Abstract
Methenyltetrahydrofolate synthetase (MTHFS) expression enhances folate-dependent de novo purine biosynthesis. In this study, the effect of increased MTHFS expression on the efficacy of the glycinamide ribonucleotide formyltransferase (GARFT) inhibitor LY309887 was investigated in SH-SY5Y neuroblastoma. GARFT catalyzes the incorporation of formate, in the form of 10-formyltetrahydrofolate, into the C8 position of the purine ring during de novo purine biosynthesis. SH-SY5Y neuroblastoma with increased MTHFS expression displayed a 4-fold resistance to the GARFT inhibitor LY309887, but did not exhibit resistance to the thymidylate synthase inhibitor Pemetrexed. This finding supports a mechanism whereby MTHFS increases the availability of 10-formyltetrahydrofolate for GARFT. MTHFS expression is elevated in animal tumor tissues compared to surrounding normal tissue, consistent with the dependence of transformed cells on de novo purine biosynthesis. The level of MTHFS expression in tumors may predict the efficacy of antipurine agents that target GARFT.
Keywords: folate metabolism, methenyltetrahydrofolate synthetase, purine biosynthesis, LY309887, antifolate, glycinamide ribonucleotide formyltransferase
Introduction
Folates coenzymes carry and chemically activate one-carbons on the N5 and/or N10 position of tetrahydrofolate (THF) for one-carbon transfer reactions. Folate coenzymes consist of a reduced pteridine, p-aminobenzoate and a glutamate polypeptide that contains 2 to 9 glutamate residues linked through γ-peptide linkages [1]. Folates and folate analogs are transported into cells as monoglutamate derivatives and then converted to polyglutamate derivatives by the enzyme folylpolyglutamate synthetase (FPGS) [1]. Polyglutamylation serves to retain the folate/folate analog within the cell, and to increase the affinity of folate derivatives for the enzymes that utilize them [1, 2]. Folate-dependent one-carbon metabolism is required for the biosynthesis of purines, thymidylate, and methionine (Figure 1). Methionine can be converted to the methyl donor S-adenosylmethionine (AdoMet), which is a cofactor for numerous methylation reactions, including the methylation of lipids, proteins, DNA and RNA.
Figure 1. Folate-dependent one-carbon metabolism in the cytoplasm and inhibition by LY309887, Lometrexol, and Pemetrexed.
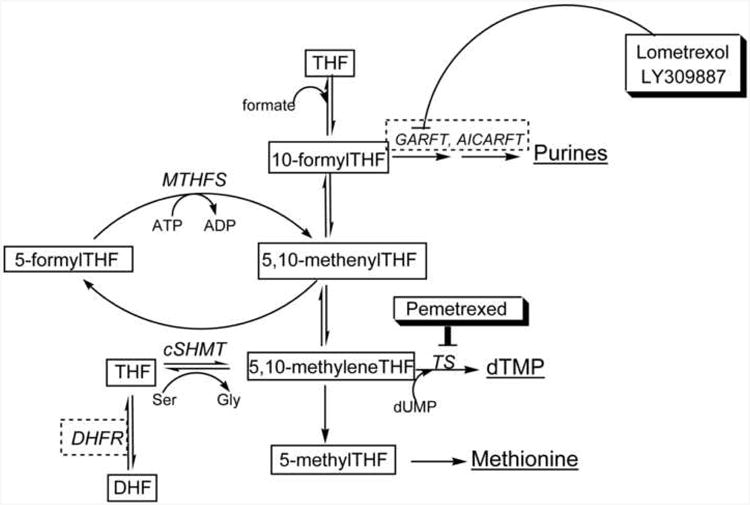
Products of one-carbon metabolism are underlined. Lometrexol and LY309887 inhibit GARFT. Pemetrexed primarily inhibits TS, but also inhibits other folate-dependent enzymes shown within dashed boxes. DHF, dihydrofolate; THF, tetrahydrofolate; MTHFS, methenyltetrahydrofolate synthetase; cSHMT, cytoplasmic serine hydroxymethyltransferase; GARFT, glycinamide ribonucleotide formyltransferase; AICARFT, phosphoribosylaminoimidazole carboxamide formyltransferase; DHFR, dihydrofolate reductase; TS, thymidylate synthase.
