Abstract
Researchers have only recently begun using functional neuroimaging to explore the human response to periods of sustained anxious anticipation, namely potential threat. Here, we investigated brain responses acquired with functional MRI during an instructed threat of shock paradigm used to create sustained periods of aversive anticipation. In this re-analysis of previously published data, we employed Quadratic Discriminant Analysis to classify the multivariate pattern of whole-brain functional connectivity and to identify connectivity changes during periods of potential threat. Our method identifies clusters with altered connectivity on a voxelwise basis, thus eschewing the need to define regions a priori. Classifier generalization was evaluated by testing on data from participants not used during training. Robust classification between threat and safe contexts was possible, and inspection of “diagnostic features” revealed altered functional connectivity involving the intraparietal sulcus, task-negative regions, striatum, and anterior cingulate cortex. We anticipate that the proposed method will prove useful to experimenters wishing to identify large-scale functional networks that distinguish between experimental conditions or groups.
Keywords: fMRI methods, functional connectivity, networks, anxiety, Nucleus accumbens
1. Introduction
Aversive processing is engaged by both transient stimuli and sustained contexts. For example, a previously shock-paired, brief tone may be encountered subsequently, thus eliciting a “fear” response; or a location where aversive events were experienced may be re-encountered, thus eliciting a sustained state of “anxious apprehension”. Indeed, the Research Domain Criteria (RDoC) developed by the National Institute of Mental Health established two constructs, namely “acute threat” (also called “fear”) and “potential threat” (also called “anxiety”), which may be subserved by different neural subsystems. The former is thought to involve a circuit centered on the amygdala, and the latter is thought to involve a circuit centered on both the bed nucleus of the stria terminalis (BNST) and the amygdala (Davis et al., 2010).
Studies of potential threat in humans have only appeared recently, but knowledge is accruing quickly (Alvarez et al., 2011; Davis et al., 2010; Hermans et al., 2014; Kim et al., 2010; McMenamin et al., 2014; Mobbs et al., 2010; Somerville et al., 2010; Vytal et al., 2014; Walker et al., 2003). Although the spatial resolution of neuroimaging studies thus far is too low for a conclusive answer, these studies have identified a site that is consistent with the location of the human BNST that is activated during potential threat. For example, in our recent study (McMenamin et al., 2014), the putative BNST area exhibited sustained responses during periods when participants could receive unpleasant, mild electric shocks.
To further understand brain processing during potential threat, in addition to studying evoked responses, it is important to unravel how the functional coupling between regions is altered by threat. In our study (McMenamin et al., 2014), we investigated changes in functional connectivity involving the amygdala, the BNST, and three large-scale networks, namely the salience, the executive, and the task-negative networks. We found that, for example, the salience network exhibited a transient increase in network efficiency followed by a period of sustained decreased efficiency.
While our previous analysis was developed to examine changes involving a priori sets of brain regions, here we develop a complementary approach and use it to re-analyze data from our previously published study (McMenamin et al., 2014). As proposed by others (Stanley et al., 2013; Zalesky et al., 2012), we employed whole-brain voxelwise functional connectivity analysis. Previous research has demonstrated that multivariate pattern analysis can be a powerful tool for inferring a participant’s cognitive state from patterns of whole-brain functional connectivity (Craddock et al., 2009; Richiardi et al., 2011; Shirer et al., 2012); unfortunately, all these approaches use regions of interest that are defined a priori and it is unclear whether they would “scale-up” to operate on a voxelwise basis. The present report identifies changes in functional connectivity between conditions using Quadratic Discriminant Analysis (QDA; (Hastie et al., 2009)), a generalization of Linear Discriminant Analysis that is able to distinguish between experimental conditions based on differences in covariance structure – here, differences in the pattern of functional connectivity. Moreover, the QDA algorithm can be applied to fMRI time series without specifying regions of interest. Thus our method is a type of multivariate pattern analysis applied to voxelwise patterns of connectivity.
2. Methods
2.1 Participants
Twenty-four right-handed participants (9 male, age 19–34 years) were recruited from the University of Maryland community. The project was approved by the University of Maryland College Park Institutional Review Board and all participants provided written informed consent prior to participation. The datasets collected from these participants were described in a previous study (McMenamin et al., 2014).
2.2 Procedure and Stimuli
The experiment used a threat of shock paradigm in which a colored (e.g., yellow) circle on the screen indicated that participants were in a “threat” block and mild electric shocks would be delivered to their left hand at random; a circle of another color (e.g., blue) indicated that participants were in a “safe” block and no shocks would be delivered. Colors were counterbalanced across participants. Each block had the average duration of 60 s (range 42.5 – 77.5 s). The whole experiment contained four “runs” resulting in a total of 16 threat and 16 safe blocks. Each threat block contained zero to four electric shocks, with five of the 16 threat blocks containing zero shocks.
Visual stimuli were presented using Presentation software (Neurobehavioral Systems, Albany, CA, USA) and viewed on a projection screen using a mirror mounted to the head coil. An electric stimulator (Coulbourn Instruments, PA, USA) delivered 500-ms stimulation to the fourth and fifth fingers of the left hand via MRI-compatible electrodes. To calibrate the intensity of the shock, each participant was asked to choose his/her own stimulation level immediately before the scanning began.
2.3 MRI data acquisition
MRI data collection used a 3 Tesla Siemens TRIO scanner (Siemens Medical Systems, Erlangen, Germany) with a 32-channel head coil. Each session began with the acquisition of a high-resolution MPRAGE anatomical scan (0.45 × 0.45 × 0.9 mm voxels). Each of the subsequent functional runs collected 201 volumes of EPI data (TR = 2.5 s, TE = 25 ms, and FOV = 192 mm). Each volume contained 44 oblique slices oriented 30° clockwise relative to the AC-PC axis with thickness 3 mm and voxels measuring 3 mm × 3 mm in plane.
2.4 Functional MRI preprocessing
Preprocessing of the functional and anatomical MRI data used the AFNI (Cox, 1996; http://afni.nimh.nih.gov/) and SPM software packages (http://www.fil.ion.ucl.ac.uk/spm/) as described in the original report (McMenamin et al., 2014). Preprocessing included slice-timing correction, rigid body transformation to correct head motion, spatial normalization to Talairach space using the TT_N27 template, spatial smoothing with a 6-mm full-width half-maximum (FWHM) Gaussian filter, and intensity normalization within each run. Analyses were restricted to cortical and subcortical grey-matter regions, excluding cerebellum, defined by the Desai anatomical atlas (Desikan et al., 2006; Destrieux et al., 2010), resulting in 34,217 gray-matter voxels for subsequent analyses.
Initially, the responses at every voxel were analyzed for each participant using multiple regression in AFNI (Cox, 1996); for further details, see McMenamin et al. (2014). Activation due to safe-block onset, threat-block onset, and physical shock delivery were modeled using cubic spline basis functions that make no assumptions about the shape of the hemodynamic response. Responses to safe- and threat-block onsets were modeled for the first 40 s of the block because that was the minimum duration of any threat or safe block. Response to physical shock delivery was modeled for 20 s. Constant, linear, and quadratic terms were included as covariates of no interest for each run to accommodate for slow-varying drifts in the MR signal. Additional covariates of no interest comprised the average signal from white matter voxels, the average signal from ventricle voxels, and six rigid-body head motion parameters. The white matter and ventricle signals were defined using the eroded maps of white matter and CSF regions from each participant’s higher-resolution anatomical scan. After accounting for all the variables above, the residuals from the overall regression analysis were used for the analyses described here.
