Abstract
Cytokines such as TNFα play an integral role in sleep/wake regulation and have recently been hypothesized to be involved in cognitive impairment due to sleep deprivation. We examined the effect of a guanine to adenine substitution at position 308 in the TNFα gene (TNFα G308A) on psychomotor vigilance performance impairment during total sleep deprivation. A total of 88 healthy women and men (ages 22–40) participated in one of five laboratory total sleep deprivation experiments. Performance on a psychomotor vigilance test (PVT) was measured every 2 to 3 h. The TNFα 308A allele, which is less common than the 308G allele, was associated with greater resilience to psychomotor vigilance performance impairment during total sleep deprivation (regardless of time of day), and also provided a small performance benefit at baseline. The effect of genotype on resilience persisted when controlling for between-subjects differences in age, gender, race/ethnicity, and baseline sleep duration. The TNFα G308A polymorphism predicted less than 10% of the overall between-subjects variance in performance impairment during sleep deprivation. Nonetheless, the differential effect of the polymorphism at the peak of performance impairment was more than 50% of median performance impairment at that time, which is sizeable compared to the effects of other genotypes reported in the literature. Our findings provided evidence for a role of TNFα in the effects of sleep deprivation on psychomotor vigilance performance. Furthermore, the TNFα G308A polymorphism may have predictive potential in a biomarker panel for the assessment of resilience to psychomotor vigilance performance impairment due to sleep deprivation.
Keywords: Sleep deprivation, Cognitive impairment, Psychomotor vigilance test (PVT), Inter-individual differences, Trait vulnerability, Phenotype, Genotype, Biomarker, Tumor necrosis factor (TNF)
1. Introduction
There are considerable inter-individual differences in cognitive performance impairment due to sleep deprivation (Wilkinson, 1961; Morgan et al., 1980; Webb and Levy, 1984; Leproult et al., 2003). These inter-individual differences are systematic over time awake and circadian rhythm, stable over repeated exposures to total sleep deprivation (TSD), and robust to variations in prior sleep/wake history. As such, inter-individual differences in vulnerability to performance impairment during sleep deprivation constitute a trait (Van Dongen et al., 2004a). Recently it was also found that this trait vulnerability to impairment due to TSD is heritable (Kuna et al., 2012) and generalizes to vulnerability to impairment due to sustained sleep restriction (Rupp et al., 2012).
The discovery of trait vulnerability to sleep loss led to a focus on the assessment of underlying mechanisms (e.g., Chee and Tan, 2010; Jackson et al., 2013) and the identification of predictors (e.g., King et al., 2009; Abe et al., 2014). Genetic markers have captured particular interest (Landolt, 2008; Goel and Dinges, 2012). A number of genetic polymorphisms differentiate, to some degree, those individuals who are more vulnerable to the effects of sleep loss from those who are more resilient. These polymorphisms include, among others, variants of the human period circadian clock 3 gene (PER3) (Lo et al., 2012), adenosine A2A receptor gene (ADORA2A) (Bodenmann et al., 2012), and adenosine deaminase gene (ADA) (Reichert et al., 2014).
As trait vulnerability appears to be expressed on a continuum, with no evidence of a bimodal or multimodal distribution, it is to be expected that many more genes are involved (King et al., 2009). Several sleep regulatory substances have been identified, including a variety of cytokines (Krueger, 2008; Opp, 2009). Cytokines have remained largely unexplored as potential predictors of trait vulnerability to performance impairment due to sleep deprivation.
Here we focus on a particular cytokine, tumor necrosis factor alpha (TNFα). The TNFα gene is located within the class III region of the major histocompatibility complex (MHC). The promoter region of the TNFα gene is highly polymorphic (Allen, 1999; Elahi et al., 2009). The TNFα G308A polymorphism – a single nucleotide polymorphism (SNP) involving a guanine to adenine substitution at position 308 in the promoter region of the TNFα gene (Wilson et al., 1992), is associated with a range of immunological diseases and disorders. The polymorphism is also implicated in the pathogenesis of obstructive sleep apnea (OSA), a sleep disorder characterized by airway obstruction during sleep, sleep fragmentation, and excessive daytime sleepiness (Jordan et al., 2014). Two studies found that OSA patients are more likely than nonapneic controls to carry the −308A allele (Riha et al., 2005; Almpanidou et al., 2012).
The TNFα G308A polymorphism has been reported to result in increased TNFα gene transcription (Kroeger et al., 2000) and TNFα cytokine production (Louis et al., 1998), which may underlie increased circulating levels of TNFα in obstructive sleep apnea patients (Vgontzas et al., 1997). TNFα is integrally involved in sleep/wake regulation (Krueger et al., 2010), but whether the TNFα G308A polymorphism is associated with a sleep/wake phenotype in healthy individuals has not been previously established. In this study, we investigated whether the TNFα G308A polymorphism is associated with phenotypic inter-individual differences in vulnerability to psychomotor vigilance performance impairment due to sleep loss.
2. Materials and methods
2.1. General overview
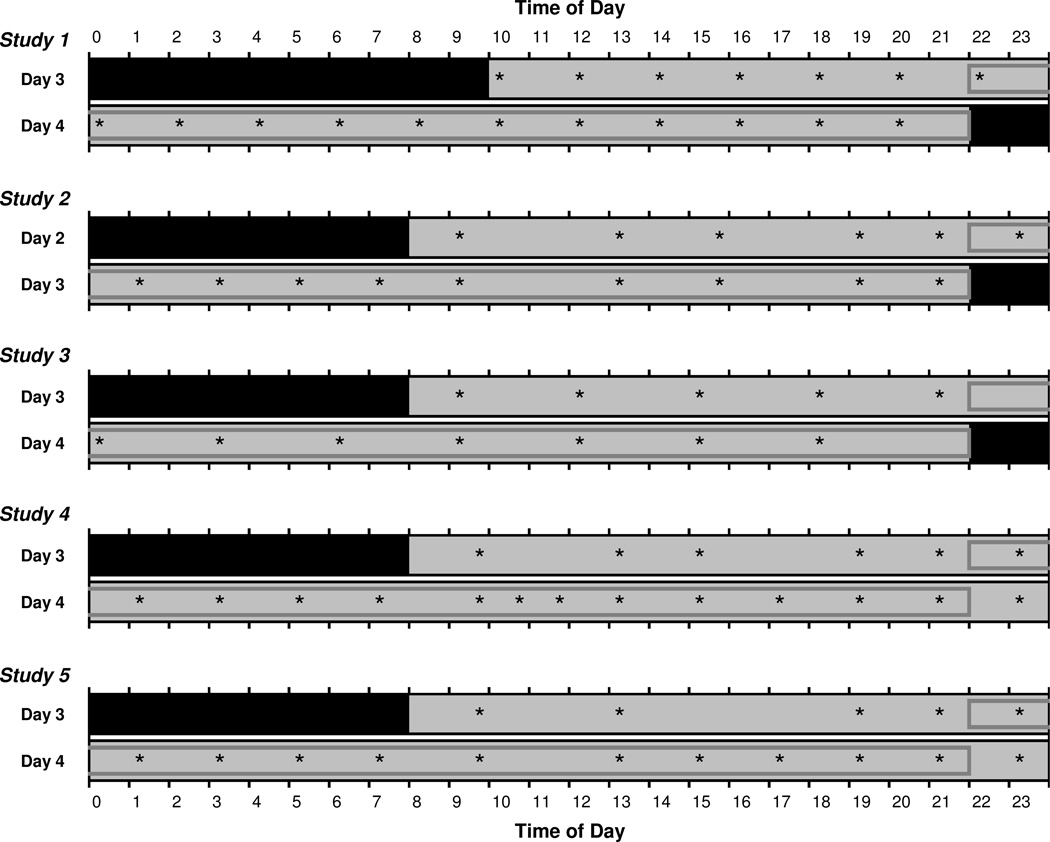
We analyzed data from 88 subjects who each participated in one of five laboratory TSD studies. During the five studies, cognitive performance was measured across 36–62 h of sustained wakefulness. As such, all five studies included at least 24 h of wake extension into the night and the following day – see Fig. 1. In all five studies, the primary performance test was a 10-min psychomotor vigilance test (PVT) (Lim and Dinges, 2008). The PVT was administered every 2–3 h over the course of scheduled wakefulness. Each subject’s vulnerability to sleep loss was quantified based on PVT performance over the 24-h period of sleep deprivation common to all five studies (Fig. 1). Subjects were grouped by genotype to determine if the TNFα G308A polymorphism predicted subject-specific PVT performance vulnerability to sleep loss.
