immunoprecipitation of endogenous ULK or DENND3, we were unable to test the ULK/DENND3 interaction at the endogenous level. However, we found that both overexpressed HA-ULK1 and HA-ULK2 are co-immunoprecipitated with Flag-tagged DENND3 (Supplementary Fig S2E and F). WD40 repeat domains often function as scaffolds for protein interactions, and this region of DENND3 is involved in the interaction with ULK (Supplementary Fig S2G). In summary, it appears likely that ULK interacts with DENND3 and mediates its starvation-induced phosphorylation.
Starvation activates Rab12
We next used a Rab12 effector-binding assay to determine whether the starvation-induced phosphorylation of DENND3 mediated by ULK changes the activation status of endogenous Rab12. Rab-interacting lysosomal protein-like 1 (RILPL1) was previously identified as a Rab12 effector 24. Consistently, GST-RILPL1 binds to Flag-Rab12 Q101L, a constitutively GTP-bound mutant, with little or no binding to Flag-Rab12 T56N, a constitutively GDP-bound mutant (Supplementary Fig S3A). GST-RILPL1 was then used in effector-binding assay with cells starved for various times. Intriguingly, as for the induction of DENND3 phosphorylation resulting from starvation (Fig2C and D), the amount of endogenous Rab12 binding to GST-RILPL1, which reflects active Rab12 and likely the GEF activity of DENND3, also peaks within 10 min and lasts for at least 120 min (Fig3A, Supplementary Fig S3B). These data support the hypothesis that phosphorylation of DENND3 enhances its GEF activity toward Rab12.
Figure 3.
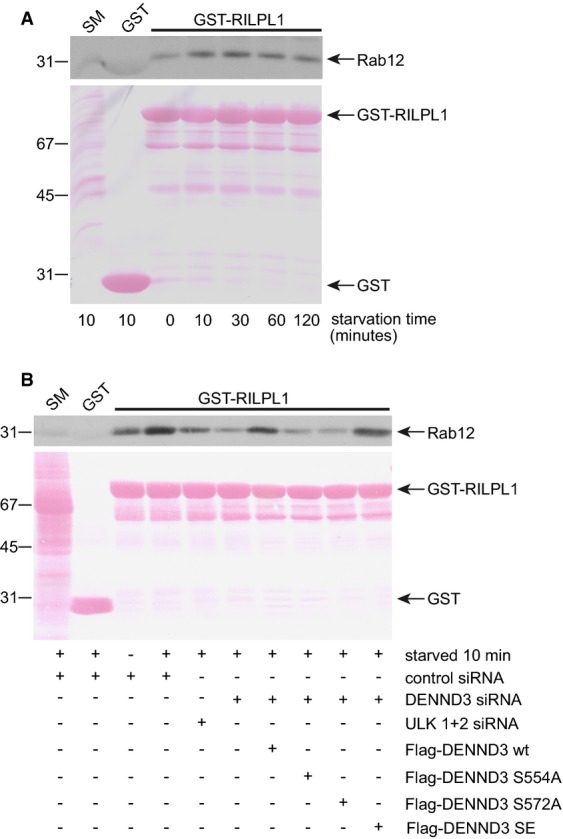
- HeLa cells were left unstarved or were starved with EBSS for the indicated time. Lysates were then prepared and incubated with GST or GST-RILPL1 coupled to glutathione-Sepharose beads. Protein specifically bound to the beads was processed for Western blot with antibody recognizing Rab12. GST and GST-RILPL1 are indicated in the Ponceau-stained transfer. An aliquot of the lysate (starting material, SM) equal to 5% of that added to the beads was analyzed in parallel (n = 3 repeats).
- HeLa cells were treated with control siRNA or with siRNA for DENND3 or ULK1 and ULK2. Additionally, cells that were treated with DENND3 siRNA were subsequently transfected with wild-type (WT) DENND3 or DENND3 with S554A, S572A, or S to E double mutations. The cells were subsequently starved with EBSS for 10 min, and then, lysates were prepared and incubated with GST or GST-RILPL1 coupled to glutathione-Sepharose beads. Protein specifically bound to the beads was processed for Western blot with antibody recognizing Rab12. GST and GST-RILPL1 are indicated in the Ponceau-stained transfer. An aliquot of the lysate (starting material, SM) equal to 2.5% of that added to the beads was analyzed in parallel (n = 3 repeats).