The original report (McMenamin et al., 2014) indicated that transitions into a threat block triggered transient activation changes lasting approximately 20 seconds, which was followed by a more sustained pattern of activation. To ensure that the classifiers were detecting connectivity changes corresponding to this period of sustained threat anticipation, we restricted our analysis to 20–40 s after block onset (nine volumes per block). Voxel time courses from these volumes were z-scored within each condition and run.
2.5 Feature selection for QDA
Our central goal was to classify experimental conditions based on the pattern of covariation across voxels. One could thus build a “connectivity” matrix (say, a correlation matrix of observed time series) and apply algorithms that would try to separate connectivity patterns observed during two conditions, for example. In general, the results obtained with statistical and machine learning classification procedures depend on the “input features” that are employed (Guyon and Elisseeff, 2003). Feature selection can greatly improve classification performance by preventing the classifier from using features that that are noisy or unreliable. Previous studies that applied QDA to fMRI data used an initial dimensionality-reduction step implemented by principal component analysis to perform feature selection that targets features that are reliable within each participant’s data (Schmah et al., 2010; Yourganov et al., 2014). Here, we employed a two-stage feature selection process to ensure that the input features were reliable both within participant and across participants. The feature selection procedure consisted of two sequential steps of standard principal component analysis.
2.5.1 First-stage feature selection (participant level)
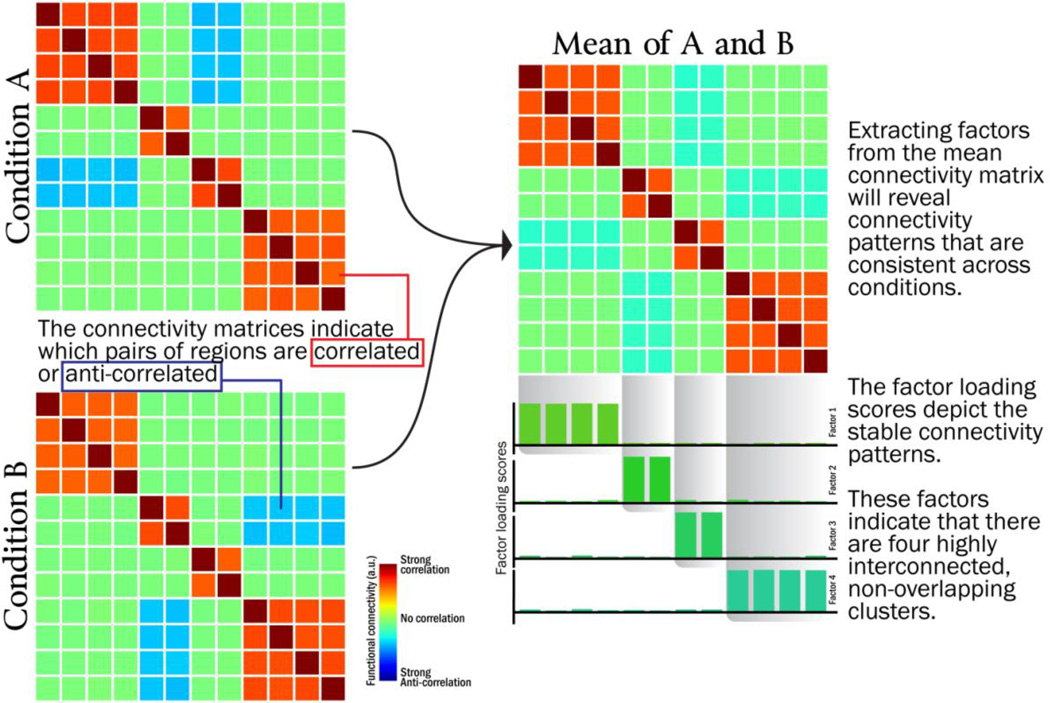
The first stage identified prominent connectivity patterns within each participant by applying principal components analysis (PCA) to each participant’s data. Each participant’s inter-voxel covariance matrix (averaged across safe and threat conditions) was decomposed in the manner depicted by Figure 1 to identify sets of voxels that reliably co-activated with one another. This PCA decomposition results in a set of components (each component is an nvoxels-by-1 vector), each of which described a pattern of co-activation across voxels. Every component is associated with an eigenvalue – a scalar values that describes how prominent each component is in the original data. Larger eigenvalues indicate that the associated component explains a greater proportion of the variance of the original dataset, whereas small eigenvalues indicate that the pattern describes relatively little from the original training data and thus may be considered to be “noise” that is likely to be specific to the training set.
Figure 1.
Illustration of how factor analysis/principal components analysis can be used to decompose functional connectivity matrices into a small number of factors/components
Previous methodological reports (Jackson, 1993; King and Jackson, 1999) have found that the distribution of eigenvalues can be compared to a “broken-stick” distribution (a distribution related to the Dirichlet process, (Ishwaran and James, 2001)) to determine which components are likely to represent a genuine source of signal and which should be labeled as noise. If the jth component’s eigenvalue is greater than the jth value in the broken-stick distribution, the component is considered to be a source of genuine signal; however if the jth component’s eigenvalue is less than the jth value in the broken-stick distribution, the component is considered to be a source of “noise”. If the broken-stick distribution indicates that the top k components should be retained for participant i, we can create a “de-noised” dataset for each participant, , where Xi is the original nvoxels-by-ntimepoints data matrix and Vi is the nvoxels-by-k matrix of retained components1. The de-noised matrix data matrix, X̃i, has the same dimension as the original data matrix but excludes the noisy patterns of voxel co-activation.
2.5.2 Second-stage feature selection (group level)
After the first stage of feature selection, every participant has a de-noised data matrix X̃i that preserves the prominent patterns of inter-voxel co-activation within each participant. The second stage of feature selection combines all of the X̃i to perform a second PCA and determine which of the remaining patterns of inter-voxel co-activation are consistent across participants. All of the X̃i are concatenated into a single nvoxels-by-(ntimepoints*nparticipants) matrix, X̃Group. A second PCA is performed on X̃Group, and once again the broken-stick distribution is used to determine the number of components to be retained. The nvoxels-by-k matrix of retained components, VGroup, is used to create second-level de-noised matrices for each participant, .
The QDA classifier was trained and tested exclusively using the de-noised datasets, namely X͌i (size nvoxels-by-ntimepoints).
2.6 Using QDA to measure changes in functional connectivity
If we were given a pattern of activation across voxels, x, and wished to determine whether it came from condition A or condition B, an intuitive approach to classification would be to simply compare the likelihood that x came from either condition. For example, if XA and XB are probability distributions that describe how data vectors are distributed in conditions A and B, respectively, the classifier could use the log-likelihood ratio as it’s classification decision value, . Large positive values w(x) of indicate that x was relatively more likely to occur in Condition A, whereas large negative values of w(x) indicate that x was relatively more likely to occur in Condition B.
Linear Discriminant Analysis (LDA) and Quadratic Discriminant Analysis (QDA) both use the maximum-likelihood framework to classify data by adding the assumption that data from each condition has a multivariate normal distribution. This assumption allows the likelihood of any input to be computed quickly with a closed-form probability density function for the multivariate normal. The LDA classifier assumes the data from Conditions A and B have different means but identical covariance structure (i.e., XA ~ 𝒩 (μA, Σ0) and XB ~ 𝒩 (μB, Σ0)), resulting in a classifier that can identify how the mean pattern of activation across voxels differs between two conditions (Misaki et al., 2010). The QDA classifier makes the more general assumption that data from Conditions A and B have different means and different covariance structures (i.e., XA ~ 𝒩 (μA, ΣA) and XB ~ 𝒩 (μB, ΣB)) (Georgiou-Karistianis et al., 2013; Hastie et al., 2009; Schmah et al., 2010).