Folate antagonists have proven to be effective in the treatment of certain cancers [3]. The efficacy of an antifolate is determined by several factors: 1) its affinity for the target enzyme, 2) its affinity for folate transporters, and 3) its ability to serve as a substrate for polyglutamylation catalyzed by FPGS [3-6]. Efficacy can also be modulated by a host of other factors including an individual's folate status and/or pharmacogenetic profile, an heritable drug response phenotype [7]. Folate antagonists that target enzymes in the de novo thymidylate biosynthesis pathway, including dihydrofolate reductase (DHFR) and thymidylate synthase (TS), have proven effective in the treatment of a variety of cancers [3]. Folate-dependent enzymes from the de novo purine nucleotide biosynthesis pathway have also been targets for the development of antineoplastic agents. Lometrexol (5,10-dideazatetrahydrofolate or DDATHF) was the first purine synthesis inhibitor to reach clinical trials. It targets glycinamide ribonucleotide formyltransferase (GARFT) and was shown to have potent antitumor activity against human xenografts in mice [3]. GARFT and aminoimidazolecarboxamide ribonucleotide formyltransferase (AICARFT) catalyze the incorporation of activated formate, from the coenzyme 10-formylTHF, into the C8 and C2 carbons of the purine ring, respectively. The clinical utility of Lometrexol is limited by its toxicity, which results from a build-up of Lometrexol polyglutamates in the liver [3, 4]. LY309887 is a second generation compound of Lometrexol and a more potent GARFT inhibitor (the Ki values for human GARFT are 60 nM and 2 nM for Lometrexol and LY309887, respectively). Moreover, LY309887 causes rapid depletion of purine pools in CCRF-CEM cells lines and more effectively inhibits tumor growth compared to Lometrexol [3, 4, 8]. LY309887 exhibits lower affinity for FPGS than Lometrexol, but was still subject to delayed toxicity during clinical trials [3]. Pemetrexed (currently in use clinically as Alimta, also known as multi-targeted antifolate) was designed as an anti-purine antifolate, but has been shown to inhibit several folate-utilizing enzymes. Pemetrexed primarily inhibits TS (Ki of 1.3 nM), but also targets DHFR and GARFT (Ki of 65 nM) (Figure 1) [3]. Pemetrexed is currently in use clinically in treatment of mesothelioma and non-small cell lung cancer [9].
Recently, 5,10-methenyltetrahydrofolate synthetase (MTHFS) was shown to enhance de novo purine biosynthesis [10]. MTHFS catalyzes the ATP-dependent conversion of 5-formylTHF to 5,10-methenylTHF, a reaction that is inhibited by 10-formylTHF, which binds tightly to MTHFS [10]. [6R,S]-10-FormylTHF tri-glutamates competitively inhibit recombinant mouse MTHFS with Ki=30 nM, indicating that 10-formylTHF is an in vivo inhibitor of MTHFS activity. 5-formylTHF does not serve as a coenzyme for any known one-carbon transfer reactions, but rather serves as a stable storage form of formyl-substituted tetrahydrofolates [11]. The inhibition of MTHFS by 10-formylTHF ensures that 5-formylTHF stores are mobilized only when 10-formylTHF pools are depleted [10]. Elevated expression of MTHFS enhances de novo purine biosynthesis relative to synthesis by the purine salvage pathway in SH-SY5Y neuroblastoma; MTHFS may facilitate the delivery of 10-formylTHF to the de novo purine synthesis pathway [10]. In this study, the ability of MTHFS to affect the efficacy of antifolates that target purine biosynthesis was investigated using three antifolate chemotherapeutic drugs: Lometrexol, LY309887, and Pemetrexed (Alimta).
Materials and Methods
Materials
MES, Thiazolyl Blue Tetrazolium Bromide or 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), and Trypan Blue Solution were purchased from Sigma. ATP was purchased from Roche Applied Science. [6S]-5-formylTHF was a generous gift from Eprova AG. Fetal bovine serum (FBS), α-minimal essential medium (α-MEM), and defined α-MEM (α-MEM lacking folate, serine, glycine, methionine, ribonucleotides, and dinucleotides) were purchased from Hyclone. Pemetrexed, Lometrexol, and LY309887 were obtained from Lilly Research Laboratories, Indianapolis, IN. All other materials were of high quality and obtained from various commercial vendors.