Fig. 1.
Schematic of the days of each laboratory study relevant to the analyses of individual subjects’ vulnerability to sleep loss. Test bouts on the PVT are indicated by asterisks. Test bouts in the 24-h period used to quantify vulnerability to sleep loss are enclosed in gray boxes. Black indicates sleep periods; light gray indicates scheduled wakefulness. Note that in studies 4 and 5, the sleep deprivation period continued beyond the days shown here.
2.2. Total sleep deprivation studies
Information relevant to the results presented here about each of the five studies is provided below and summarized in Fig. 1 and Table 1.
Table 1.
Characteristics of the five laboratory total sleep deprivation studies.
| Study 1 | Study 2 | Study 3 | Study 4 | Study 5 | |
|---|---|---|---|---|---|
| Number of Subjects | 25 | 23 | 14 | 13 | 13 |
| TSD Period (h) | 36 | 38 | 38 | 62 | 62 |
| Baseline Time in Bed | 22:00–10:00 | 22:00–08:00 | 22:00–08:00 | 22:00–08:00 | 22:00–08:00 |
| Number of Baseline Nights | 2 | 1 | 2 | 2 | 2 |
| Number of PVT Bouts per Subject for Estimation of Vulnerability | 12 | 10 | 7 | 13 | 11 |
Study 1: n=25 healthy young adults (ages 22–40, 16 females) lived in the laboratory for 12 days (11 nights), during which time they underwent 36 h of TSD three times. Prior to each 36-h TSD period, subjects had two baseline days, each with 12 h time in bed (TIB) for sleep from 22:00 until 10:00. Following the third TSD period, subjects had two recovery days, each with 12 h TIB (22:00–10:00). Each TSD period was assigned either a moderate or a high workload for performance testing. The high workload condition occurred only once, in randomized order. For the present analyses, only performance data from the first TSD period were used. Subjects whose first TSD period was designated as high workload were not included. During TSD, the 10-min PVT was administered every 2 h (Fig. 1). Study 1 was first described in Tucker et al. (2007).
Studies 2 and 3: These two studies were similar in design. In study 2, n=23 healthy young adults (ages 22–36, 11 females) lived in the laboratory for 4 days (3 nights). In study 3, n=14 healthy young adults (ages 22–40, 7 females) lived in the laboratory for 5 days (4 nights). Both studies included a 38-h TSD period. Prior to TSD, subjects had one (study 2) or two (study 3) baseline days with 10 h TIB for sleep (22:00–08:00). Following TSD, subjects had a recovery day with 10 h TIB (22:00–08:00). During TSD, the 10-min PVT was administered approximately every 2 h (study 2) or 3 h (study 3) (Fig. 1). Studies 2 and 3 were first described in Grant et al. (2013b) and Grant et al. (2013a), respectively.
Studies 4 and 5: These two studies were similar in design. In study 4, n=13 healthy young adults (ages 22–37, 7 females) lived in the laboratory for 7 days (6 nights). Likewise, in study 5, n=13 healthy young adults (ages 22–40, 6 females) lived in the laboratory for 7 days (6 nights). Both studies included a 62-h TSD period. Prior to TSD, subjects had two baseline days with 10 h TIB for sleep (22:00–08:00). Following TSD, subjects had two recovery days with 10 h TIB (22:00–08:00). During TSD, the 10-min PVT was administered approximately every 2 h (Fig. 1). Studies 4 and 5 are described in Whitney et al. (in press) and Tucker et al. (2010), respectively.
2.3. Subjects
For each of the five studies, volunteers were recruited through newspaper and internet advertisements and posted flyers. Subjects eligible for study participation reported habitually sleeping between 6 and 10 h (for study 1: between 7 and 9 h) and getting up between 06:00 and 09:00 (for study 1: between 06:30 and 08:30).
Subjects eligible for study participation also met the following criteria: age 22–40 years (for study 1: 21–40 years); physically and psychologically healthy; no clinically significant abnormalities in blood and urine; no current medical or drug treatment (except contraceptives); no sleep or circadian disorders; free of alcohol and drugs, and not a current smoker; no history of alcohol abuse in the past year, no history of drug abuse (for studies 2–5: in the past year), and no history of methamphetamine abuse; not an extreme morning- or evening-type (studies 1 and 5 only); not pregnant; and no past adverse neuropsychiatric reactions to sleep deprivation.
For studies 2–5, subjects met the following additional criteria: no travel across times zones within 1 month of entering the study; no shift work within 3 months (for study 4: 1 month) of entering the study; no history of moderate to severe brain injury; no history of learning disabilities; not vision impaired unless corrected to normal; not hearing impaired unless corrected to normal (studies 2, 3 and 5); and proficient (for studies 4 and 5: native) speaker of English. For study 4, which included procedures involving intravenous blood sampling and performance testing on a high-fidelity driving simulator during TSD, the following criteria also applied: suitable veins for intravenous catheter insertion; no history of problems with blood draws or blood donation; not having donated blood within 2 months of entering the study; not susceptible to simulator adaptation syndrome; and valid driver’s license.
For the week prior to each laboratory experiment, subjects were instructed to abstain from using caffeine, tobacco, alcohol, and drugs, and they were not allowed to nap. Subjects were also instructed to maintain their habitual sleep/wake times. Compliance was verified by means of wrist actigraphy and sleep diary. In addition, subjects called a time-stamped voice recorder each day to report their sleep and wake times. Upon arrival at the laboratory for the experiment, subjects were checked for drug and alcohol use by means of urine and breathalyzer testing (studies 2–5).
All studies were approved by the Institutional Review Board (IRB) of Washington State University. Study 1 was also approved by the IRB of the University of Pennsylvania. All subjects gave written informed consent.
2.4. Experimental procedures
Studies 2–5 were conducted at the Sleep and Performance Research Center at Washington State University Spokane. Study 1 was conducted partially at the General Clinical Research Center of the Hospital of the University of Pennsylvania (15 subjects) and partially at the Sleep and Performance Research Center of Washington State University Spokane (10 subjects).
Laboratory conditions were strictly controlled throughout the experimental phase of each study. Light levels were fixed below 100 lux (for study 1: below 50 lux) during scheduled wakefulness and below 1 lux during scheduled sleep periods. Ambient temperature was maintained at 21–22 °C (± 1 °C). While in the laboratory, subjects were not allowed to engage in strenuous physical activity. They did not have contact with individuals outside the laboratory, and did not have access to live radio or television, phones, personal computers, the internet, or video games. Trained research assistants monitored subjects’ behavior continuously.
Baseline sleep was recorded polysomnographically in all five laboratory studies. Baseline polysomnograms were scored visually and checked to confirm the absence of any evidence for sleep disorders, including OSA.
For the present analyses, total sleep time in the baseline night immediately preceding the TSD period was recorded. Due to equipment failure, total sleep time records were missing for seven subjects (out of 25) in study 1 and one subject (out of 13) in study 4.