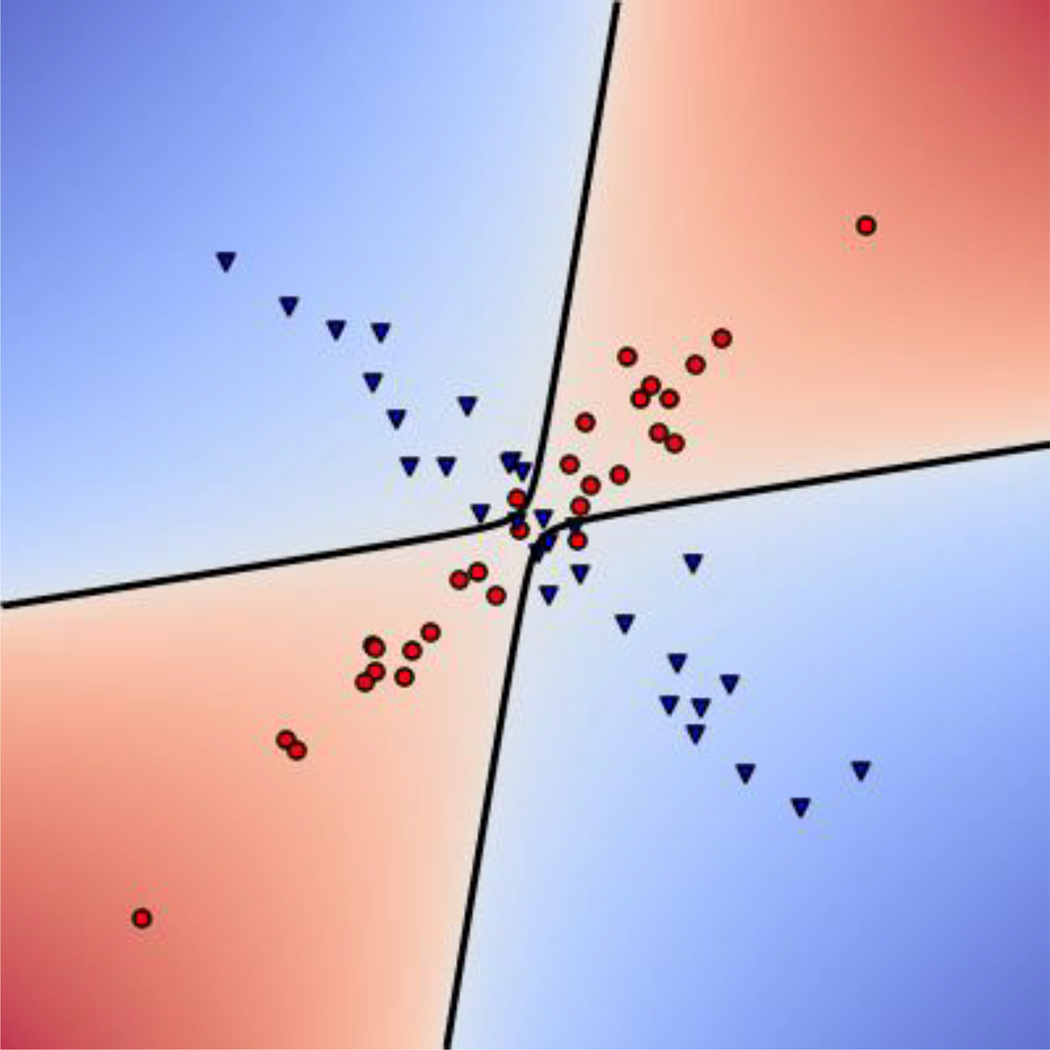
The QDA classifier applied to fMRI data can not only learn to distinguish two conditions based on differences in the mean activation patterns across voxels (e.g., comparing nvoxels-by-1 activation patterns μA and μB), but also based on differences in the functional connectivity patterns across voxels (e.g., comparing nvoxels-by-nvoxels connectivity matrices ΣA and ΣB). If two conditions evoked identical activation patterns, an LDA classifier would be unable to distinguish them even if their covariance differed (i.e., μA = μB, ΣA ≠ ΣB); however, a QDA classifier would be able to distinguish these two conditions based solely on the difference in covariance by implementing a non-linear decision boundary (Figure 2).
Figure 2.
QDA can learn to discriminate two categories based on differences in covariance patterns. Here, Conditions A and B have the same mean so they are linearly inseparable. However, Condition A has a positive correlation between X and Y whereas Condition B has a negative correlation. The QDA classifier can use this covariance difference to create a non-linear decision boundary for separating the groups.
Classifiers are trained to determine condition labels from the inputs; but we can also use the classifiers to identify which input patterns are maximally diagnostic of membership to either condition. The nvoxels-by-1 input pattern that maximizes w(x) is the diagnostic feature vector for Condition A, dA = argmaxx,‖x‖=1[w(x)], and corresponds to the input pattern that the classifier would have the greatest confidence in labeling as Condition A. Conversely, the input pattern which minimizes w(x) is the diagnostic feature vector for Condition B, dB = argmaxx,‖x‖=1[w(x)].
Interpreting results from non-linear classifiers can be more difficult than interpreting the results from a linear classifier (Norman et al., 2006), but the implementation of QDA in the present report was developed to make the extraction and interpretation of diagnostic feature vectors straightforward. Because the activation time course of every voxel is z-scored within each condition, the mean activation pattern for each condition is a vector of all zeroes (i.e., μA = μB = 0). This simplifies the multivariate normal probability density functions, so w(x) can be re-written as:
From this equation, we can find the diagnostic feature vectors for each condition (dA and dB) by finding the unit-norm vector that maximizes or minimizes the value of . The solution to this maximization/minimization problem is well known and based on the eigendecomposition of (Appendix 1): The value of w(x) is maximized when x is the eigenvector with largest associated eigenvalue, and minimized when x is the eigenvector with smallest (oftentimes negative) associated eigenvalue. These two eigenvalues are the diagnostic feature vectors and are labelled as dA and dB, respectively. These diagnostic feature vectors are each nvoxels-by-1 vectors that can be plotted as a pattern of positive and negative values across the brain to understand patterns of whole-brain connectivity that distinguish the two conditions.2.
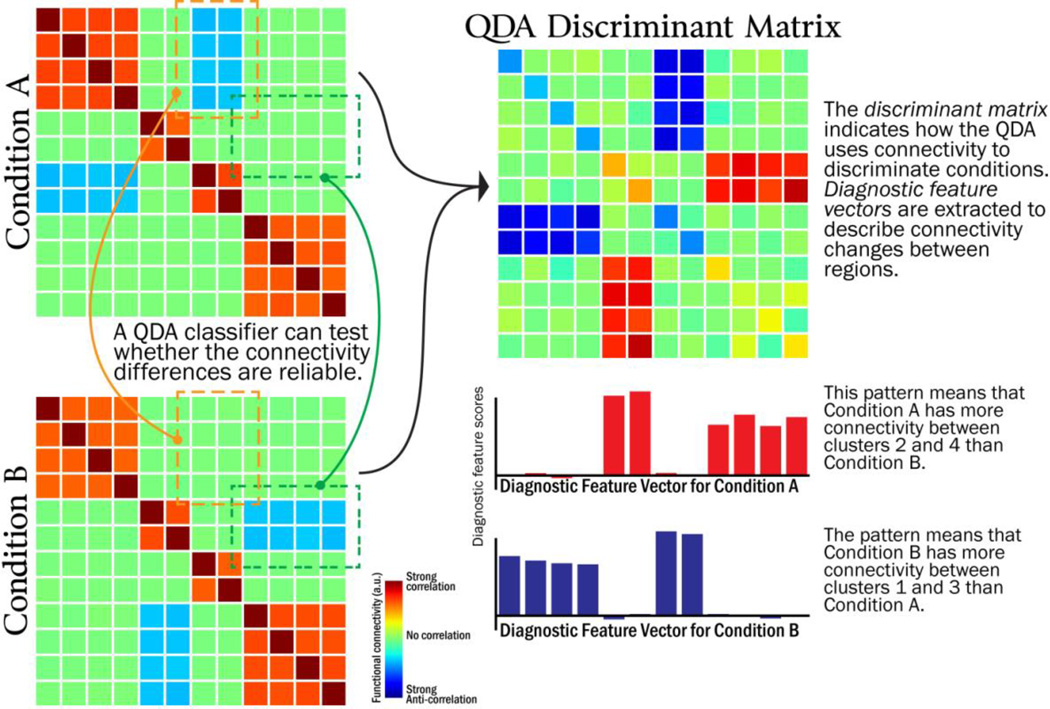
Figure 3 illustrates how the pattern of positive and negative values in each diagnostic feature vector can be interpreted. Voxels with the same sign in the diagnostic feature vector become more connected with one another (i.e., the voxels with positive values connect more strongly with other positive-valued voxels; voxels with negative values connect more strongly with other negative-valued voxels). Voxels with opposite signs “disconnect” with one another, that is, voxels with positive values connect less strongly from voxels with negative values).