We next performed knockdown/rescue experiments to confirm that phosphorylation of DENND3 contributes to the enhanced Rab12 activity resulting from starvation. Knockdown of DENND3 suppresses starvation-induced increases in Rab12 activity (Fig3B, Supplementary Fig S3C). Moreover, knockdown of ULK1/2 also suppresses starvation-induced Rab12 activation (Fig3B, Supplementary Fig S3C). The decrease in starvation-induced Rab12 activation resulting from DENND3 knockdown is rescued with wild-type DENND3 and with a phosphomimetic mutant in which both S554 and S572 are mutated to E. Importantly, DENND3 S554A and S572A phosphorylation-defective mutants fail to rescue the decrease in starvation-induced Rab12 activation resulting from DENND3 knockdown (Fig3B). Thus, ULK-mediated phosphorylation of DENND3 resulting from starvation causes increased activation of Rab12.
Our work not only identifies DENND3 as a new target for ULK, for which there are currently a limited number of substrates 7, 8, 9, 10, 11, but also demonstrates that ULK phosphorylation creates 14-3-3 docking sites on a substrate. One unresolved question is the role of 14-3-3 binding in the regulation of DENND3 function. Interaction with 14-3-3 proteins can alter protein localization or catalytic activity of a binding partner 23. Given that phosphorylation of DENND3, which is required for 14-3-3 binding, appears to enhance the GEF activity of DENND3 toward Rab12, we speculate that 14-3-3 binding influences the catalytic activity of DENND3. Perhaps 14-3-3 binding functions to shield the phosphate groups from phosphatases to facilitate the change in GEF activity. Future studies will address these issues.
Rab12 interacts with LC3
A recent proteomic analysis of the autophagy interaction network revealed that DENND3 associates with LC3 25. In examining this interaction, we made the surprising discovery that Rab12 also interacts with LC3 (Fig4A). We thus asked whether LC3 is an effector for Rab12. Since Rabs bind to their effectors when they are in the GTP-bound form, we performed a pull-down experiment using Rab12 loaded with GTPγS or rendered nucleotide-free through the addition of EDTA. Endogenous LC3 did not show a preference for either of the forms of Rab12 (Fig4A). In contrast, the DENN domain of DENND3 prefers the nucleotide-free form of Rab12, a hallmark feature of GEFs (Fig4B). This not only validates the nucleotide status of the Rab12 protein under the different treatment conditions, but it also indicates that the interaction between LC3 and Rab12 is unlikely to be mediated indirectly via DENND3. Moreover, there was no preference of Rab12 for LC3-1 versus LC3-II (Fig4A). However, considering that GTP-loaded Rabs associate with membranes and that starvation leads to enhanced levels of active Rab12 (Fig3, Supplementary Fig S3B), Rab12 should have a greater opportunity to interact with membrane-associated LC3-II if it localizes to autophagosome membranes after it is activated by DENND3 upon starvation.
Figure 4.