The PVT was administered repeatedly over the course of scheduled wakefulness (Fig. 1). The task is considered a gold standard measure of behavioral alertness (Dorrian et al., 2005). In studies 1, 2, 4 and 5, the PVT was administered on a desktop computer; in study 3 it was administered on a laptop computer. Both computer types were calibrated for accurate measurement of reaction time. Subjects were asked to respond, by pressing a button on a response box (study 3: by pressing the space bar on the keyboard), to the appearance of a visual stimulus (a millisecond counter) as quickly as possible without making false starts. The stimulus was presented in random intervals between 2 and 10 s over the course of the 10-min test duration.
The number of PVT lapses of attention, defined as reaction times > 500 ms, was used as the primary measure of psychomotor vigilance performance impairment. This measure captures the right tail of the PVT reaction time distribution, which is most affected by sleep deprivation (Doran et al., 2001). As secondary measures, the mean of the 10% fastest reaction times and the median reaction time were analyzed, in order to also examine the left tail and the central tendency of the reaction time distribution.
For 12 of the 88 subjects, one or more test bouts on the PVT were confounded by brief instances of distraction, non-compliance, or microsleeps, as documented by the research assistants. These PVT bouts were removed from the dataset prior to analysis. Of the 12 individuals, eight had 1 test bout removed, five had 2 test bouts removed, and one had 3 test bouts removed. There were 1,371 PVT bouts (98.5% of the original total) left in the overall dataset.
2.5. Genotyping
During pre-study screening sessions, venous whole blood samples were collected in Vacutainer tubes coated with ethylenediaminetetraacetic acid dipotassium dihydrate (K2EDTA). The samples were aliquoted and stored at −80 °C until analysis. After the completion of the studies, the samples were analyzed blind to study and subject.
For each subject, 100 µl of whole blood was red-cell depleted and genomic DNA was extracted and used for assaying. Genotyping for the TNFα G308A polymorphism (SNP rs1800629, chromosome 6) was performed using published procedures for the analysis of SNPs (Fargion et al., 2001; Schofield et al., 2009; Napolioni et al., 2011; Manjari et al., 2014) involving standard polymerase chain reaction (PCR) and restriction enzyme digestion. Our specific assay procedures were based on those described by Ozen et al. (2002).
Samples were amplified with 20 µM forward primer 5’– GAG GCA ATA GGT TTT GAG GGC CAT – 3’ and 20 µM reverse primer 5’ – GGG ACA CAC AAG CAT CAAG – 3’. PCR procedures were carried out in a final reaction volume of 20 µl containing 14 µl PCR master polymerase mix (Custom Genome Services, Pullman, WA), 1 µl of each primer (forward and reverse), 2 µl nuclease free water, and 2 µl genomic DNA. PCR conditions involved denaturation at 94 °C for 3 min, followed by 35 cycles of: denaturation at 94 °C for 30 s, annealing at 59 °C for 1 min, and extension at 72 °C for 2 min. After another 5 min of extension at 72 °C the reaction ended.
Amplified products were digested with NcoI (Invitrogen by Life Technologies, Grand Island, NY), a mutation-specific restriction enzyme. The restriction enzyme recognized a restriction site on the G allele. Digestion of the PCR fragments yielded products of 117 bp (A allele) and 97 bp and 20 bp (G allele). The digestion reactions were carried out in a final volume of 11 µl containing 1 µl NcoI, 1 µl 10× Buffer K, 1 µl 0.1% BSA, and 8 µl PCR product. Products were digested for 3.5 h at 37 °C, followed by 10 min at 65 °C to inactivate the enzyme. The final digested products were electrophoresed on a 3% agarose gel stained with ethidium bromide and visualized under UV light to determine genotypes: G/G, A/G or A/A.
Samples identified as A/G or A/A were subjected to a reverse (control) assay (Vinasco et al., 1997), involving amplification with 20 µM forward primer 5’ – GAG GCA ATA GGT TTT GAG GGT CAT – 3’ and the above-mentioned reverse primer, using the PCR procedures described above with an annealing temperature of 57°C. Amplified products were digested with another restriction enzyme, BspHI (New England Biolabs, Inc., Ipwsich, MA), which recognized a restriction site on the A allele. Digestion of the PCR fragments yielded products of 117 bp (G allele) and 97 bp and 20 bp (A allele). The digestion reactions were carried out in a final volume of 10 µl containing 1 µl BspHI, 1 µl 10× NEBuffer, and 8 µl PCR product. Products were digested for 1 h at 37 °C, followed by 20 min at 80 °C to inactivate the enzyme. The final digested products were visualized in the same manner as described above. After running this control assay, we found that one sample had been misclassified as A/A instead of A/G based on the NcoI digest.
2.6. Statistical analyses
The genotype distribution across the combined sample from the five studies was examined for deviation from Hardy-Weinberg equilibrium using a α2 goodness-of-fit test.
Each subject’s vulnerability to sleep loss was quantified by averaging the number of PVT lapses over the 24-h period (i.e., one circadian cycle) of sleep deprivation common to all five studies (from 22:00 until 22:00 the next day) (Fig. 1). This 24-h period was also divided into two 12-h blocks (from 22:00 until 10:00 and from 10:00 until 22:00) to examine nighttime and daytime performance separately. In addition, each subject’s baseline performance level was quantified by averaging the number of PVT lapses over the 12 h of wakefulness immediately preceding the 24-h period of sleep deprivation (from 10:00 until 22:00) (Fig. 1). The first test bout of study 1 in this baseline period was not included because it occurred immediately after awakening and could therefore have been affected by sleep inertia. Data reduction of secondary PVT measures (mean of 10% fastest reaction times and median reaction time) paralleled that of PVT lapses.
The subject-specific averages for vulnerability to sleep loss were analyzed using nonparametric one-way analysis of rank scores with genotype as independent variable and controlling for study. Secondary analyses also controlled for baseline performance, baseline sleep duration, age, sex, and race/ethnicity (in addition to study). One-way analysis of variance (ANOVA) was used to determine the percentage of variance in performance impairment during TSD that was explained by genotype, and to estimate Cohen’s local effect size f2. One-way ANOVA was used to test for differences between genotypes in baseline performance, baseline sleep duration and age; logistic regression was used to test for differences between genotypes in sex and race/ethnicity distributions.
For interpretation of results, PVT lapses were also analyzed as a function of time. To handle the differences in test bout times among the five studies (Fig. 1), the data of the 12-h baseline and 24-h sleep deprivation periods (i.e., 36-h period of wakefulness) were binned into six consecutive 6-h time intervals. Average PVT lapses per 6-h interval were calculated for each subject. These subject-specific averages were analyzed using mixed-effects ANOVA with genotype and time and their interaction as independent variables (controlling for study). For reference, the sample was also divided into tertiles based on subjects’ rank order of vulnerability (as previously quantified by averaging the number of PVT lapses over the 24-h period of sleep deprivation). The temporal profiles of the genotypes as revealed by the mixed-effects ANOVA were visually compared to the group-average temporal profiles of each of the tertiles.
3. Results
A total of N=88 subjects (ages 22–40; 47 females) participated in one of the five inlaboratory TSD studies. Their demographics are shown in Table 2, and their genotypes are summarized in Tables 2 and 3. The allele frequencies in our sample were 0.8580 for the G allele and 0.1420 for the A allele. The genotypes were found to be in Hardy-Weinberg equilibrium (χ21=0.46, P=0.50), and were comparable to previously published studies reporting on the same polymorphism (Beste et al., 2010; Almpanidou et al., 2012).
Table 2.