Figure 3.
Illustration of how Quadratic Discriminant Analysis (QDA) can be used to identify connectivity differences between conditions.
The diagnostic feature vectors from QDA bear a similarity to the outputs from several mathematically related methods for unsupervised decomposition of fMRI time series into “intrinsic connectivity networks” (ICNs), such as factor analysis, and independent components analysis (Calhoun et al., 2001). The ICNs generated by these algorithms indicate which clusters of voxels consistently co-activate/deactivate in the fMRI time series, and are useful for identifying large-scale network structure. However, because these algorithms are unsupervised, the ICNs that are extracted may not have a clear relationship with experimental manipulations. By contrast, QDA is a supervised algorithm that ignores structure that is present across multiple conditions to specifically identify the differences between conditions.
2.6.1 Classifier training and testing
The QDA classifier was implemented using the SciKit-Learn package in Python (http://scikit-learn.org/) and file I/O was performed using the NiBabel package (http://nipy.org/nibabel/). Classifier performance was measured as the mean classifier accuracy in a leave-one-subject-out cross validation scheme. Each iteration of cross-validation divided participants into training and test datasets: the training dataset contained 23 participants and the test dataset contained a single held-out participant. This process was repeated 24 times so that each participant would serve as the test dataset. On every iteration, the two-stage feature selection (Section 2.5) was performed using the training dataset, and then QDA classifier was trained to discriminate between threat and safe conditions using the training dataset participants (23 subjects * 144 timepoints/subject = 3312 timepoints/condition). The continuous-valued classifier outputs, w(x), were calculated for each volume of data from the remaining held-out participant in the test dataset. These outputs were averaged across the timepoints within each threat or safe block, resulting in an average decision value for each block of the held-out participant (sixteen threat and sixteen safe blocks). This process was iterated so each participant served as the held-out participant to have their classification accuracy measured.
2.6.2 Estimating classifier chance performance
To determine if the classifier was operating above chance accuracy, a permutation test was used to simulate the null distribution of accuracy scores. We expected that the null distribution would be centered at 50% given the goal to classify between two equiprobable categories. Nevertheless, we simulated the null distribution to estimate the variability in “chance” performance and to determine whether the observed classification accuracy was robustly greater than chance performance. On each iteration of the permutation test, the threat and safe labels were randomly reassigned within each participant, and the cross-validated accuracy was recalculated. This was repeated 10,000 times to create a distribution of classifier accuracy scores expected under the assumption that threat and safe blocks are exchangeable.
2.6.3 Extracting discriminant vectors
If the classifier successfully discriminated the safe and threat conditions, diagnostic feature vectors were extracted to explain which patterns of functional connectivity the classifier used to make the discrimination. The classifier was trained using data from all participants, and two diagnostic feature vectors were extracted from the classifier, dThreat and dsafe. These vectors are each size nvoxelsby-1 with positive and negative values placed on each voxel to describe the patterns of functional connectivity diagnostic of each condition.
A bootstrapping procedure, analogous to that used by Woo et al (2014), was used to estimate the confidence intervals for each of the dimensions (i.e., voxels) in dThreat and dsafe to determine which ones were reliably greater-than or less-than zero. The bootstrapping procedure draws a random sample (with replacement) from the set of twenty-four participants in our sample, trains the QDA classifier to discriminate threat and safe blocks, and extracts two new diagnostic feature vectors. By repeating this 10,000 times, we can estimate the variability of each value in dThreat and dsafe as the standard deviation in feature scores across bootstrap iterations. Voxels were considered reliably positive/negative if their mean value across bootstrap iterations was at least 3.09 standard deviations away from zero (e.g., corresponding to uncorrected p < 0.001).
3. Results
3.1 Feature selection
Stimulus time series were pre-processed with PCA to derive a “de-noised” time series that emphasized patterns of voxel co-activation that could be identified within each participant (see Section 2.5). The analysis indicated that each participant should retain between 14 – 21 principal components (M = 17.8). The second-stage of feature selection identified which of the first-stage patterns were consistent across participants. Twenty-two group-level principal components were retained for each iteration of the cross-validation analysis. Each participant’s data was “de-noised” using the 22 group-level patterns identified on each iteration, and these data (with the same dimensionality as the initial time series) were then used as the input of the classification procedure, outlined next.
3.2 Classifier performance
We used quadratic discriminant analysis (QDA) to investigate the functional connectivity structure of the data under the safe and threat conditions. To evaluate classifier performance, we employed a leave-one-participant-out cross-validation procedure. The classification accuracy achieved by QDA was 62.9% correct. To assess the robustness of this value, we determined the null distribution for classifier performance by randomly flipping the condition labels (Section 2.6.2). According to this null distribution, the mean (i.e., “chance”) performance was 50.0% and classification accuracy exceeding 55.2% was very unlikely (p < 0.01). The probability of the actual classification accuracy observed (63% correct) or higher was very small (p < 1e-4), indicating that QDA classification reliably identified safe and threat conditions based on inter-voxel connectivity patterns.
3.3.1 Diagnostic feature vectors for threat
To better understand the patterns of functional connectivity that contributed to classification, we extracted the diagnostic feature vectors for the safe condition (dsafe) and the threat condition (dthreat) (see Section 2.6.3). These nvoxels-by-1 vectors contained positive or negative values at each voxel that described the pattern of functional connectivity for each condition. Voxels with the same sign in the diagnostic feature vector become more connected with one another (i.e., the voxels with positive values connect more strongly with other positive-valued voxels; voxels with negative values connect more strongly with other negative-valued voxels). Voxels with opposite signs “disconnect” with one another, that is, voxels with positive values connect less strongly from voxels with negative values).