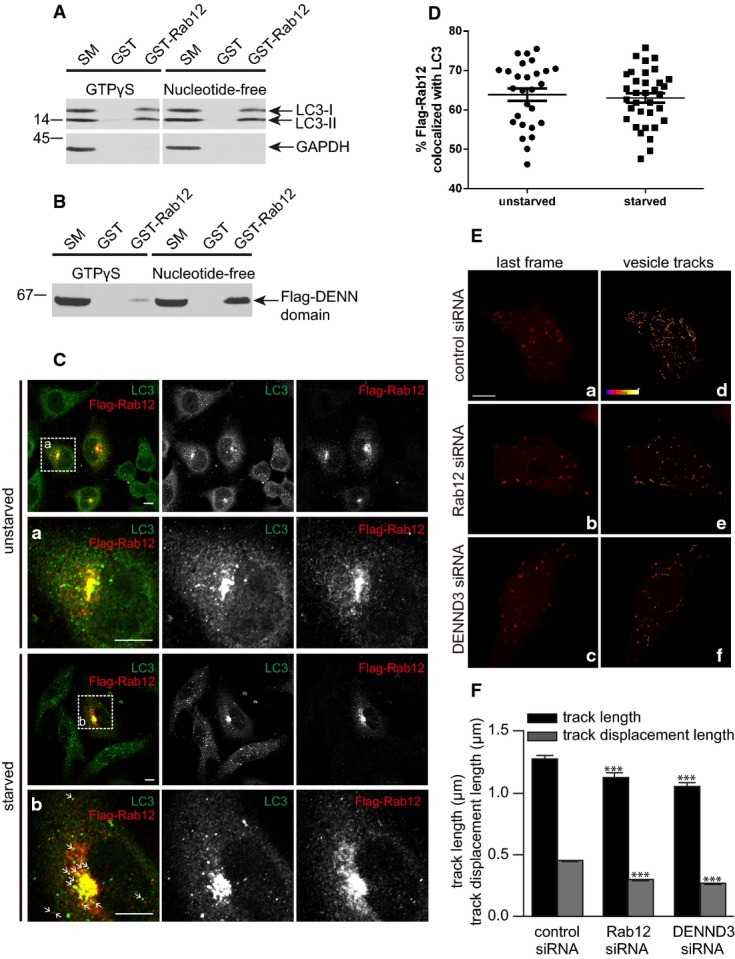
- Lysates prepared from HeLa cells starved with EBSS for 4 h were incubated with GST or GST-Rab12 pre-bound to glutathione-Sepharose beads. GST-Rab12 was loaded with GTPγS and stabilized with the addition of MgCl2 or was rendered nucleotide-free by the addition of EDTA. GST was processed in the same manner. Proteins specifically bound to the beads were processed for Western blot with antibodies against the indicated proteins. An aliquot of the cell lysate (starting material, SM) equal to 1% of that added to the beads was analyzed in parallel (n = 3 repeats).
- Lysates from HEK-293T cells transfected with Flag-tagged DENN domain of DENND3 were incubated with GST or GST-Rab12 pre-bound to glutathione-Sepharose beads and treated as described in (A). Proteins bound specifically to the beads were processed for Western blot with anti-Flag antibody. An aliquot of the cell lysate (starting material, SM) equal to 10% of that added to the beads was analyzed in parallel (n = 3 repeats).
- HeLa cells transfected with Flag-Rab12 were left unstarved or were starved with EBSS for 40 min and were subsequently processed for immunocytochemistry with antibodies recognizing Flag and LC3. The second rows of panels under each condition are a magnified view of the region labeled by the white boxes on the lower magnification image on the top. The scale bar represents 5 μm.
- Co-localization of overexpressed Flag-Rab12 and LC3 was quantified using Imaris. Experiments were repeated 3 times, n = 61 cells. Bars represent mean ± SEM. Statistical analysis employed unpaired t-test.
- HeLa cells, transfected with control siRNA or siRNAs targeting Rab12 or DENND3, were subsequently transfected with mCherry-LC3, starved with EBSS for 1 h and imaged live. Panels a–c (with increased brightness to highlight the puncta) show the last frames of image sequences from cells treated with control siRNA, Rab12 siRNA, and DENND3 siRNA, respectively. The scale bar represents 5 μm. Panels d–f show the same images as in a–c, respectively, overlaid with mCherry-LC3 vesicle tracks recorded during 1 min of imaging. The color of the tracks represents the time when the tracks occurred, as indicated in the color scale bar in d, which represents a time line from 0 to 60 s. The images shown here correspond to Supplementary Videos S2, S3 and S4.
- LC3 vesicle track length and track displacement length were quantified from over 40 cells and 9,000 tracks. The experiments were repeated 3 times. The bars represent mean ± SEM. Statistical analysis employed one-way ANOVA followed by Dunnett's post-test. ***P < 0.001.