MTT assay
SH-SY5Y cells expressing the human MTHFS cDNA (SH-SY5YMTHFS) cells have been described elsewhere [12, 13]. Cells were maintained in α-MEM prior to assay. For IC50 determinations, cells were cultured with defined α-MEM (α-MEM lacking folate, serine, glycine, methionine, ribonucleotides, deoxynucleotides, ribonucleosides, and deoxyribonucleosides) supplemented with 200 μM methionine and 25 nM 5-formyltetrahydrofolate and 11% dialyzed fetal bovine serum. SH-SY5Y cells were plated at a density of 5 × 103 cells/well in V-bottom 96-well plates (Corning) in quadruplicate. 24 h later, cells were treated with drug (prepared in defined α-MEM) at final concentrations ranging from 100 nM to 250 μM for Lometrexol and 10 nM to 5 μM for Pemetrexed and LY309887. After 72 h exposure to drug, 30 μL of 5 mg/mL MTT in phosphate-buffered saline was added to each well and the cells were incubated for 4 h at 37°C. Tetrazolium Bromide (MTT) is reduced to an insoluble formazan product in mitochondria of living cells. The insoluble formazan product was pelleted at 4000 rpm in a table top centrifuge, the supernatant was removed, and the insoluble formazan was resuspended in 100 μL DMSO. The absorbance at λ=540 nm was quantified using a Dynex MRXTC II microplate reader. IC50 values were determined from concentration-response curves generated using Prism software (GraphPad Software, Inc). Results are expressed as average and standard deviation of three independent experiments.
Trypan blue exclusion
SH-SY5Y and SH-SY5YMTHFS cells were plated in duplicate at 2 × 105 cells/well in 6-well plates in a total volume of 2 mL defined α-MEM/well (the same defined α-MEM as used for the MTT assay). After 24h, 1 mL of the antifolate prepared in defined α-MEM was added at final concentrations ranging from 100 nM to 250 μM for Lometrexol and 10 nM to 5 μM for Pemetrexed and LY309887. After 72 h, medium was removed, cells were trypsinized and pelleted. After resuspension in defined α-MEM, cells were mixed with an equal volume of 0.4% Trypan Blue. Viable cells (cells which exclude trypan blue) and nonviable cells were counted using a hemacytometer. IC% (viable cells/total cells) was calculated and IC50 values were determined from concentration-response curves generated using Prism software (GraphPad Software, Inc). Results are expressed as average and standard error of the mean from measurements made in triplicate as generated by Prism software.
Animal tumor sample collection
Tumor and normal tissue samples were collected at the time of surgery from client-owned dogs and cats presenting for management of cancer at the Cornell University Hospital for Animals. Core tissue samples were removed from the excised tumor and surrounding normal tissue bed using a 4-6 mm punch biopsy after dissection to identify the tumor – normal tissue interface. Normal tissue samples were selected by visual inspection to be 0.5-1.0 cm from the tumor interface and to avoid epidermis and fat. Samples were stored at -80°C. Histopathologic diagnoses were subsequently made for each tumor.
GARFT western blot
Cell pellets from 100 mm culture dishes were collected and lysed in 10 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5 mM DTT, 0.1 mM PMSF, and 1% Triton X-100. Cell lysates were loaded onto 12% SDS-PAGE gels with 30 μg total protein/lane (protein concentrations were determined using the Lowry-Bensadoun method [14]). Proteins were then transferred to an Immobilon-P PVDF Membrane (Millipore). The membrane was blocked overnight at 4°C in phosphate-buffered saline with 5% nonfat dry milk and 1% NP40. The membrane was incubated at room temperature for 1 hour in 1 μg/mL α-GARFT antibody (Abnova). After four washes of 10 minutes each, the membrane was incubated for 1 hour in 1:10,000 HRP-conjugated goat α-mouse antibody (Pierce). After four washes of 10 minutes each, membranes were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and subjected to autoradiography. Densitometry was performed using ChemiImager 4400 software (Alpha Innotech Corp.).
MTHFS activity assays
Tissue samples were sonicated 4× for 15 s in a buffer containing 100 mM HEPES pH 7.0, 100 mM sodium chloride, 5 mM EDTA, 1% Tween-20. The solution was clarified by centrifugation. MTHFS activity was determined using a spectrophotometer by monitoring the appearance of 5,10-methenylTHF, which has an absorbance maximum at 355 nm. For a typical assay, 50 μL of clarified tissue supernatant was added to a quartz cuvette containing 100 μM [6S]-5-formylTHF, 1 mM Mg-ATP and 100 mM MES, pH 6.0 and the rate of 5,10-methenylTHF formation was quantified. Activity measurements were normalized to total protein that was quantified using the Lowry-Bensadoun method [14].
For all inhibition experiments, 70 nM recombinant murine MTHFS (purified as described elsewhere [15]) was preincubated with an inhibitor for 4 minutes in the assay buffer prior to initiating the reaction by adding [6S]-5-formylTHF. Ki values were determined from Dixon plots, and are reported the average and standard deviation of at least two independent determinations.