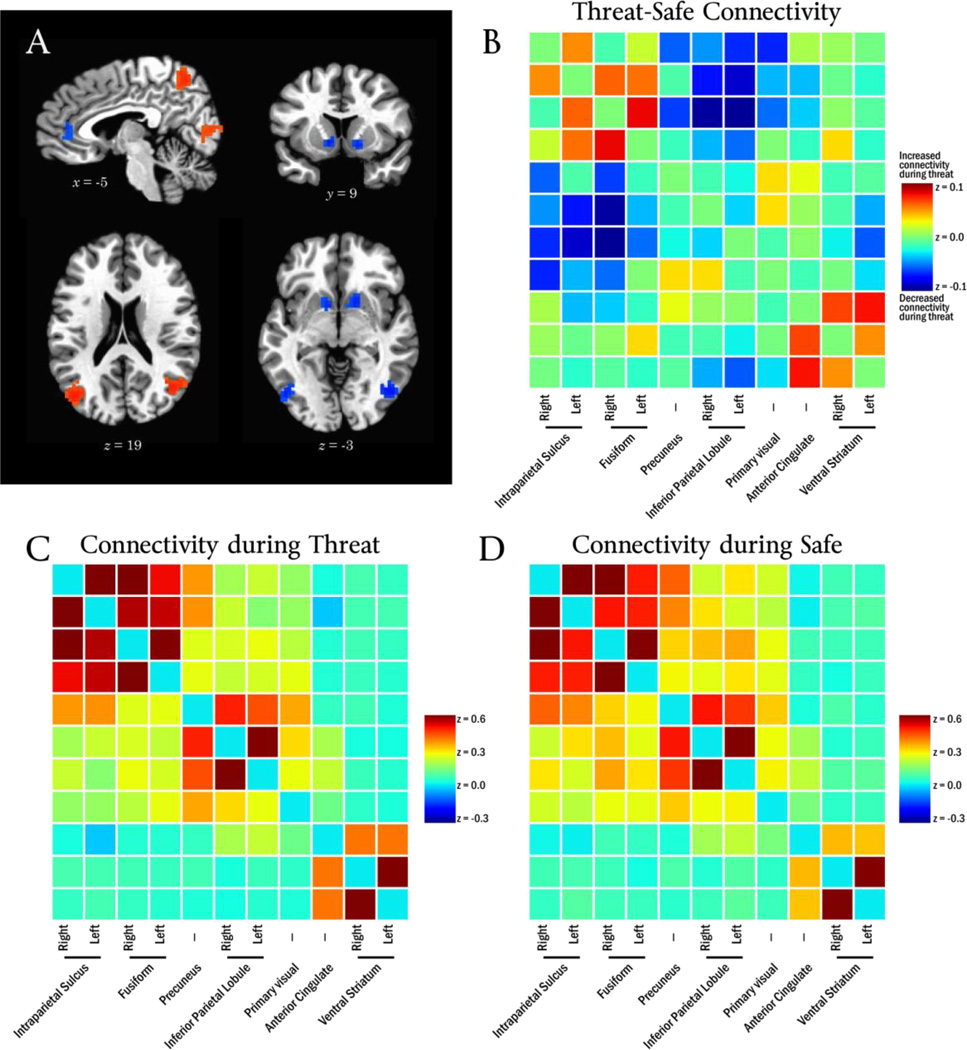
The diagnostic feature vector for the threat condition, dThreat, described a pattern of connectivity that occurred during threat conditions which involved multiple brain regions (Figure 4A; Table 1), including precuneus, bilateral inferior parietal lobule, bilateral intraparietal sulcus, bilateral fusiform gyrus, anterior cingulate cortex, and ventral parts of the striatum.
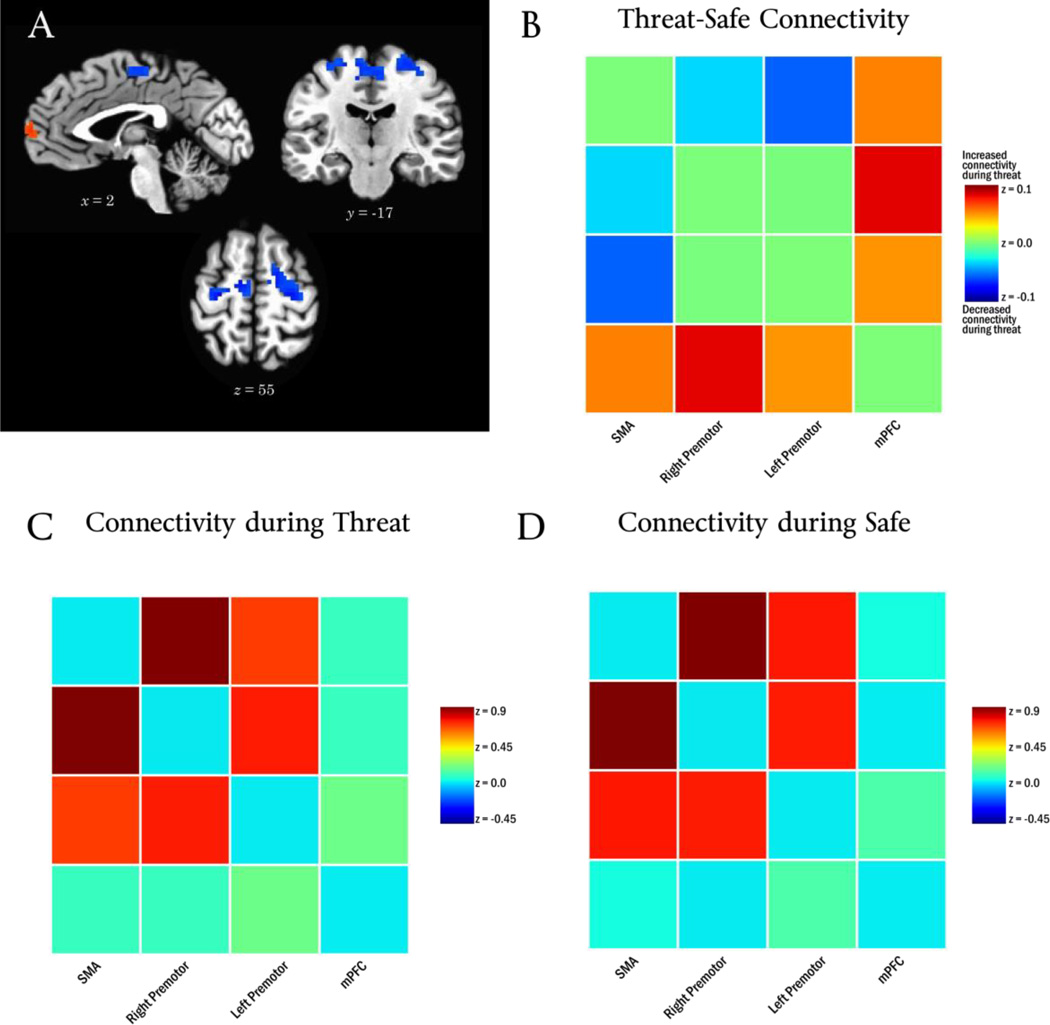
Figure 4.
Panel A depicts the diagnostic feature vector for threat. Panel B depicts the observed Threat-Safe connectivity change between every pair of regions in the diagnostic feature vector. Panels C and D depict connectivity during Threat and Safe conditions, respectively.
Table 1.
Regions identified on diagnostic feature vector from threat.
| Region | Hemisphere | Cluster extent (voxels) |
Center of Mass Coordinates |
z-score at Center of Mass |
||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Intraparietal Sulcus | L | 34 | 28 | −63 | 52 | −3.88 |
| R | 254 | −29 | −63 | 46 | −4.57 | |
| Fusiform Gyrus | L | 41 | 42 | −67 | −5 | −3.63 |
| R | 112 | −45 | −62 | −7 | −3.63 | |
| Anterior Cingulate | - | 29 | −2 | 40 | 8 | −3.55 |
| Ventral Striatum | L | 22 | 10 | 8 | −1 | −3.84 |
| R | 28 | −14 | 9 | −3 | −3.79 | |
| Primary Visual Cortex | - | 307 | −5 | −64 | 8 | |
| 15 | −56 | 7 | 3.53 | |||
| −19 | −55 | 8 | 3.52 | |||
| Precuneus | - | 2 | −58 | 50 | 3.97 | |
| Inferior Parietal Lobule | L | 66 | 39 | −70 | 23 | 4.25 |
| R | 65 | −44 | −63 | 22 | 3.96 | |
To help illustrate the pattern of threat-evoked connectivity change depicted by the threat diagnostic feature vector, dThreat, the change in functional connectivity was explicitly calculated between every pair of regions in Table 1. The average timeseries was extracted from each of the 11 regions of interest, and temporal correlations were calculated between every pair of regions for each participant and condition. Correlations were Fisher-transformed, averaged across participants, and the Threat-Safe connectivity difference determined (Figure 4B).