We thus tested for association of Rab12 with autophagosome membranes. HeLa cells transfected with Flag-Rab12 were processed for immunofluorescence with or without starvation. Flag-Rab12 partially co-localizes with endogenous LC3 at the perinuclear region (Fig4C and D). In fact, LC3 staining becomes more prominent in the perinuclear region in the Flag-Rab12-transfected cells, providing additional evidence to support the interaction between Rab12 and LC3. We quantified the extent of co-localization of Flag-Rab12 and LC3 with and without starvation and found that the co-localization was independent of starvation (Fig4D). Consistent with a previous report 26, the perinuclear pool of Rab12 corresponds to Rab11- and transferrin receptor-positive recycling endosomes (Supplementary Fig S4). Interestingly, upon starvation, a portion of Flag-Rab12 redistributes to LC3-positive puncta away from the perinuclear region, likely corresponding to LC3-positive autophagosomes (indicated by the arrows in Fig4D). Thus, Rab12 and LC3 co-localize at recycling endosomes, a source of autophagosome membranes 27 and potentially also in autophagosomes.
DENND3 and Rab12 mediate autophagosome trafficking
Finally, we wondered whether Rab12 was involved in trafficking of autophagosomes. We thus performed live cell imaging using spinning disk confocal microscopy in starved HeLa cells transfected with GFP-Rab12 and mCherry-LC3. These studies reveal that a portion of GFP-Rab12 puncta co-traffic with mCherry-LC3, suggesting a role of Rab12 in autophagosome trafficking (Supplementary Video S1). To test this more directly, we performed Rab12 and DENND3 knockdown in HeLa cells expressing mCherry-LC3. Live cell imaging reveals that the track length and track displacement length of LC3-labeled autophagosomes are decreased in both Rab12 and DENND3 knockdown cells compared to control (Fig4E and F, Supplementary Videos S2, S3 and S4). This indicates that Rab12 is involved in the transport of autophagosomes and, moreover, that activation of Rab12 by DENND3 is likely also necessary for this process.
Rab12 has been reported to induce autophagy indirectly by promoting trafficking of the proton-coupled amino acid transporter 4 (PAT4) from recycling endosomes to lysosomes leading to PAT4 downregulation at steady state, such that Rab12 activation leads to decreased amino acid transport, inhibition of mTOR, and activation of autophagy 15. However, recycling endosomes are a key site where ATG16L1- and mATG9-containing autophagic membranes coalesce 27 and membrane from recycling endosomes is delivered to and incorporates into forming autophagosomes following cell starvation 28. These results are consistent with a more direct role for Rab12 in autophagy. Our data suggest a model in which ULK, activated by starvation, phosphorylates DENND3 to upregulate DENND3 GEF activity and to activate Rab12, with the activated GTPase on the membranes of the recycling endosome subsequently integrated into autophagosomes. From there, Rab12 plays a role in facilitating autophagosome trafficking. It is possible that the interaction between Rab12 and LC3 aids in the incorporation of either protein into the forming autophagosomes and that Rab12 and DENND3 ablation suppresses the formation of autophagosome due to accumulation of autophagosomes with less mobility.
Rab12 has been reported to be involved in transferrin receptor trafficking from recycling endosomes to lysosomes 26 and in retrograde trafficking of Shiga toxin 29. Given that transferrin receptor can be localized to ULK-positive autophagosomes under amino acid starvation 28 and that Shiga toxin can induce autophagy and localize to autophagosomes 30, our data suggest the role of Rab12 in autophagosome trafficking accounts for the phenomena of the involvement of Rab12 in transferrin receptor and Shiga toxin transport. Considering the motor proteins are prominent among Rab effectors 31, Rab12 facilitates autophagosome trafficking likely through interacting with certain motor protein. The following work searching for the motor protein may shed light on the mechanism underlying autophagosome trafficking.