Results
Murine MTHFS is inhibited by antifolates
Pemetrexed, Lometrexol, and LY309887 competitively inhibited murine MTHFS in vitro with Ki values of 48 ± 11 uM, 9 ± 3 uM, 6 ± 1 uM, respectively. This is consistent with previous findings that MTHFS has affinity for a wide range of natural and synthetic folates [10, 16, 17]. Polyglutamate derivatives of these compounds were not available, but considering that folate polyglutamylation is an important determinant of substrate and inhibitor affinity for murine MTHFS [10], these compounds are expected to have higher affinity for MTHFS in vivo after polyglutamylation. The Ki values for these antifolates were markedly higher than that determined for the natural folate, [6R]-10-formylTHF (Ki = 150 nM) [10].
Effect of MTHFS expression on sensitivity to antifolates in SH-SY5Y neuroblastoma
SH-SY5Y cells expressing the MTHFS cDNA, SH-SY5YMTHFS, have been described elsewhere [12, 13]. SH-SY5YMTHFS cells were shown to exhibit enhanced de novo purine biosynthesis relative to control cells [10]. To test whether MTHFS expression modulates the cytotoxicity of anti-purine drugs, the IC50 values for Pemetrexed, Lometrexol, and LY309887 were determined in SH-SY5Y and SH-SY5YMTHFS using the MTT Assay [18]. The IC50 value for LY309887 was significantly higher in SY-SY5YMTHFS compared to SH-SY5Y, but was not different between the two cell lines for Lometrexol and Pemetrexed; the IC50 for LY309887 in SH-SY5YMTHFS (IC50=29 nM) was 2-fold higher than for SH-SY5Y (IC50=14 nM)(Table 1). To validate findings from the MTT assay, and because decreased respiration observed in the MTT assay may not always coincide with cell death [18, 19], IC50 values were verified using the trypan blue exclusion viability assay. In this assay, SH-SY5YMTHFS were 4-fold resistant to LY309887 relative to SH-SY5Y, and this difference was statistically significant (p=0.03 in students unpaired, two-tailed t-test). Similar to the results obtained from the MTT assay, SH-SY5YMTHFS was not resistant to either Pemetrexed or Lometrexol relative to SH-SY5Y (Table 1).
Table 1. IC50 for SH-SY5Y neuroblastoma.
MTHFS expression diminishes the efficacy of LY309887. MTT Assay results represent average and standard deviation of three experiments. Trypan Blue Exclusion assay results are presented as the IC50 and the standard error of the mean from the concentration response curve as determined using Prism software with each measurement made in triplicate.
GARFT expression in SH-SY5YMTHFS
Immunoblots were performed on cell extracts from SH-SY5Y and SH-SY5YMTHFS to ensure that resistance to LY309887 in SH-SY5YMTHFS was not the result of increased GARFT expression. Surprisingly, GARFT protein levels (normalized to GAPDH) were 60% lower in SH-SY5YMTHFS than in SH-SY5Y (Figure 2).
Figure 2. Western blot of GARFT in SH-SY5Y and SH-SY5YMTHFS.
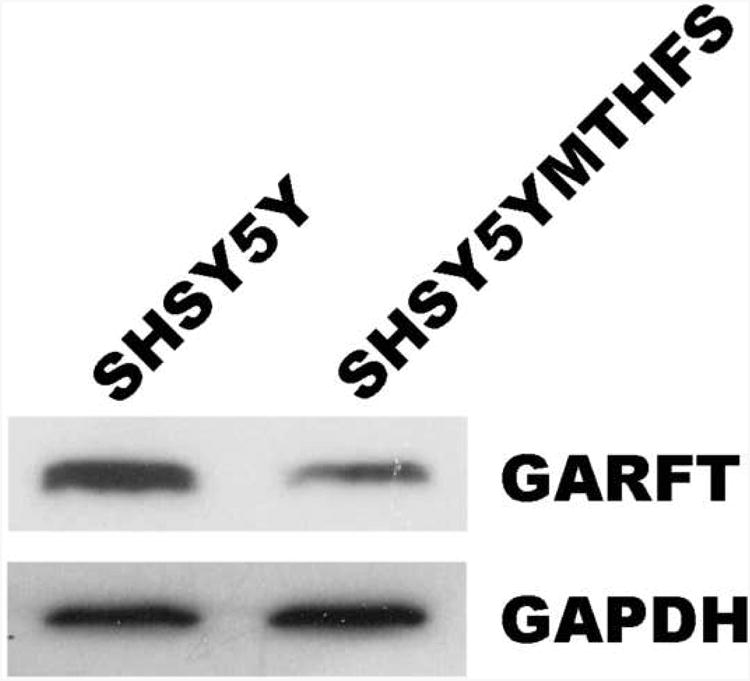
Cell lysates were loaded onto 12% SDS-PAGE gels with 30 μg total protein/lane. The western blot was carried out as described in Materials and Methods using 1 μg/mL α-GARFT antibody (Abnova).