The effect of threat on connectivity among these regions can be summarized by three general changes: a) regions often associated with the “task-negative network” (i.e., precuneus, bilateral inferior parietal lobule) become “disconnected” from the other regions (that is, threat decreases connectivity between them); b) connectivity increased among regions often associated with the “executive network” (i.e., bilateral intraparietal sulcus, bilateral fusiform gyrus); and c) connectivity increased between the cingulate and striatum regions.
3.3.2 Diagnostic feature vectors for safety
The bootstrapping analysis indicated that the diagnostic feature vector for the safe condition, dsafe, contained four regions reliably different from zero (Table 2; Figure 5A). In particular, the threat period changed the pattern of connectivity for bilateral premotor cortex, such that it “disconnected” from the supplementary motor area (SMA).
Table 2.
Regions identified on diagnostic feature vector from safe.
| Region | Hemisphere | Cluster extent (voxels) |
Center of Mass Coordinates |
z-score at Center of Mass |
||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| Supplemental Motor Area | - | 120 | 10 | −13 | 56 | −3.52 |
| Premotor cortex | L | 25 | 27 | −19 | 55 | −3.81 |
| R | 117 | −23 | −12 | 57 | −3.30 | |
| Medial Prefrontal Cortex | - | 62 | 9 | 62 | 17 | 3.25 |
Figure 5.
Panel A depicts the diagnostic feature vector for safety. Panel B depicts the observed Threat-Safe connectivity change between every pair of regions in the diagnostic feature vector. Panels C and D depict connectivity during Threat and Safe conditions
4. Discussion
The present study used Quadratic Discriminant Analysis to discover changes to whole-brain functional connectivity during periods of sustained threat. The analysis identified patterns of functional connectivity that a classifier could use to predict whether individuals were currently in a safe or threat condition. To identify whether an individual was in a safe or threat period, the classifier used a pattern of connectivity between regions from the putative task-negative network (e.g., precuneus), the putative executive control network (e.g., bilateral IPS), cingulate, and striatum. The patterns of connectivity that were used to distinguish threat and safety can be summarized by four general changes: 1) threat increased connectivity among “executive” regions, 2) threat increased connectivity between cingulate and striatum regions, 3) threat decreased connectivity between both “executive” and cingulate/striatum regions and task-negative regions, and 4) threat decreased connectivity between the supplementary motor area and bilateral premotor regions.
Why is the methodology involving QDA developed here needed? In a nutshell, it can be used as a type of “localizer” method for detecting functional connectivity differences. Importantly, our method can be used to determine changes in functional connectivity associated with experimental conditions in a voxel-wise manner without a priori regions having to be defined. In addition, if a given set of regions is of priori interest, our method can be used to test for changes in functional connectivity between those regions.
Of particular interest was the involvement of territories in the vicinity of the ventral caudate including a putative accumbens region of interest. The ventral portions of the caudate, including the accumbens, have traditionally been associated with transient, reward-related processes (Haber and Knutson, 2009), but recent more research has suggests a potential role for the nucleus accumbens during aversive processing (Becerra et al., 2001; Cabib and Puglisi-Allegra, 1994, 1996; Delgado et al., 2008; Jensen et al., 2003; McMenamin et al., 2014; Oleson et al., 2012; Robinson et al., 2013; Salamone, 1994; Schoenbaum and Setlow, 2003). Critically, this accumbens region of interest may have gone unnoticed if common data analysis strategies were used to search for connectivity changes (such as using amygdala-based seed analysis).
If a more lenient statistical threshold were applied, the accumbens region extends medially/dorsally into a region consistent with the BNST. The BNST plays an important role during sustained threat states (Davis and Shi, 1999; Walker et al., 2003), and has strong anatomical connections to the accumbens (Alheid et al., 1998; Brog et al., 1993; Delfs et al., 1998; Dong et al., 2001; Dong and Swanson, 2004; Georges and Aston-Jones, 2001; Krüger et al., 2015). Because the spatial resolution employed in our study was relatively coarse (3 mm isotropic voxels), the signals arising from accumbens and the BNST cannot be dissociated and we cannot determine whether or not the functional connectivity changes we have identified are specific to either particular brain structure. In any case, the discovery of altered connectivity under threat demonstrates the importance of methods that include a data-driven component for identifying changes that may occur in unanticipated locations.
4.1 Alternative functional connectivity methods
Seed-based functional connectivity analyses are often used for testing changes in connectivity from a “seed” region to other voxels in the brain. The seed is generally selected based on a predefined anatomical structure, so the experimenter is limited to finding connectivity changes involving regions with which they have a priori theoretical interest. Unfortunately, this approach means that contributions from unanticipated (or hard to define anatomically) structures will be overlooked. For example, studies of negative affect often place their seed region in the amygdala (Hahn et al., 2011; Kim et al., 2010; Vytal et al., 2014), which may lead to an amygdala-centric view of aversive processing that overlooks contributions from other structures.
An alternative method for defining seed regions is to use localizer tasks to identify regions with activation differences across conditions, and then test how their connectivity patterns differ across conditions (Rissman et al., 2004). Unfortunately, this approach will overlook regions that exhibit changes to connectivity but not activation level – precisely the pattern expected for key information processing “hubs” that are activated by many tasks but reconfigure their pattern of connectivity to alter network structure (Cole et al., 2013; Pessoa, 2014; van den Heuvel and Sporns, 2011). Moreover, this approach requires a well-defined task with well-defined stimulus time courses to measure evoked responses, so it cannot be used on many datasets (e.g., resting-state data or free viewing of movies).
Closer to the methods developed here, another approach to measuring changes in functional connectivity between conditions would be to apply traditional multivariate statistical tests for differences in covariance, such as Wilks’ Lambda or Box's M (Nagarsenker and Pillai, 1973; Seber, 1984). Conventional multivariate statistics are often ill-suited to the dimensionality of functional neuroimaging problems (i.e., the socalled “large p, small n” problem), but the use of feature selection in the present report maps the problem into a sufficiently low-dimensional space, where these statistics could be used to test whether covariance (i.e., connectivity) differs between conditions. Unfortunately, these tests are difficult to apply even after dimensionality reduction because dependence between successive samples of fMRI data (i.e., the time series exhibit autocorrelation), which make it difficult to determine the correct degrees of freedom to be employed. By formulating the problem in terms of out-of-sample classification accuracy, we bypass distributional assumptions related to multivariate statistics and instead perform a direct test of the reliability of covariance differences that can easily accommodate a repeated-measures design, for instance.
Given the limitations of seed-based connectivity analyses and traditional multivariate statistics, it is promising to see the development of machine learning algorithms for detecting and interpreting large-scale connectivity changes (Craddock et al., 2009; Richiardi et al., 2011; Shirer et al., 2012; Zalesky et al., 2012). However, these multivariate methods still require some form of a priori region definition to make their implementation computationally tractable. In this respect, existing methods perform an initial data-reduction step that partitions the brain into regions of interest (usually on the order of 100 regions) using anatomical atlases or clustering based approaches. While valuable, unfortunately, this strategy will likely overlook smaller structures (e.g., the striatum cluster observed here). By contrast, the QDA method developed here showed that it can scale up to operate effectively on a voxelwise basis.