Subject demographics for the five laboratory total sleep deprivation studies.
| Study 1 n=25 (28.4%) |
Study 2 n=23 (26.1%) |
Study 3 n=14 (15.9%) |
Study 4 n=13 (14.8%) |
Study 5 n=13 (14.8%) |
Total N=88 (100.0%) |
|
|---|---|---|---|---|---|---|
| Sex | ||||||
| Female (%) | 16 (64.0%) | 11 (47.8%) | 7 (50.0%) | 7 (53.8%) | 6 (46.2%) | 47 (53.4%) |
| Male (%) | 9 (36.0%) | 12 (52.2%) | 7 (50.0%) | 6 (46.2%) | 7 (53.8%) | 41 (46.6%) |
| Age (mean ± SD) | 28.8 ± 5.8 | 26.7 ± 4.8 | 29.6 ± 6.4 | 26.8 ± 4.3 | 28.4 ± 5.6 | 28.0 ± 5.4 |
| Race/Ethnicity | ||||||
| Caucasian (%) | 13 (52.0%) | 22 (95.7%) | 11 (78.6%) | 8 (61.5%) | 12 (92.3%) | 66 (75.0%) |
| African-American (%) | 9 (36.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 9 (10.2%) |
| Hispanic (%) | 0 (0.0%) | 0 (0.0%) | 1 (7.1%) | 1 (7.7%) | 0 (0.0%) | 2 (2.3%) |
| Asian (%) | 1 (4.0%) | 0 (0.0%) | 0 (0.0%) | 1 (7.7%) | 1 (7.7%) | 3 (3.4%) |
| American Indian (%) | 1 (4.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (1.1%) |
| Mixed (%) | 1 (4.0%) | 1 (4.3%) | 0 (0.0%) | 3 (23.1%) | 0 (0.0%) | 5 (5.7%) |
| Undisclosed (%) | 0 (0.0%) | 0 (0.0%) | 2 (14.3%) | 0 (0.0%) | 0 (0.0%) | 2 (2.3%) |
| Genotype | ||||||
| G/G (%) | 21 (84.0%) | 14 (60.9%) | 7 (50.0%) | 11 (84.6%) | 11 (84.6%) | 64 (72.7%) |
| A/G (%) | 4 (16.0%) | 9 (39.1%) | 6 (42.9%) | 2 (15.4%) | 2 (15.4%) | 23 (26.1%) |
| A/A (%) | 0 (0.0%) | 0 (0.0%) | 1 (7.1%) | 0 (0.0%) | 0 (0.0%) | 1 (1.2%) |
Table 3.
Genotype counts and frequencies.
| G/G | A/G | A/A | |
|---|---|---|---|
| Genotype Count | |||
| Expecteda | 64.78 | 21.45 | 1.78 |
| Observed | 64 | 23 | 1 |
| Genotype Frequency | |||
| Expecteda | 0.7361 | 0.2437 | 0.0202 |
| Observed | 0.7273 | 0.2614 | 0.0114 |
| Publishedb | 0.5729 | 0.4063 | 0.0208 |
| Publishedc | 0.6928 | 0.2633 | 0.0439 |
Based on Hardy-Weinberg equilibrium.
Beste et al. (2010), 96 healthy subjects.
Almpanidou et al. (2012), 319 healthy control subjects.
Each subject’s vulnerability to sleep loss was quantified by averaging the number of PVT lapses over a 24-h period (i.e., a circadian cycle) of sleep deprivation, from 22:00 until 22:00 the next day (Fig. 1). Non-parametric one-way analysis of rank scores, controlling for study, revealed a significant effect of genotype (F2,81=5.49, P=0.006). There was no significant difference between the A/G and A/A genotypes (F1,81=1.04, P=0.31). Therefore, since there was only one subject with the A/A genotype, the analysis was repeated with the A/A and A/G genotypes combined. The significant effect of genotype persisted (F1,82=9.94, P=0.002). There was no significant effect of which study the subjects participated in (F4,82=0.83, P=0.51).
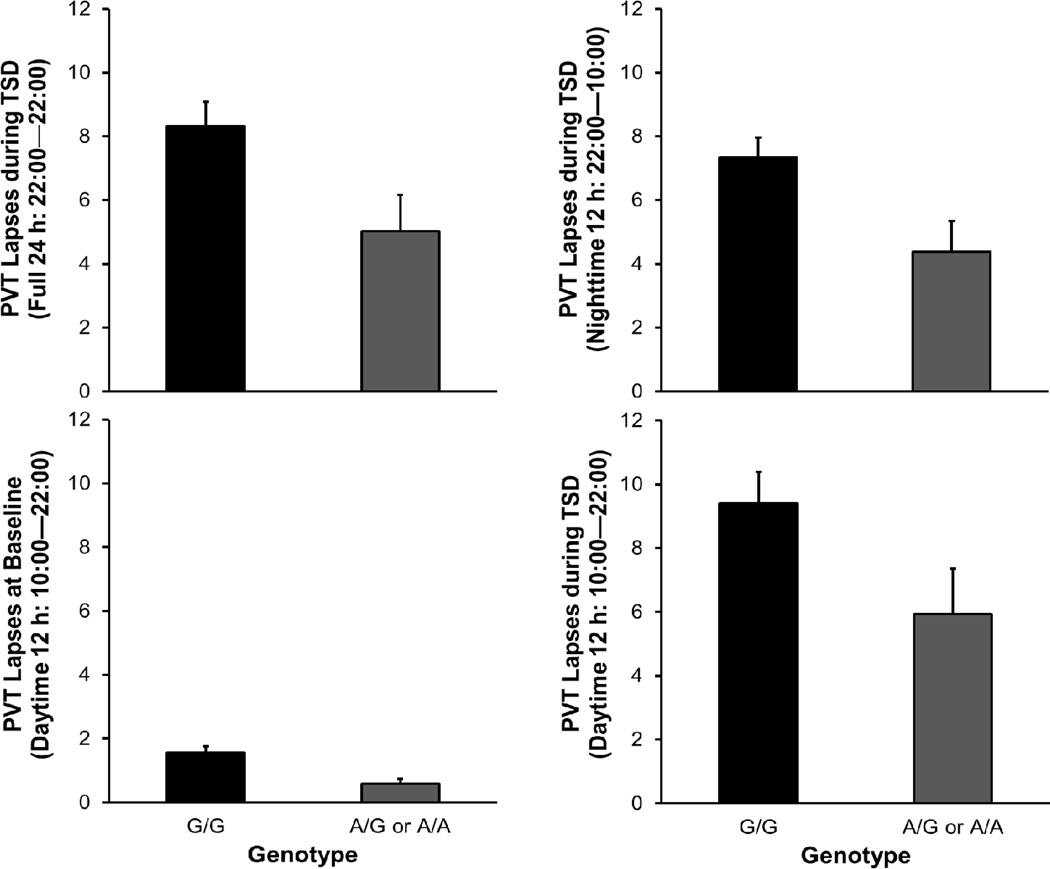
Individuals homozygous or heterozygous for the A allele exhibited fewer PVT lapses during TSD than individuals homozygous for the G allele – see Fig. 2 (top left). The proportion of variance in psychomotor vigilance performance impairment during TSD that was explained by genotype was 6.4% (correlation r=0.25). The local effect size of genotype was f2=0.071, which is small (but not negligible) according to the guidelines of Cohen (1988). See the supplemental material to compare with other genes previously found to be associated with vulnerability to sleep loss (ADORA2A, PER3, TLR4, and DQB1*0602).
Fig. 2.
Average number of PVT lapses as a function of genotype. The panels on the left show performance across 24 h of TSD (top) and at baseline (bottom). The panels on the right show performance during the first 12-h period of TSD (22:00–10:00; top) and the second 12-h period of TSD (10:00–22:00; bottom), where times of day during the second 12-h period correspond to those of the baseline period. Error bars denote standard error.