Analysis of MTHFS levels in animal tissue samples
MTHFS activity normalized to total soluble protein was examined in sets of canine and feline normal and tumor tissue biopsies. MTHFS specific activity was increased in all tumor samples relative to surrounding normal tissues in all paired sets of tumor and normal tissue samples (Table 2). The specific activity values ranged from 0.07 to 1.83 pmoles of 5,10-methenylTHF/min/μg protein for the normal tissue samples, and 0.03 to 3.4 pmoles of 5,10-methenylTHF/min/μg protein for the tumor tissues; 9 of 15 tumors demonstrated a greater than 5-fold increase in MTHFS specific activity relative to surrounding normal tissue.
Table 2. MTHFS activity in normal and tumor tissue biopsies.
The specific activities of MTHFS (in units of pmoles 5,10-methenylTHF/min/μg protein) were determined for the feline and canine tissue samples. Specific activity values shown represent the average of triplicate measurements and variation is expressed as standard deviations of the mean. The (*) indicates two independent biopsies from the same animal. For all tumors, one-tailed student's t-tests were performed assuming equal variance (p< 0.003).
| Normal Tissue | Tumor Tissue | Fold | ||
|---|---|---|---|---|
| Species | Tumor type | Specific Activity | Specific Activity | Increase |
| canine | mastocytoma | 0.07 ± 0.03 | 0.93 ± 0.01 | 13.30 |
| canine | mastocytoma | 0.08 ± 0.02 | 1.97 ± 0.08 | 24.63 |
| canine | mastocytoma | 0.18 ± 0.01 | 1.77 ± 0.09 | 12.64 |
| canine | mastocytoma | 0.33 ± 0.10 | 1.42 ± 0.04 | 4.30 |
| canine | mastocytoma | 0.06 ± 0.01 | 0.30 ± 0.01 | 5.00 |
| canine | sarcoma | 1.83 ± 0.08 | 3.38 ± 0.10 | 1.85 |
| canine | sarcoma | 0.36 ± 0.07 | 2.02 ± 0.02 | 5.61 |
| canine | sarcoma | 0.22 ± 0.01 | 1.47 ± 0.26 | 6.68 |
| feline | sarcoma | 0.09 ± 0.05 | 1.35 ± 0.04 | 15.00 |
| canine | melanoma | 1.43 ± 0.05 | 1.86 ± 0.09 | 1.31 |
| canine | osteosarcoma | 0.20 ± 0.03 | 1.29 ± 0.06 | 6.45 |
| canine | rectal carcinoma | n.d. | 0.03 ± 0.001 | \ |
| *feline | hemangiosarcoma | 0.57 ± 0.03 | 1.72 ± 0.13 | 3.02 |
| *feline | hemangiosarcoma | 0.49 ± 0.04 | 3.43 ± 0.10 | 7.00 |
| feline | breast | n.d. | 0.12 ± 0.01 | \ |
Discussion
Physiological states associated with increased rates of cell proliferation and DNA replication have increased demands for folate cofactors for thymidylate and purine biosynthesis [8, 20]. The de novo purine biosynthetic pathway consists of ten enzyme-catalyzed reactions, and two of these enzymes require 10-formylTHF as a coenzyme: GARFT and AICARFT [2]. Although most normal tissues synthesize purines preferentially from the purine salvage pathway, tumor cells do not effectively salvage purines and display increased activity of folate-dependent de novo purine synthesis enzymes [3, 21, 22]. Our previous findings that MTHFS enhanced de novo purine biosynthesis indicated that MTHFS expression may influence the efficacy of antifolates that target purine biosynthesis.