Acknowledgments
We would like to thank Mahshid Najafi for useful comments and feedback. Support for this work was provided in part by the National Institute of Mental Health (MH071589).
Appendix 1: Use of eigendecomposition to maximize/minimize the QDA decision function
For a real-valued square matrix, M, and vector, x, the Rayleigh quotient is defined as . Ostrowski (1959) shows that for a given M, the local extrema of the Rayleigh quotient occur when x equals an eigenvector of M. Moreover, the global maximum occurs when x is the eigenvector with the largest associated eigenvalue, and the global minimum when the x equals the eigenvector with the smallest associated eigenvalue. This property of the Rayleigh quotient can be used to find the extrema of the QDA decision function, w(x) by setting . This results in the numerator of the Rayleigh quotient being equal to the first term in w(x), and the denominator serves as a scaling factor that enforces the constraint that ‖x‖ = 1.
Appendix 2: Connection between diagnostic feature vectors and derivative-based voxel importance maps
Yourganov et al. (2014) developed a general framework for understanding which spatial activation patterns classifiers use to make category decisions. In this framework, given a particular classifier’s decision function w(x), a voxelwise an “importance map” is defined as the voxel-wise partial derivatives of the classifier’s decision function. This results in a map across voxels that can be used to illustrate how a change in activation at each voxel changes the classifier output – for example, a voxel with a positive value on the importance map means that increased activity in that voxel would push the classifier toward labelling a pattern “Category A”, and decreased activity in that voxel would push the classifier toward labelling a pattern “Category B”. Yourganov et al. report that the importance map for a QDA can be calculated by averaging the value of across all of the x values used in classifier training. Therefore, the final nvoxels-by-1 derivative-based importance map, dMap, can be rewritten as:
where μA,B is the grand mean across both conditions.
For the analyses carried out in the present paper, we know that μA = μB = 0, which results in dMap being a vector of all zeros. This is expected because without any systematic differences in the mean activation between conditions, a change in the activation level at any individual voxel would not affect the classifier’s output. However, a change in co-activation between pairs of voxels can convey useful information about the covariance structure that the QDA classifier uses for classification. To measure the effect of pairwise voxel co-activation on classifier output, we can calculate the nvoxels-by-nvoxels Hessian matrix of w(x), H. This matrix contains all second-order partial derivatives of w(x), calculated as:
The value of H(x) does not depend on x, so this matrix does not need to be averaged over the values x the classifier used for training. Analogous to how the ith entry of the derivative-based importance map tells us how a change in activation of the ith unit affects classification, the ijth entry of the matrix H tells us how an increase in connectivity between the ith and jth units affects classification.
Furthermore, we can use the Hessian matrix to find which nvoxels-by-1 pattern across voxels exhibits the co-activation pattern with the largest effect on classifier output. The scalar value xTHx indicates how much the input pattern x would alter the classifier output, so we can define the patterns which maximize and minimize the change in classifier output as:
Note that these Hessian-based importance maps are equivalent to the diagnostic feature vectors, dA and dB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no competing financial interests
Many applications use PCA to perform “dimensionality reduction” by identifying a small set of components that explain a large proportion of the overall variance in Xi, for example the nvoxels-by-k matrix of retained components. Dimensionality reduction can be used to transform the original nvoxels-dimensional dataset into a k-dimensional dataset with the matrix multiplication . The “denoised” matrix, X̃i, is simply the projection of that low-dimensional dataset back into the original high- dimensional space.
This framework for calculating the diagnostic feature vector is not unique to QDA or nonlinear classifiers. For example, a linear classifier uses the decision function w(x) = bTx to distinguish two conditions by placing a decision weight on each voxel (stored in the nvoxels-by-1 vector). The inner-product of the decision function can be rewritten as w(x) = ‖b‖‖x‖ cos α, where α is the angle between b and x. Subject to the contraint ‖x‖ = 1, w(x), is maximized when α = 0° and minimized when α = 180°. This means that the two diagnostic feature vectors for a linear classifier correspond to the pattern of decision weights across voxels given by .
References
- Alheid G, Beltramino C, De Olmos J, Forbes M, Swanson D, Heimer L. The neuronal organization of the supracapsular part of the stria terminalis in the rat: the dorsal component of the extended amygdala. Neuroscience. 1998;84:967–996. doi: 10.1016/s0306-4522(97)00560-5. [DOI] [PubMed] [Google Scholar]
- Alvarez RP, Chen G, Bodurka J, Kaplan R, Grillon C. Phasic and sustained fear in humans elicits distinct patterns of brain activity. Neuroimage. 2011;55:389–400. doi: 10.1016/j.neuroimage.2010.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra L, Breiter HC, Wise R, Gonzalez RG, Borsook D. Reward circuitry activation by noxious thermal stimuli. Neuron. 2001;32:927–946. doi: 10.1016/s0896-6273(01)00533-5. [DOI] [PubMed] [Google Scholar]
- Brog JS, Salyapongse A, Deutch AY, Zahm DS. The patterns of afferent innervation of the core and shell in the “Accumbens” part of the rat ventral striatum: Immunohistochemical detection of retrogradely transported fluoro-gold. Journal of Comparative Neurology. 1993;338:255–278. doi: 10.1002/cne.903380209. [DOI] [PubMed] [Google Scholar]
- Cabib S, Puglisi-Allegra S. Opposite responses of mesolimbic dopamine system to controllable and uncontrollable aversive experiences. J Neurosci. 1994;14:3333–3340. doi: 10.1523/JNEUROSCI.14-05-03333.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib S, Puglisi-Allegra S. Stress, depression and the mesolimbic dopamine system. Psychopharmacology (Berl) 1996;128:331–342. doi: 10.1007/s002130050142. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Human brain mapping. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nature Neuroscience. 2013;16:1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Craddock RC, Holtzheimer PE, Hu XP, Mayberg HS. Disease state prediction from resting state functional connectivity. Magnetic Resonance in Medicine. 2009;62:1619–1628. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M, Shi C. The extended amygdala: are the central nucleus of the amygdala and the bed nucleus of the stria terminalis differentially involved in fear versus anxiety? Ann N Y Acad Sci. 1999;877:281–291. doi: 10.1111/j.1749-6632.1999.tb09273.x. [DOI] [PubMed] [Google Scholar]
- Davis M, Walker DL, Miles L, Grillon C. Phasic vs sustained fear in rats and humans: role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology. 2010;35:105–135. doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfs JM, Zhu Y, Druhan JP, Aston-Jones GS. Origin of noradrenergic afferents to the shell subregion of the nucleus accumbens: anterograde and retrograde tract-tracing studies in the rat. Brain research. 1998;806:127–140. doi: 10.1016/s0006-8993(98)00672-6. [DOI] [PubMed] [Google Scholar]
- Delgado MR, Li J, Schiller D, Phelps EA. The role of the striatum in aversive learning and aversive prediction errors. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:3787–3800. doi: 10.1098/rstb.2008.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Petrovich GD, Watts AG, Swanson LW. Basic organization of projections from the oval and fusiform nuclei of the bed nuclei of the stria terminalis in adult rat brain. Journal of Comparative Neurology. 2001;436:430–455. doi: 10.1002/cne.1079. [DOI] [PubMed] [Google Scholar]