For secondary PVT measures – mean of the 10% fastest reaction times and median reaction time – the averages over the 24-h period of sleep deprivation did not differ significantly by genotype, with A/A and A/G combined (fastest 10%: F1,82=0.69, P=0.41; median: F1,82=2.76, P=0.10). This indicates that the effect of genotype did not involve a general difference in speed of cognitive processing. In contrast, the significant effect of genotype on PVT lapses indicates that the TNFα G308A polymorphism affected primarily the right tail of the reaction time distribution, pointing to an effect that is linked to sleep deprivation (Doran et al., 2001). All further results presented here are focused on PVT lapses, with the A/A and A/G genotypes combined.
There were no significant differences between the genotypes for age (F1,86=1.43, P=0.24), sex (χ21=1.80, P=0.18), and race/ethnicity distribution (χ21=0.92, P=0.34). Indeed, inter-individual differences in performance impairment during TSD were not predicted by age (F1,81=0.23, P=0.63), sex (F1,81=0.87, P=0.35), or race/ethnicity (F6,76=1.73, P=0.13).
There was also no significant difference between the genotypes for total sleep time (TST) in the baseline night immediately preceding the TSD period (F1,78=0.50, P=048). Even so, inter-individual differences in performance impairment during TSD were predicted by baseline TST (F1,73=5.16, P=0.026), with greater baseline TST corresponding to higher rankings for vulnerability to performance impairment during TSD. The effect of genotype remained significant when controlling for baseline TST (F1,73=6.53, P=0.013). There was no significant interaction between genotype and baseline TST (F1,72=0.97, P=0.33), indicating that the association between baseline TST and vulnerability to performance impairment in this dataset was unrelated to the TNFα G308A polymorphism.
The genotypes differed significantly in terms of baseline levels of psychomotor vigilance performance (F1,86=7.46, P=0.008). Individuals homozygous or heterozygous for the A allele exhibited fewer PVT lapses at baseline than individuals homozygous for the G allele – see Fig. 2 (bottom left). These baseline differences were significantly predictive of performance impairment during TSD (F1,81=31.47, P<0.001) (cf. Chua et al., 2014).
To examine nighttime and daytime performance during sleep deprivation, PVT lapses during the TSD periods from 22:00 until 10:00 and from 10:00 until 22:00 were analyzed separately. For both 12-h periods, individuals carrying the A allele exhibited fewer PVT lapses than individuals homozygous for the G allele (Fig. 2, right). Non-parametric analysis of rank order scores, controlling for study, showed that the genotypes differed with statistical significance during the first 12-h TSD interval (22:00–10:00; F1,82=6.20, P=0.015) and during the second 12- h TSD interval (10:00–22:00; F1,82=4.02, P=0.048).
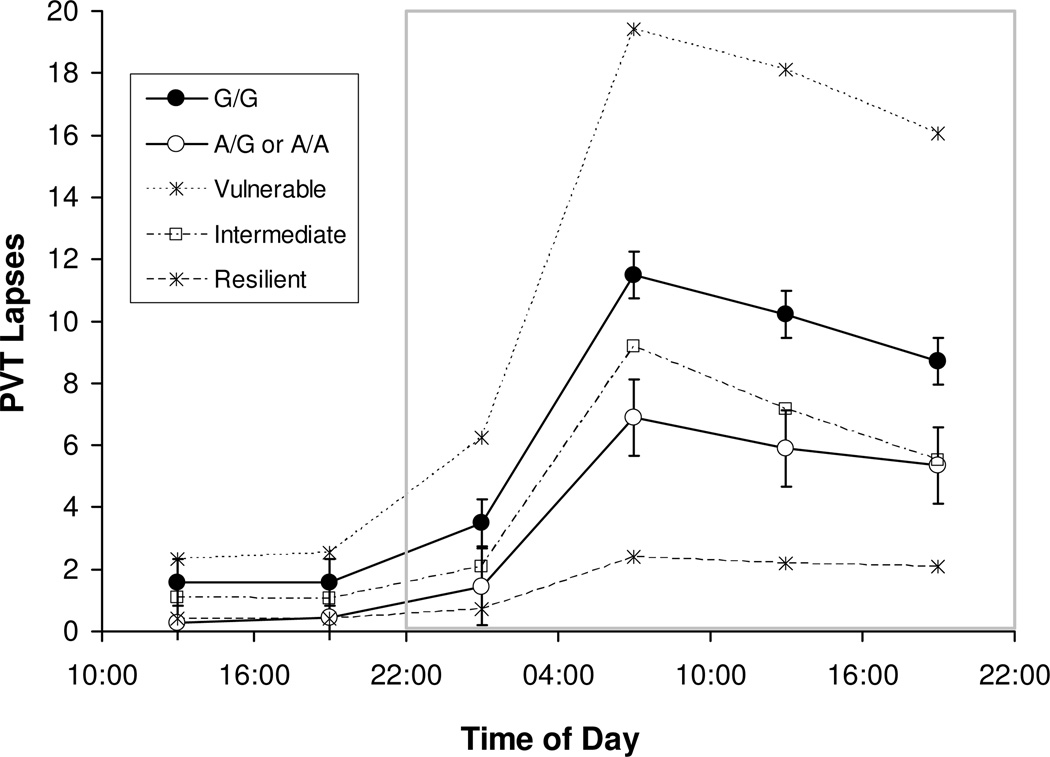
Investigating the temporal profiles of psychomotor vigilance performance changes further, mixed-effects ANOVA of PVT lapses over time – i.e., analysis of subject-specific averages over six consecutive 6-h intervals of wakefulness – yielded significant main effects of genotype (F1,428=6.63, P=0.010) and time interval (F5,428=50.70, P<0.001) and a trend for the interaction (F5,428=2.07, P=0.068).
Fig. 3 shows the temporal profiles of PVT lapses for the subjects with the A/A or A/G genotype as compared to those with the G/G genotype. Both groups exhibited the characteristic profile of increased psychomotor vigilance performance impairment with the progression of time awake, modulated by circadian rhythm (Van Dongen and Dinges, 2005b). However, the A/A and A/G genotype group displayed consistently better PVT performance than the G/G genotype group. The difference between the two groups was small, but not insignificant, at baseline (10:00–22:00) and grew during sleep deprivation (from 22:00 until 22:00 the next day) in proportion to overall level of impairment. At the peak of performance impairment (04:00–10:00 interval), the differential effect of the polymorphism was 4.6 PVT lapses (standard error: 1.5). This difference amounted to 57.0% of median performance impairment at that time expressed relative to median performance at baseline (10:00–22:00 average).
Fig. 3.
Average number of PVT lapses across consecutive 6-h intervals of sustained wakefulness for subjects with the G/G genotype versus subjects with the A/G or A/A genotypes. Error bars denote standard error. Intervals that are part of the 24-h period used to quantify vulnerability to sleep loss are enclosed in the gray box (cf. Fig. 1). For reference, the average number of PVT lapses across consecutive 6-h intervals is also shown for the most vulnerable, intermediate, and most resilient tertiles of our sample irrespective of genotype.
Comparison to the temporal profiles of the most vulnerable and resilient tertiles of the sample regardless of genotype (Fig. 3), which were similar to previously observed tertile profiles (Van Dongen et al., 2004b), revealed that TNFα G308A genotype captured only a modest portion of the observed inter-individual differences. Nonetheless, our results (Figs. 2 and 3) indicate that the A allele conferred an advantage in psychomotor vigilance performance, even at baseline, as well as relative resilience to performance degradation due to sleep deprivation and circadian rhythm.