5,10-Dideazatetrahydrofolic acid (DDATHF or Lometrexol), discovered as a potent GARFT inhibitor in the mid 1980's, was the first purine biosynthesis inhibitor to reach clinical trials [3, 8, 23]. LY309887 proved to be a more potent GARFT inhibitor both in vitro and in vivo; LY309887 exhibited a Ki 30-fold lower than Lometrexol for recombinant human GARFT. Moreover, LY309887 was a more effective suppressor of tumor growth in a number of murine tumors and human xenografts [3, 23]. In this study, Pemetrexed, Lometrexol, and LY309887 were all found to be competitive inhibitors of murine recombinant MTHFS with Ki values in the low micromolar range. In two distinct assays, MTHFS expression in SH-SY5Y neuroblastoma conferred resistance only to LY309887 (Table 1). MTHFS expression in SH-SY5Y did not alter the efficacy of Pemetrexed or Lometrexol (Table 1). Pemetrexed, also known as mutli-targeted antifolate, primarily inhibits TS and DHFR, and to a much lesser extent GARFT. The lack of resistance to Lometrexol compared to LY309887 in SH-SY5YMTHFS relative to SH-SY5Y may be a result of the increased specificity of LY309887 for GARFT, or because SH-SY5Y were relatively resistant to Lometrexol; much higher concentrations were required (up to 250 uM) to achieve cytotoxicity. At this level, Lometrexol has been shown to inhibit other folate-dependent enzymes such as DHFR [24] and will inhibit MTHFS. Taken together, these data suggest that the resistance to LY309887 (and not Pemetrexed or Lometrexol) in SH-SY5YMTHFS is due the specific targeting of de novo purine synthesis by LY309887 under these conditions. The degree of the resistance to LY309887 conferred by MTHFS is particularly striking considering that SH-SY5YMTHFS cells contain 60% less of the drug target GARFT than SH-SY5Y cells. De novo purine biosynthesis is often upregulated in tumor cells [3, 21, 22]. However, little is known about the regulation of GARFT expression.
The protection against LY309887 conferred by MTHFS indicates that MTHFS may serve as a sink by sequestering this antifolate and thereby protect GARFT from inhibition (Figure 3, Mechanism 1). However, this is unlikely because all three antifolates competitively inhibited MTHFS with Ki values in the low micromolar range, but MTHFS was only able to significantly modify the efficacy of LY309887 in neuroblastoma (Table 1). Alternatively, MTHFS may modify the efficacy of LY309887 by making 10-formylTHF more available to GARFT. This could be achieved through the MTHFS-induced increase in 10-formylTHF levels observed previously in SH-SY5Y cells [10] or potentially by channeling 10-formylTHF directly to GARFT (Figure 3, Mechanism 2), assuming that 10-formylTHF in the MTHFS-10-formylTHF complex has higher affinity for GARFT than free 10-formylTHF. In support of this mechanism, a recent study has demonstrated that the enzymes involved in de novo purine biosynthesis are present in a multi-enzyme complex termed a “purinosome” [25]. The enzyme C1-tetrahydrofolate synthetase, which is responsible for the synthesis of 10-formylTHF, was not found in this “purinosome” complex [25]. MTHFS-mediated channeling of 10-formylTHF to this complex or simply to GARFT could account for the diminished efficacy of GARFT inhibitors in SH-SY5Y neuroblastoma expressing MTHFS. Support for such a mechanism is present in the literature. The concentration of folate-binding proteins within the cell is much higher than the intracellular concentration of folate, such that the concentration of unbound folate cofactors is negligible [26, 27] and folate-dependent anabolic reactions must compete for a limited amount of folate cofactors [26]. This competition leads to “substrate channeling” between enzymes in a complex or between active-sites of multi-functional enzymes [28].
Figure 3. Possible mechanisms of MTHFS-mediated resistance to LY309887.
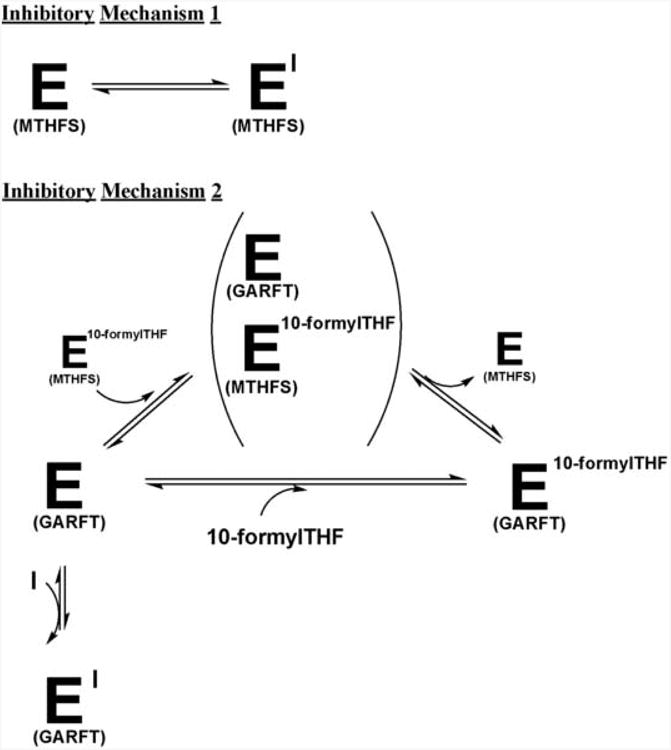
Inhibitory Mechanism 1 indicates that MTHFS alters resistance to LY309887 by directly binding inhibitors. Inhibitory Mechanism 2 demonstrates that MTHFS-bound 10- formylTHF may be more readily available to GARFT than is free 10-formylTHF, thus altering the cytotoxicity of inhibitor.