- Dong HW, Swanson LW. Organization of axonal projections from the anterolateral area of the bed nuclei of the stria terminalis. Journal of Comparative Neurology. 2004;468:277–298. doi: 10.1002/cne.10949. [DOI] [PubMed] [Google Scholar]
- Georges F, Aston-Jones G. Potent regulation of midbrain dopamine neurons by the bed nucleus of the stria terminalis. J Neurosci. 2001;21:RC160. doi: 10.1523/JNEUROSCI.21-16-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou-Karistianis N, Gray M, Domínguez DJ, Dymowski A, Bohanna I, Johnston L, Churchyard A, Chua P, Stout J, Egan G. Automated differentiation of pre-diagnosis Huntington's disease from healthy control individuals based on quadratic discriminant analysis of the basal ganglia: the IMAGE-HD study. Neurobiology of disease. 2013;51:82–92. doi: 10.1016/j.nbd.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. Journal of Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- Haber SN, Knutson B. The Reward Circuit: Linking Primate Anatomy and Human Imaging. Neuropsychopharmacology. 2009;35:4–26. doi: 10.1038/npp.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Stein P, Windischberger C, Weissenbacher A, Spindelegger C, Moser E, Kasper S, Lanzenberger R. Reduced resting-state functional connectivity between amygdala and orbitofrontal cortex in social anxiety disorder. Neuroimage. 2011;56:881–889. doi: 10.1016/j.neuroimage.2011.02.064. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J, Hastie T, Friedman J, Tibshirani R. The elements of statistical learning. Springer; 2009. [Google Scholar]
- Hermans EJ, Henckens MJ, Joëls M, Fernández G. Dynamic adaptation of large-scale brain networks in response to acute stressors. Trends in neurosciences. 2014;37:304–314. doi: 10.1016/j.tins.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, James LF. Gibbs sampling methods for stick-breaking priors. Journal of the American Statistical Association. 2001;96 [Google Scholar]
- Jackson DA. Stopping rules in principal components analysis: a comparison of heuristical and statistical approaches. Ecology. 1993:2204–2214. [Google Scholar]
- Jensen J, McIntosh AR, Crawley AP, Mikulis DJ, Remington G, Kapur S. Direct activation of the ventral striatum in anticipation of aversive stimuli. Neuron. 2003;40:1251–1257. doi: 10.1016/s0896-6273(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Gee DG, Loucks RA, Davis FC, Whalen PJ. Anxiety Dissociates Dorsal and Ventral Medial Prefrontal Cortex Functional Connectivity with the Amygdala at Rest. Cerebral cortex. 2010;21:1667–1673. doi: 10.1093/cercor/bhq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JR, Jackson DA. Variable selection in large environmental data sets using principal components analysis. Environmetrics. 1999;10:67–77. [Google Scholar]
- Krüger O, Shiozawa T, Kreifelts B, Scheffler K, Ethofer T. Three distinct fiber pathways of the bed nucleus of the stria terminalis to the amygdala and prefrontal cortex. Cortex; 2015. [DOI] [PubMed] [Google Scholar]
- McMenamin BW, Langeslag SJ, Sirbu M, Padmala S, Pessoa L. Network Organization Unfolds over Time during Periods of Anxious Anticipation. The Journal of Neuroscience. 2014;34:11261–11273. doi: 10.1523/JNEUROSCI.1579-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaki M, Kim Y, Bandettini PA, Kriegeskorte N. Comparison of multivariate classifiers and response normalizations for pattern-information fMRI. Neuroimage. 2010;53:103–118. doi: 10.1016/j.neuroimage.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobbs D, Yu R, Rowe JB, Eich H, FeldmanHall O, Dalgleish T. Neural activity associated with monitoring the oscillating threat value of a tarantula. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20582–20586. doi: 10.1073/pnas.1009076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarsenker B, Pillai K. Distribution of the likelihood ratio criterion for testing a hypothesis specifying a covariance matrix. Biometrika. 1973;60:359–364. [Google Scholar]
- Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: multivoxel pattern analysis of fMRI data. Trends in cognitive sciences. 2006;10:424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Oleson EB, Gentry RN, Chioma VC, Cheer JF. Subsecond dopamine release in the nucleus accumbens predicts conditioned punishment and its successful avoidance. J Neurosci. 2012;32:14804–14808. doi: 10.1523/JNEUROSCI.3087-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski A. On the convergence of the Rayleigh quotient iteration for the computation of the characteristic roots and vectors. III. Archive for Rational Mechanics and Analysis. 1959;3:325–340. [Google Scholar]
- Pessoa L. Understanding brain networks and brain organization. 2014 doi: 10.1016/j.plrev.2014.03.005. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Eryilmaz H, Schwartz S, Vuilleumier P, Van De Ville D. Decoding brain states from fMRI connectivity graphs. Neuroimage. 2011;56:616–626. doi: 10.1016/j.neuroimage.2010.05.081. [DOI] [PubMed] [Google Scholar]
- Rissman J, Gazzaley A, D'Esposito M. Measuring functional connectivity during distinct stages of a cognitive task. Neuroimage. 2004;23:752–763. doi: 10.1016/j.neuroimage.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Robinson OJ, Overstreet C, Charney DR, Vytal K, Grillon C. Stress increases aversive prediction error signal in the ventral striatum. Proc Natl Acad Sci U S A. 2013;110:4129–4133. doi: 10.1073/pnas.1213923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamone JD. The involvement of nucleus accumbens dopamine in appetitive and aversive motivation. Behavioural brain research. 1994;61:117–133. doi: 10.1016/0166-4328(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Schmah T, Yourganov G, Zemel RS, Hinton GE, Small SL, Strother SC. Comparing classification methods for longitudinal fMRI studies. Neural computation. 2010;22:2729–2762. doi: 10.1162/NECO_a_00024. [DOI] [PubMed] [Google Scholar]
- Schoenbaum G, Setlow B. Lesions of nucleus accumbens disrupt learning about aversive outcomes. Journal of Neuroscience. 2003;23:9833–9841. doi: 10.1523/JNEUROSCI.23-30-09833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seber G. Multivariate observations. New York: John Wiley & Sons; 1984. [Google Scholar]
- Shirer W, Ryali S, Rykhlevskaia E, Menon V, Greicius M. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cerebral cortex. 2012;22:158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville LH, Whalen PJ, Kelley WM. Human bed nucleus of the stria terminalis indexes hypervigilant threat monitoring. Biological psychiatry. 2010;68:416–424. doi: 10.1016/j.biopsych.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley ML, Moussa MN, Paolini BM, Lyday RG, Burdette JH, Laurienti PJ. Defining nodes in complex brain networks. Frontiers in computational neuroscience. 2013;7 doi: 10.3389/fncom.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. Journal of Neuroscience. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytal KE, Overstreet C, Charney DR, Robinson OJ, Grillon C. Sustained anxiety increases amygdala–dorsomedial prefrontal coupling: a mechanism for maintaining an anxious state in healthy adults. Journal of psychiatry & neuroscience: JPN. 2014;39:130145. doi: 10.1503/jpn.130145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Toufexis DJ, Davis M. Role of the bed nucleus of the stria terminalis versus the amygdala in fear, stress, and anxiety. Eur J Pharmacol. 2003;463:199–216. doi: 10.1016/s0014-2999(03)01282-2. [DOI] [PubMed] [Google Scholar]
- Woo C-W, Koban L, Kross E, Lindquist MA, Banich MT, Ruzic L, Andrews-Hanna JR, Wager TD. Separate neural representations for physical pain and social rejection. Nature communications. 2014;5 doi: 10.1038/ncomms6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yourganov G, Schmah T, Churchill NW, Berman MG, Grady CL, Strother SC. Pattern classification of fMRI data: Applications for analysis of spatially distributed cortical networks. Neuroimage. 2014;96:117–132. doi: 10.1016/j.neuroimage.2014.03.074. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Cocchi L, Fornito A, Murray MM, Bullmore E. Connectivity differences in brain networks. Neuroimage. 2012;60:1055–1062. doi: 10.1016/j.neuroimage.2012.01.068. [DOI] [PubMed] [Google Scholar]