4. Discussion
Our findings indicate that the A allele at the TNFα 308 locus – in comparison with the more common G allele – is associated with a degree of resilience to psychomotor vigilance performance impairment during total sleep deprivation, and even provides a small performance advantage at baseline (Fig. 2). There was only one subject in our sample who was homozygous for the A allele (Table 3). Although this subject’s performance impairment during sleep deprivation was similar to that of the 23 subjects who were heterozygous, our sample was too small to draw firm conclusions about the A/A genotype. Furthermore, the association between the TNFα G308A polymorphism and resilience to performance impairment due to sleep deprivation may be limited to the population we drew from, consisting of healthy adult women and men aged 22–40. Our findings are also bound to be limited to a specific cognitive domain that encompasses psychomotor vigilance performance impairment (as measured by PVT), as studies of inter-individual differences in responses to sleep deprivation have shown these to be dependent on the type of performance task used to measure impairment (Frey et al., 2004; Van Dongen et al., 2004a; Franzen et al., 2008; Van Dongen et al., 2011b).
While our primary results were based on PVT performance impairment averaged over a full circadian cycle (Fig. 2), follow-up analyses by time of day showed that the effect of the TNFα G308A polymorphism were not specific to any part of the circadian cycle (Fig. 3). Rather, the differential effect of the polymorphism appeared to be tied to homeostatic sleep pressure built up over time awake in interaction with circadian rhythm (Fig. 2; cf. Van Dongen and Dinges, 2003, 2005a). This is in agreement with the hypothesized role of TNFα in sleep/wake regulation (Krueger et al., 2010) and waking cognitive performance (Van Dongen et al., 2011a).
The effect of genotype on subjects’ rank order of vulnerability to sleep loss was statistically significant and robust to variance associated with subjects’ baseline sleep duration, age, gender, race/ethnicity, as well as the study they participated in. As shown in the supplemental material, the TNFα G308A polymorphism had no significant associations with other genetic variants implicated in vulnerability to sleep loss, such as ADORA2A (Bodenmann et al., 2012), PER3 (Lo et al., 2012), TLR4 (Wisor et al., 2011), and a gene polymorphism located in the MHC nearby that of TNFα, DQB1*0602 (Goel et al., 2010).
The TNFα G308A polymorphism predicted less than 10% of the overall variance from inter-individual differences in psychomotor vigilance performance impairment during sleep deprivation. Yet, the differential effect of the polymorphism (Fig. 3, closed versus open circles) at the peak of performance impairment was greater than 50% of median performance impairment at that time. Although use of different PVT metrics and variable transformations in the literature generally makes it difficult to compare directly, it seems that to date, a similarly sizeable effect on resilience to performance impairment due to sleep loss has only been reported for a polymorphism of the basic helix-loop-helix family, member e41 gene (BHLHE41) in a single twin pair (Pellegrino et al., 2014). See the supplemental material to compare with the effect sizes for ADORA2A, PER3, TLR4 and DQB1*0602, which were all smaller than the effect size for TNFα in our sample.
Perhaps more important than the predictive potential of the TNFα G308A polymorphism is what its role may be in the mechanisms underlying psychomotor vigilance performance impairment due to sleep deprivation. In this regard it is noteworthy that the polymorphism also predicted baseline performance (Fig. 2), and that the genotype-dependent difference at baseline became amplified during total sleep deprivation (Fig. 3). A priori, the genotype effect might be explained by a difference in motor response speed from differential neuromuscular transmission due to a TNFα-mediated effect on myelination (Briones and Woods, 2014). However, this explanation is inconsistent with the lack of an effect of genotype on the fastest 10% of reaction times, and would also not be a plausible reason for the amplified genotype effect seen during sleep deprivation as compared to baseline.
Local sleep theory (Krueger and Obál, 1993; Krueger et al., 2008) and its extension to the effects of sleep deprivation on cognitive performance (Van Dongen et al., 2011a) may help to elucidate our findings. In this theoretical framework it is posited that neuronal activity from extended wakefulness and especially from intensive use during task performance leads to release of adenosine triphosphate (ATP) into the extracellular space. Binding of ATP to the purine type 2 receptor X7 triggers activation of a cascade of cytokines, including relatively rapid release of TNFα from nearby glia (Hide et al., 2000). TNFα promotes a local sleep-like state (Churchill et al., 2008; Krueger, 2012), which is hypothesized to interfere with neuronal information processing and thereby cause cognitive instability (Van Dongen et al., 2011a), which in turn can be observed as lapses of attention on the PVT (Doran et al., 2001).
Resilience to performance impairment due to sleep deprivation is found in those whose activation of task-relevant neuronal pathways (as measured with functional magnetic resonance imaging) is less reduced during sleep deprivation compared to baseline (Chee and Tan, 2010). This may be seen as evidence that in resilient individuals, information processing capacity is less degraded by local sleep (Chee and Van Dongen, 2013). In terms of cognitive performance, this means greater signal-to-noise ratio or, in the context of a diffusion cognitive model for PVT performance, higher diffusion drift rate for information processing (Ratcliff and Van Dongen, 2011). Furthermore, inter-individual differences in drift rate at baseline predict inter-individual differences in drift rate and PVT performance impairment during sleep deprivation (Patanaik et al., in press).
Collectively, these theoretical considerations and empirical findings make sense with respect to the present results if inter-individual differences in TNFα production are associated with interindividual differences in the occurrence of local sleep in task-relevant neuronal networks. Local sleep theory makes a specific prediction regarding the direction of this relationship, that is, those individuals who produce the most TNFα in response to neuronal use should be the most vulnerable to performance impairment (Van Dongen et al., 2011a; Krueger et al., 2013). Although the current literature on TNFα production is inconclusive (Hajeer and Hutchinson, 2001), this is a falsifiable prediction of the theory.
In conclusion, our data provide evidence for the involvement of the cytokine TNFα in determining a level of resilience to psychomotor vigilance performance impairment due to sleep deprivation. Prospective studies in diverse study populations are needed to investigate to what extent the TNFα G308A polymorphism is a sensitive and reliable biomarker of resilience in response to sleep deprivation.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contributions of David Dinges and the staff of the Unit for Experimental Psychiatry in the University of Pennsylvania Perelman School of Medicine and the Clinical and Translational Research Center at the Hospital of the University of Pennsylvania (study 1), Gregory Belenky, Matthew Layton, Melinda Jackson, Devon Grant and the staff of the Human Sleep and Cognition Laboratory in the Sleep and Performance Research Center at Washington State University Spokane and John Hinson and Paul Whitney in the Department of Psychology at Washington State University Pullman (studies 2–5), and Daniel Mollicone and Christopher Mott of Pulsar Informatics, Inc. (study 3), for their contributions to conducting the experiments. The experiments were funded by NIH grant R01HL070154 (study 1), NIH grant R21CA167691 (study 2), ONR grant N00014-13-C-0063 (study 3), NIH grant R01HL105768 and FAA grant DTFAAC-11-A-00003 (study 4), and CDMRP grant W81XWH-05-1-0099 (study 5). Genotyping was supported by ONR grant N00014-13-1-0302 and by an Elliot D. Weitzman, M.D. Research Grant from the Sleep Research Society Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Mollicone D, Basner M, Dinges DF. Sleepiness and safety: where biology needs technology. Sleep Biol. Rhythms. 2014;12:74–84. doi: 10.1111/sbr.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. Polymorphism of the human TNF-α promoter – random variation or functional diversity? Mol. Immunol. 1999;36:1017–1027. doi: 10.1016/s0161-5890(99)00127-3. [DOI] [PubMed] [Google Scholar]