These data demonstrate that MTHFS is a modifier of LY309887 efficacy. The expression of MTHFS varies widely among neoplastic cell lines [13], and MTHFS expression may modify the effectiveness of other GARFT inhibitors still under clinical investigation such as AG2037, a GARFT inhibitor that shows less toxicity than Lometrexol or LY309887 [3, 4]. The increased and variable MTHFS expression demonstrated here in animal tumors indicates that the efficacy of GARFT inhibitors may vary among tumors as a function of MTHFS expression.
Acknowledgments
This work was supported by PHS HD35687 to PJS.
Abbreviations
- MTHFS
methenyltetrahydrofolate synthetase
- GARFT
glycinamide ribonucleotide formyltransferase
- AICARFT
aminoimidazolecarboxamide ribonucleotide formyltransferase
- THF
tetrahydrofolate
- AdoMet
S-adenosylmethionine
- AdoHcy
S-adenosylhomocysteine
- TS
thymidylate synthase
- FPGS
folylpolyglutamate synthetase
- RFC
reduced folate carrier
- FR
folate receptor
- PCFT/HCP1
proton-coupled electrogenic folate transporter
- DDATHF
5,10-dideazatetrahydrofolate
- DHFR
dihydrofolate reductase
- MTT
Thiazolyl Blue Tetrazolium Bromide MTT, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- α-MEM
α-minimal essential media
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shane B. Folate Chemistry and Metabolism. In: Bailey LB, editor. Folate in Health and Disease. Marcel Dekker, Inc; New York: 1995. pp. 1–22. [Google Scholar]
- 2.Kisliuk RL. Folate Biochemistry in Relation to Antifolate Selectivity. In: Jackman AL, editor. Anticancer Drug Development Guide: Antifolate Drugs in Cancer Therapy. Humana Press Inc.; Totawa, NJ: 1999. pp. 13–36. [Google Scholar]
- 3.McGuire JJ. Anticancer antifolates: current status and future directions. Curr Pharm Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- 4.Kisliuk RL. Deaza analogs of folic acid as antitumor agents. Curr Pharm Des. 2003;9:2615–2625. doi: 10.2174/1381612033453695. [DOI] [PubMed] [Google Scholar]
- 5.Walling J. From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Invest New Drugs. 2006;24:37–77. doi: 10.1007/s10637-005-4541-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhao R, Goldman ID. Resistance to antifolates. Oncogene. 2003;22:7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- 7.Relling MV, Dervieux T. Pharmacogenetics and cancer therapy. Nat Rev Cancer. 2001;1:99–108. doi: 10.1038/35101056. [DOI] [PubMed] [Google Scholar]
- 8.Mendelsohn LG, Shih C, Schultz RM, Worzalla JF. Biochemistry and pharmacology of glycinamide ribonucleotide formyltransferase inhibitors: LY309887 and lometrexol. Invest New Drugs. 1996;14:287–294. doi: 10.1007/BF00194532. [DOI] [PubMed] [Google Scholar]
- 9.Zhao R, Zhang S, Hanscom M, Chattopadhyay S, Goldman ID. Loss of reduced folate carrier function and folate depletion result in enhanced pemetrexed inhibition of purine synthesis. Clin Cancer Res. 2005;11:1294–1301. [PubMed] [Google Scholar]
- 10.Field MS, Szebenyi DM, Stover PJ. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem. 2006;281:4215–4221. doi: 10.1074/jbc.M510624200. [DOI] [PubMed] [Google Scholar]
- 11.Stover P, Schirch V. The metabolic role of leucovorin. Trends Biochem Sci. 1993;18:102–106. doi: 10.1016/0968-0004(93)90162-g. [DOI] [PubMed] [Google Scholar]
- 12.Girgis S, Suh JR, Jolivet J, Stover PJ. 5-Formyltetrahydrofolate regulates homocysteine remethylation in human neuroblastoma. J Biol Chem. 1997;272:4729–4734. doi: 10.1074/jbc.272.8.4729. [DOI] [PubMed] [Google Scholar]