It has been reported that the phosphorylation of some ULK substrates has relevance for starvation-induced autophagy, such as Beclin-1 and Atg9 9, 10. However, few studies have provided detailed information regarding the exact mechanism linking ULK activation to autophagy initiation. The interface between signal transduction and membrane trafficking is an important and emerging area in cell biology. By uncovering the role of Rab12 and DENND3 in autophagy, our study presents a novel pathway from starvation-induced ULK signaling through DENND3 to Rab12-mediated membrane trafficking required for autophagy, providing a vital new understanding of ULK function in autophagy.
Materials and Methods
Antibodies and reagents
Rabbit polyclonal antibodies against Rab12 and ULK1 were from Proteintech and Santa Cruz, respectively. Rabbit polyclonal antibodies recognizing LC3 were from Cell Signaling and Medical and Biological Laboratories. Mouse monoclonal pan-14-3-3 and GAPDH antibodies were from Santa Cruz. Mouse monoclonal Flag antibody was from Sigma. Rat monoclonal antibody against HA was from Roche. The rabbit polyclonal antibodies against pS554 or pS572 of DENND3 were custom made under contract with Phosphosolutions. H1299 cells stably expressing GFP-LC3 were a gift from Dr. Gordon Shore, McGill University. Production of various constructs is described in Supplementary Materials and Methods.
Plasmid overexpression and siRNA-mediated knockdown
Constructs and siRNAs were expressed in cell lines using jetPRIME (Polyplus transfection) or Lipofectamine RNAiMAX (Life Technologies), according to the manufacturer's instructions. AllStars Negative Control siRNA (Qiagen) served as non-targeting negative control siRNA. siRNA sequences used are described in Supplementary Materials and Methods.
Immunoprecipitation and pull-down assays
For immunoprecipitation assays, cells were collected in HEPES lysis buffer (20 mM HEPES, pH 7.4, 10 mM sodium fluoride, 0.5 mM sodium orthovanadate, 60 nM okadaic acid, 100 mM sodium chloride, 1% Triton X-100, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mM benzamidine, and 0.23 mM phenylmethylsulfonyl fluoride). Following 10 min at 4°C, lysates were spun at 20,000 g for 15 min. The supernatant was incubated for ∽3 h at 4°C with antibodies coupled to protein A/G-Sepharose. Beads were subsequently washed three times with HEPES lysis buffer and analyzed by SDS–PAGE. Samples were then processed for Western blot, or bands co-immunoprecipitating with Flag-DENND3, revealed by Coomassie staining, were analyzed by mass spectrometry. For pull-down assays, lysates prepared as described above were incubated for ∽3 h at 4°C with GST or GST fusion proteins coupled to glutathione-Sepharose. The samples were washed and prepared for Western blot as described above.
In vitro kinase assay
HEK-293T cells were transfected with HA-ULK1, wild-type (WT) or a kinase-inactive mutant, or HA-ULK2, wild-type (WT) or a kinase-inactive mutant. Lysates were prepared, and HA-tagged ULK proteins were immunoprecipitated from the lysates with anti-HA antibody coupled to protein G-Sepharose beads. The purified proteins on beads were washed in kinase buffer (25 mM Tris–HCl, pH 7.5, 0.1 mM sodium orthovanadate, 10 mM magnesium chloride, 5 mM β-glycerophosphate, 2 mM dithiothreitol) and added to Eppendorf tubes containing purified GST fusion protein from E. coli in kinase buffer encoding a fragment of DENND3 from residue 538 to 973 after supplementing ATP (final 200 μM). The samples were incubated at 37°C for 30 min with vigorous shaking and subsequently processed for SDS–PAGE.
Effector-binding assay
Cells were collected in Tris lysis buffer (pH 7.4, 50 mM Tris, 10 mM sodium fluoride, 0.5 mM sodium orthovanadate, 60 nM okadaic acid, 100 mM sodium chloride, 3.3 mM magnesium chloride, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 5% glycerol, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mM benzamidine, and 0.23 mM phenylmethylsulfonyl fluoride). Following 10 min at 4°C, lysates were spun at 20,000 g for 15 min. Samples from different treatments were equalized to the same protein concentration after Bradford assay, then incubated with GST or GST-RILPL1 coupled to glutathione-Sepharose for ∽3 h at 4°C. After three washes, samples were analyzed by Western blot.