- Almpanidou P, Hadjigeorgiou G, Gourgoulianis K, Papadimitriou A. Association of tumor necrosis factor-α gene polymorphism (−308) and obstructive sleep apnea-hypopnea syndrome. Hippokratia. 2012;16:217–220. [PMC free article] [PubMed] [Google Scholar]
- Beste C, Baune BT, Falkenstein M, Konrad C. Variations in the TNF-α gene (TNF-α −308GαA) affect attention and action selection mechanisms in a dissociated fashion. J. Neurophysiol. 2010;104:2523–2531. doi: 10.1152/jn.00561.2010. [DOI] [PubMed] [Google Scholar]
- Bodenmann S, Hohoff C, Freitag C, Deckert J, Rétey JV, Bachmann V, et al. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep EEG after sleep deprivation. Brit. J. Pharmacol. 2012;165:1904–1913. doi: 10.1111/j.1476-5381.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones TL, Woods J. Dysregulation in myelination mediated by persistent neuroinflammation: possible mechanisms in chemotherapy-related cognitive impairment. Brain Behav. Immun. 2014;35:23–32. doi: 10.1016/j.bbi.2013.07.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MWL, Tan JC. Lapsing when sleep deprived: neural activation characteristics of resistant and vulnerable individuals. NeuroImage. 2010;51:835–843. doi: 10.1016/j.neuroimage.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Chee MWL, Van Dongen HPA. Functional imaging of inter-individual differences in response to sleep deprivation. In: Nofzinger E, Maquet P, Thorpy MJ, editors. Neuroimaging of Sleep and Sleep Disorders. Cambridge, UK: Cambridge University Press; 2013. pp. 154–162. [Google Scholar]
- Chua EC, Yeo SC, Lee IT, Tan LC, Lau P, Cai S, et al. Sustained attention performance during sleep deprivation associates with instability in behavior and physiologic measures at baseline. Sleep. 2014;37:27–39. doi: 10.5665/sleep.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill L, Rector DM, Yasuda K, Fix C, Rojas MJ, Yasuda T, et al. Tumor necrosis factor alpha: activity dependent expression and promotion of cortical column sleep in rats. Neurosci. 2008;156:71–80. doi: 10.1016/j.neuroscience.2008.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JE. Statistical Power Analysis for the Behavioral Sciences. Hillsdale, NJ: Lawrence Erlbaum Associates, Inc.; 1988. [Google Scholar]
- Doran SM, Van Dongen HPA, Dinges DF. Sustained attention performance during sleep deprivation: evidence of state instability. Arch. Ital. Biol. 2001;139:253–267. [PubMed] [Google Scholar]
- Dorrian J, Rogers NL, Dinges DF. Psychomotor vigilance performance: a neurocognitive assay sensitive to sleep loss. In: Kushida CA, editor. Sleep Deprivation. Clinical Issues, Pharmacology, and Sleep Loss Effects. New York, NY: Marcel Dekker, Inc.; 2005. [Google Scholar]
- Elahi MM, Asotra K, Matata BM, Mastana SS. Tumor necrosis factor alpha −308 gene locus promoter polymorphism: An analysis of association with health and disease. BBA-Mol. Basis Dis. 2009;1792:163–172. doi: 10.1016/j.bbadis.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Fargion S, Valenti L, Dongiovanni P, Scaccabarozzi A, Fracanzani AL, Taioli E, et al. Tumor necrosis factor alpha promoter polymorphisms influence the phenotypic expression of hereditary hemochromatosis. Blood. 2001;97:3707–3712. doi: 10.1182/blood.v97.12.3707. [DOI] [PubMed] [Google Scholar]
- Franzen PL, Siegle GJ, Buysse DJ. Relationships between affect, vigilance, and sleepiness following sleep deprivation. J. Sleep Res. 2008;17:34–41. doi: 10.1111/j.1365-2869.2008.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey DJ, Badia P, Wright KP. Inter- and intra-individual variability in performance near the circadian nadir during sleep deprivation. J. Sleep Res. 2004;13:305–315. doi: 10.1111/j.1365-2869.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Goel N, Banks S, Mignot E, Dinges DF. DQB1*0602 predicts interindividual differences in physiologic sleep, sleepiness and fatigue. Neurology. 2010;75:1509–1519. doi: 10.1212/WNL.0b013e3181f9615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Dinges DF. Predicting risk in space: genetic markers for differential vulnerability to sleep restriction. Acta Astronaut. 2012;77:207–213. doi: 10.1016/j.actaastro.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant DA, Honn KA, Kogan CJ, Van Dongen HPA. Unobtrusive, Wearable Sensor Array to Collect Actigraphy, Ship Motion, Vibration, Noise and Temperature – Phase II: PVT Validation. Philadelphia, PA: Pulsar Informatics, Inc.; 2013a. [Google Scholar]
- Grant DA, Whitney P, Hinson JM, Layton ME, Van Dongen HPA. The effect of total sleep deprivation on semantic encoding. Sleep. 2013b;36:A76–A77. doi: 10.1080/07420528.2017.1411361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajeer AH, Hutchinson IV. Influence of TNF gene polymorphisms on TNF production and disease. Hum. Immunol. 2001;62:1191–1199. doi: 10.1016/s0198-8859(01)00322-6. [DOI] [PubMed] [Google Scholar]
- Hide I, Tanaka M, Inoue A, Nakajima K, Kohsaka S, Inoue K, et al. Extracellular ATP triggers tumor necrosis factor-alpha release from rat microglia. J. Neurochem. 2000;75:965–972. doi: 10.1046/j.1471-4159.2000.0750965.x. [DOI] [PubMed] [Google Scholar]
- Jackson ML, Gunzelmann G, Whitney P, Hinson JM, Belenky G, Rabat A, et al. Deconstructing and reconstructing cognitive performance in sleep deprivation. Sleep Med. Rev. 2013;17:215–225. doi: 10.1016/j.smrv.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan AS, McSharry DG, Malhotra A. Adult obstructive sleep apnoea. Lancet. 2014;383:736–747. doi: 10.1016/S0140-6736(13)60734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, Belenky G, Van Dongen HPA. Performance impairment consequent to sleep loss: determinants of resistance and susceptibility. Curr. Opin. Pulm. Med. 2009;15:555–564. doi: 10.1097/MCP.0b013e3283319aad. [DOI] [PubMed] [Google Scholar]
- Kroeger KM, Steer JH, Joyce DA, Abraham LJ. Effects of stimulus and cell type on the expression of the −308 tumour necrosis factor promoter polymorphism. Cytokine. 2000;12:110–119. doi: 10.1006/cyto.1999.0529. [DOI] [PubMed] [Google Scholar]
- Krueger JM. The role of cytokines in sleep regulation. Curr. Pharm. Des. 2008;14:3408–3416. doi: 10.2174/138161208786549281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM. Translation of brain activity into sleep. Hirosaki Igaku. 2012;63:S1–S16. [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Huang YH, Rector DM, Buysse DJ. Sleep: a synchrony of cell activity-driven small network states. Eur. J. Neurosci. 2013;38:2199–2209. doi: 10.1111/ejn.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Obál F. A neuronal group theory of sleep function. J. Sleep Res. 1993;2:63–69. doi: 10.1111/j.1365-2869.1993.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Krueger JM, Taishi P, De A, Davis CJ, Winters BD, Clinton J, et al. ATP and the purine type 2 X7 receptor affect sleep. J. Appl. Physiol. 2010;109:1318–1327. doi: 10.1152/japplphysiol.00586.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger JM, Rector DM, Roy S, Van Dongen HPA, Belenky G, Panksepp J. Sleep as a fundamental property of neuronal assemblies. Nat. Rev. Neurosci. 2008;9:910–919. doi: 10.1038/nrn2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuna ST, Maislin G, Pack FM, Staley B, Hachadoorian R, Coccaro EF, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35:1223–1233. doi: 10.5665/sleep.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt H-P. Sleep homeostasis: a role for adenosine in humans? Biochem. Pharmacol. 2008;75:2070–2079. doi: 10.1016/j.bcp.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Leproult R, Colecchia EF, Berardi AM, Stickgold R, Kosslyn SM, van Cauter E. Individual differences in subjective and objective alertness during sleep deprivation are stable and unrelated. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;284:R280–R290. doi: 10.1152/ajpregu.00197.2002. [DOI] [PubMed] [Google Scholar]