- 13.Anguera MC, Suh JR, Ghandour H, Nasrallah IM, Selhub J, Stover PJ. Methenyltetrahydrofolate synthetase regulates folate turnover and accumulation. J Biol Chem. 2003;278:29856–29862. doi: 10.1074/jbc.M302883200. [DOI] [PubMed] [Google Scholar]
- 14.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 15.Anguera MC, Liu X, Stover PJ. Cloning, expression, and purification of 5,10-methenyltetrahydrofolate synthetase from Mus musculus. Protein Expr Purif. 2004;35:276–283. doi: 10.1016/j.pep.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand R, Jolivet J. Methenyltetrahydrofolate synthetase prevents the inhibition of phosphoribosyl 5-aminoimidazole 4-carboxamide ribonucleotide formyltransferase by 5-formyltetrahydrofolate polyglutamates. J Biol Chem. 1989;264:8843–8846. [PubMed] [Google Scholar]
- 17.Bertrand R, MacKenzie RE, Jolivet J. Human liver methenyltetrahydrofolate synthetase: improved purification and increased affinity for folate polyglutamate substrates. Biochim Biophys Acta. 1987;911:154–161. doi: 10.1016/0167-4838(87)90004-5. [DOI] [PubMed] [Google Scholar]
- 18.Mosmann T. Rapid Colorimetric Assay for Cellular Growth and Survival -Application to Proliferation and Cyto-Toxicity Assays. Journal of Immunological Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 19.Tonkinson JL, Marder P, Andis SL, Schultz RM, Gossett LS, Shih C, Mendelsohn LG. Cell cycle effects of antifolate antimetabolites: Implications for cytotoxicity and cytostasis. Cancer Chemotherapy and Pharmacology. 1997;39:521–531. doi: 10.1007/s002800050608. [DOI] [PubMed] [Google Scholar]
- 20.Novakovic P, Stempak JM, Sohn KJ, Kim YI. Effects of folate deficiency on gene expression in the apoptosis and cancer pathways in colon cancer cells. Carcinogenesis. 2006;27:916–924. doi: 10.1093/carcin/bgi312. [DOI] [PubMed] [Google Scholar]
- 21.McLeod HL, Cassidy J, Powrie RH, Priest DG, Zorbas MA, Synold TW, Shibata S, Spicer D, Bissett D, Pithavala YK, Collier MA, Paradiso LJ, Roberts JD. Pharmacokinetic and pharmacodynamic evaluation of the glycinamide ribonucleotide formyltransferase inhibitor AG2034. Clinical Cancer Research. 2000;6:2677–2684. [PubMed] [Google Scholar]
- 22.Jackson RC, Harkrader RJ. The Contributions of de novo and Salvage Pathways of Nucleotide Biosynthesis in Normal and Malignant Cells. In: Tattersall MNH, Fox RM, editors. Nucleosides and Cancer Treatment. Academic Press; Sydney: 1981. pp. 18–31. [Google Scholar]
- 23.Habeck LL, Leitner TA, Shackelford KA, Gossett LS, Schultz RM, Andis SL, Shih C, Grindey GB, Mendelsohn LG. A novel class of monoglutamated antifolates exhibits tight-binding inhibition of human glycinamide ribonucleotide formyltransferase and potent activity against solid tumors. Cancer Res. 1994;54:1021–1026. [PubMed] [Google Scholar]
- 24.Beardsley GP, Moroson BA, Taylor EC, Moran RG. A New Folate Antimetabolite, 5,10-Dideaza-5,6,7,8-Tetrahydrofolate Is a Potent Inhibitor of De novo Purine Synthesis. Journal of Biological Chemistry. 1989;264:328–333. [PubMed] [Google Scholar]
- 25.An SG, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 26.Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- 27.Matherly LH, Czajkowski CA, Muench SP, Psiakis JT. Role for cytosolic folate-binding proteins in the compartmentation of endogenous tetrahydrofolates and the 5-formyl tetrahydrofolate-mediated enhancement of 5-fluoro-2′-deoxyuridine antitumor activity in vitro. Cancer Res. 1990;50:3262–3269. [PubMed] [Google Scholar]
- 28.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5:2645–2651. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]