Quantitative real-time PCR
Quantitative real-time PCR was performed through qPCR service from Institute for Research in Immunology and Cancer (IRIC), Montreal. DENND3 oligos: forward: tcatgggagcatcacctactc, reverse: gagctggaggctctgcac; ACTB control oligos: forward: attggcaatgagcggttc, reverse: tgaaggtagtttcgtggatgc; GAPDH control oligos: forward: agccacatcgctcagacac, reverse: gcccaatacgaccaaatcc.
Statistical evaluation
Values, expressed as mean ± SEM, were obtained from not less than three independent experiments. Statistical analysis of the results was carried out by t-test or one-way analysis of variance, followed by Dunnett's or Tukey's multiple comparison test when appropriate. P < 0.05 was considered significant.
Acknowledgments
We thank Jacynthe Philie for technical assistance and Heidi McBride and Ayumu Sugiura for access to and assistance with spinning disk confocal microscope. We also thank Gordon Shore for the gift of H1299 cells stably expressing GFP-LC3. This work was supported by a grant from the Canadian Institutes of Health Research (MOP-62684) to PSM. JX was supported by a Fonds de la recherche en santé du Québec (FRSQ) fellowship, and PSM is a James McGill Professor.
Author contributions
JX conceived and performed the experiments and wrote the manuscript. MF performed the experiments. PSM conceived the experiments and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Materials and Methods, Supplementary Figures, Supplementary Figure and Video Legends
Supplementary Table S1
Supplementary Video S1
Supplementary Video S2
Supplementary Video S3
Supplementary Video S4
Review Process File
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci USA. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Lee JY, Tang HW, Debnath J, Thomas SM, Settleman J. Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy. 2008;4:37–45. doi: 10.4161/auto.5141. [DOI] [PubMed] [Google Scholar]
- Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–651. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari Z, Sass M. The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy. 2014;10:1154–1166. doi: 10.4161/auto.29395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Puri C, Moreau K, Rubinsztein DC. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci. 2013;126:1059–1069. doi: 10.1242/jcs.123075. [DOI] [PubMed] [Google Scholar]
- Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14:450–457. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. Discovery of Novel DENN Proteins: implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Noguchi K, Fukuda M. Dennd3 Functions as a Guanine Nucleotide Exchange Factor for Small GTPase Rab12 in Mouse Embryonic Fibroblasts. J Biol Chem. 2014;289:13986–13995. doi: 10.1074/jbc.M113.546689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. How do 14-3-3 proteins work?– Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Itoh T, Fukuda M. Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic. 2011;12: 1432–1443. doi: 10.1111/j.1600-0854.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell GE, Renard HF, Garcia-Castillo MD, Dingli F, Loew D, Lamaze C, Romer W, Johannes L. Rab12 localizes to Shiga toxin-induced plasma membrane invaginations and controls toxin transport. Traffic. 2014;15:772–787. doi: 10.1111/tra.12173. [DOI] [PubMed] [Google Scholar]
- Lee MS, Cherla RP, Jenson MH, Leyva-Illades D, Martinez-Moczygemba M, Tesh VL. Shiga toxins induce autophagy leading to differential signalling pathways in toxin-sensitive and toxin-resistant human cells. Cell Microbiol. 2011;13:1479–1496. doi: 10.1111/j.1462-5822.2011.01634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer JA, 3rd, Wu XS. Rabs grab motors: defining the connections between Rab GTPases and motor proteins. Curr Opin Cell Biol. 2002;14:69–75. doi: 10.1016/s0955-0674(01)00296-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods, Supplementary Figures, Supplementary Figure and Video Legends
Supplementary Table S1
Supplementary Video S1
Supplementary Video S2
Supplementary Video S3
Supplementary Video S4
Review Process File