- Lim J, Dinges DF. Sleep deprivation and vigilant attention. Ann. N. Y. Acad. Sci. 2008;1129:305–322. doi: 10.1196/annals.1417.002. [DOI] [PubMed] [Google Scholar]
- Lo JCY, Groeger JA, Santhi N, Arbon EL, Lazar AS, Hasan S, et al. Effects of partial and acute total sleep deprivation on performance across cognitive domains, individuals and circadian phase. PLoS ONE. 2012;7:e45987. doi: 10.1371/journal.pone.0045987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, Roland S, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-α production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin. Exp. Immunol. 1998;113:401–406. doi: 10.1046/j.1365-2249.1998.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjari KS, Jyothy A, Kumar PS, Prabhakar B, Devi MU, Ramanna M, et al. A single-nucleotide polymorphism in tumor necrosis factor-α (−308 G/A) as a biomarker in chronic pancreatitis. Gene. 2014;539:186–189. doi: 10.1016/j.gene.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Morgan BB, Winne PS, Dugan J. The range and consistency of individual differences in continuous work. Hum. Factors. 1980;22:331–340. [Google Scholar]
- Napolioni V, Carpi FM, Giannì P, Sacco R, Blasio LD, Mignini F, et al. Age- and gender-specific epistasis between ADA and TNF-α influences human life-expectancy. Cytokine. 2011;56:481–488. doi: 10.1016/j.cyto.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Opp MR. Sleep and psychoneuroimmunology. Immunol. Allergy Clin. North Am. 2009;29:295–307. doi: 10.1016/j.iac.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Ozen S, Alikasifoglu M, Bakkaloglu A, Duzova A, Jarosova K, Nemcova D, et al. Tumour necrosis factor α Gα A −238 and Gα A −308 polymorphisms in juvenile idiopathic arthritis. Rheumatology. 2002;41:223–227. doi: 10.1093/rheumatology/41.2.223. [DOI] [PubMed] [Google Scholar]
- Patanaik A, Zagorodnov V, Kwoh CK, Chee MWL. Predicting vulnerability to sleep deprivation using diffusion model parameters. J. Sleep Res. 2014 doi: 10.1111/jsr.12166. in press. [DOI] [PubMed] [Google Scholar]
- Pellegrino R, Halil Kavakli I, Goel N, Cardinale CJ, Dinges DF, Kuna ST, et al. A novel BHLHE41 variant is associated with short sleep and resistance to sleep deprivation in humans. Sleep. 2014;37:1327–1336. doi: 10.5665/sleep.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff R, Van Dongen HPA. Diffusion model for one-choice reaction-time tasks and the cognitive effects of sleep deprivation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11285–11290. doi: 10.1073/pnas.1100483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert CF, Maire M, Gabel V, Viola AU, Kolodyazhniy V, Strobel W, et al. Insights into behavioral vulnerability to differential sleep pressure and circadian phase from a functional ADA polymorphism. J. Biol. Rhythms. 2014;29:119–130. doi: 10.1177/0748730414524898. [DOI] [PubMed] [Google Scholar]
- Riha RL, Brander P, Vennelle M, McArdle N, Kerr SM, Anderson NH, et al. Tumor necrosis factor-α (−308) gene polymorphism in obstructive sleep apnoea-hypopnea syndrome. Eur. Respir. J. 2005;26:673–678. doi: 10.1183/09031936.05.00130804. [DOI] [PubMed] [Google Scholar]
- Rupp LT, Wesensten NJ, Balkin TJ. Trait-like vulnerability to total and partial sleep loss. Sleep. 2012;35:1163–1472. doi: 10.5665/sleep.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, et al. Disturbances in selective information processing associated with the BDNF Val66Met polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biol. Psychol. 2009;80:176–188. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tucker AM, Dinges DF, Van Dongen HPA. Trait interindividual differences in the sleep physiology of healthy young adults. J. Sleep Res. 2007;16:170–180. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- Tucker AM, Whitney P, Belenky G, Hinson JM, Van Dongen HPA. Effects of sleep deprivation on dissociated components of executive functioning. Sleep. 2010;33:47–57. doi: 10.1093/sleep/33.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen HPA, Baynard MD, Maislin G, Dinges DF. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004a;27:423–433. [PubMed] [Google Scholar]
- Van Dongen HPA, Belenky G, Krueger JM. A local, bottom-up perspective on sleep deprivation and neurobehavioral performance. Curr. Top. Med. Chem. 2011a;11:2414–2422. doi: 10.2174/156802611797470286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen HPA, Caldwell JA, Caldwell JL. Individual differences in cognitive vulnerability to fatigue in the laboratory and in the workplace. Prog. Brain Res. 2011b;190:145–153. doi: 10.1016/B978-0-444-53817-8.00009-8. [DOI] [PubMed] [Google Scholar]
- Van Dongen HPA, Dinges DF. Investigating the interaction between the homeostatic and circadian processes of sleep-wake regulation for the prediction of waking neurobehavioural performance. J. Sleep Res. 2003;12:181–187. doi: 10.1046/j.1365-2869.2003.00357.x. [DOI] [PubMed] [Google Scholar]
- Van Dongen HPA, Dinges DF. Circadian rhythms in sleepiness, alertness, and performance. In: Kryger MH, Roth T, Dement WC, editors. Principles and Practice of Sleep Medicine. 4th ed. Philadelphia, PA: Elsevier Saunders; 2005a. pp. 435–443. [Google Scholar]
- Van Dongen HPA, Dinges DF. Sleep, circadian rhythms, and psychomotor vigilance. Clin. Sports Med. 2005b;24:237–249. doi: 10.1016/j.csm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Van Dongen HPA, Maislin G, Dinges DF. Dealing with inter-individual differences in the temporal dynamics of fatigue and performance: importance and techniques. Aviat. Space Environ. Med. 2004b;75:A147–A154. [PubMed] [Google Scholar]
- Vinasco J, Beraún Y, Nieto A, Fraile L, Mataran E, Pareja J, et al. Polymorphism at the TNF loci in rheumatoid arthritis. Tissue Antigens. 1997;49:74–78. doi: 10.1111/j.1399-0039.1997.tb02715.x. [DOI] [PubMed] [Google Scholar]
- Vgontzas AN, Papanicolaou DA, Bixler EO, Kales A, Tyson K, Chrousos GP. Elevation of plasma cytokines in disorders of excessive daytime sleepiness: role of sleep disturbance and obesity. J. Clin. Endocr. Metab. 1997;82:1313–13116. doi: 10.1210/jcem.82.5.3950. [DOI] [PubMed] [Google Scholar]
- Webb WB, Levy CM. Effects of spaced and repeated total sleep deprivation. Ergonomics. 1984;27:45–58. doi: 10.1080/00140138408963462. [DOI] [PubMed] [Google Scholar]
- Whitney P, Hinson JM, Jackson ML, Van Dongen HPA. Feedback is lost on the sleep-deprived. Sleep. 2014 in press. [Google Scholar]
- Wilkinson RT. Interaction of lack of sleep with knowledge of results, repeated testing, and individual differences. J. Exp. Psychol. 1961;62:263–271. doi: 10.1037/h0048787. [DOI] [PubMed] [Google Scholar]
- Wilson AG, di Giovine FS, Blakemore AIF, Duff GW. Single base change in the human tumor necrosis factor alpha (TNF-α) gene detectable by NcoI restriction of PCR product. Human Mol. Genet. 1992;1:353–358. doi: 10.1093/hmg/1.5.353. [DOI] [PubMed] [Google Scholar]
- Wisor JP, Clegern WC, Schmidt MA. Toll-like receptor 4 is a regulator of monocyte and electroencephalographic responses to sleep loss. Sleep. 2011;34:1335–1345. doi: 10.5665/SLEEP.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.