Abstract
TaGW2 is an orthologue of rice gene OsGW2, which encodes E3 RING ubiquitin ligase and controls the grain size in rice. In wheat, three copies of TaGW2 have been identified and mapped on wheat homoeologous group 6 viz. TaGW2-6A, TaGW2-6B and TaGW2-6D. In the present study, using as many as 207 Indian wheat genotypes, we identified four SNPs including two novel SNPs (SNP-988 and SNP-494) in the promoter sequence of TaGW2-6A. All the four SNPs were G/A or A/G substitutions (transitions). Out of the four SNPs, SNP-494 was causal, since it was found associated with grain weight. The mean TGW (41.1 g) of genotypes with the allele SNP-494_A was significantly higher than mean TGW (38.6 g) of genotypes with the allele SNP-494_G. SNP-494 also regulates the expression of TaGW2-6A so that the wheat genotypes with SNP-494_G have higher expression and lower TGW and the genotypes with SNP-494_A have lower expression but higher TGW. Besides, SNP-494 was also found associated with grain length-width ratio, awn length, spike length, grain protein content, peduncle length and plant height. This suggested that gene TaGW2-6A not only controls grain size, but also controls other agronomic traits. In the promoter region, SNP-494 was present in ‘CGCG’ motif that plays an important role in Ca2+/calmodulin mediated regulation of genes. A user-friendly CAPS marker was also developed to identify the desirable allele of causal SNP (SNP-494) for use in marker-assisted selection for improvement of grain weight in wheat. Using four SNPs, five haplotypes were identified; of these, Hap_5 (G_A_G_A) was found to be a desirable haplotype having significantly higher grain weight (41.13g) relative to other four haplotypes (36.33-39.16 g).
Introduction
Improvement in average grain yield in wheat has attracted the attention of wheat breeders all over the world, and several initiatives in this direction have recently been taken, both at the national and international levels. These include Wheat Inititaive (www.wheatinitiative.org), Wheat Yield Network (WYN; www.bbsrc.ac.uk/web/FILES/Resources/wheat-yield-network.pdf) and its International Wheat Yield Partnership (IWYP; iwyp.org) program and 20:20 program of the UK (www.rothamsted.ac.uk/ our-science/2020-wheat). One of the most important traits contributing to grain yield is grain size (thousand grain weight = TGW), which is also related to higher flour recovery [1–2] and milling quality of grain. Keeping this in view, grain size in wheat has been one of the targets for selection both during domestication and modern wheat breeding [3–4].
Grain weight is a polygenic trait and is controlled by a large number of genes/QTL that are distributed on all wheat chromosomes [2, 5–21]. Among these genes, three genes that are orthologous to rice gene OsGW2 were earlier identified and mapped on three chromosomes of the homoeologous group 6; these were described as TaGW2-6A, TaGW2-6B, TaGW2-6D [22]. Two SNPs (-593A/G and -739G/A) were also earlier reported in the promoter region of the gene TaGW2-6A. One of the two SNPs (-593A/G) in the promoter region of TaGW2-6A, and an insertion of a single T-base in the eighth exon of this gene (detected in a large kernel wheat variety Lankaodali) were shown to be associated with grain size [22–23]. Although, a negative correlation between expression of TaGW2 and grain size was observed in two earlier studies [22–23], a positive correlation was suggested in another study, where knocking out of the gene using RNA interference (RNAi) involving reduction in TaGW2 transcript levels, led to reduction in endosperm cell number associated with reduction in grain size [24]. Further studies may be needed to resolve this apparent contradiction in the results. Biochemical and molecular analyses revealed that TaGW2-6A encodes a functional E3 RING ubiquitin ligase with nucleocytoplasmic subcellular partitioning.
In the present study, we analysed sequence polymorphism in the promoter region of TaGW2-6A in a collection of 207 Indian wheat genotypes. Interestingly, we found two novel SNPs (one SNP present in CGCG motif) in the promoter region along with two other SNPs that were also reported by Su et al. [22]. A study of association of these SNPs and that of the corresponding haplotypes with TGW in Indian wheat genotypes allowed identification of a novel causal SNP and a causal haplotype. The causal SNP also modulated the expression of the gene TaGW2 in developing grains so that the negative regulation of the gene expression was associated with higher grain weight. A functional marker (cleaved amplified polymorphic sequence—CAPS) was also developed for identification of individual alleles of causal SNP for use in wheat breeding programs aimed at grain weight improvement.
Materials and Methods
Plant material and recording of data on grain size and other agronomic traits
The plant material used in this study comprised as many as 207 Indian wheat genotypes, released during 1910–2006 for commercial cultivation in different agro-climatic regions of India. The seed of the above genotypes was procured from the Indian Institute of Wheat and Barley Research (IIWBR), Karnal (India). The data on TGW, grain width, grain length, length-width ratio, and five other agronomic traits recorded on the above 207 Indian wheat genotypes were used in the present study.
Each metric observation was based on an average of 10 randomly selected plants. The observations and data on different traits were recorded in the following manner: (i) 1000-grain weight (TGW); weight of 1000 grains expressed in grams; (ii) grain-length; recorded in milimeter using software SmartGrain, (iii) grain-width; recorded in milimeter using software SmartGrain (iv) grain length-width ratio; recorded using software SmartGrain; (v) awn length; measured in cm from middle one-third region of the ear; (vi) spike length; measured in cm from the base of the ear to the tip of the apical spikelet (excluding awns); (vii) grain protein content: estimated using Food and Feed Analyzer NIR 1255; (viii) peduncle length: measured in cm from base (collar) of the spike to the first node; and (ix) plant height: measured in centimetre (cm) from base of the plant to the tip of the spike (excluding awns) of the longest tiller. Data for grain length, width and length-width ratio were recorded during present study, and those for the remaining traits were procured from IIWBR, Karnal [25]; the data at IIWBR was generated in evaluation trials, conducted for DUS traits, during three consecutive years (2003–04 to 2005–06) at Karnal, India.
DNA isolation and PCR amplification
For each genotype, genomic DNA was extracted from the leaves of one month-old plants using a modified CTAB method [26]. Isolated DNA was purified by RNase A treatment and phenol: chloroform: isoamyl alcohol precipitation following Sambrook et al. [27]. The quality and quantity of DNA were checked on agarose gel through a comparison with known quantities of λ Hind III DNA marker. The gene-primers that were specific for the sub-genome A (Hap-6A-P1_For and Hap-6A-P1_Rev) reported earlier were used to amplify the promoter region of gene TaGW2-6A [22]. PCR reactions were performed using a total volumes of 15 μl, with 3 pmol of each primer, 120 μM of each dNTP, 80 ng genomic DNA, 0.75 unit Jumpstart Accu Taq La DNA polymerase and 2 μl 10× buffer (Catalog number B0174), Sigma, USA. The PCR was carried out using Veriti Thermal Cycler, Applied Biosystem using the following profile with a ramp rate of 3.35°C/second: initial denaturation at 95°C for 3 min, followed by 32 cycles at 95°C for 30s, annealing at 58°C for 30s, and extension at 72°C 30s, with a final extension at 72°C for 10 min. PCR products were resolved by electrophoresis on 2% agarose gels.
Sequencing of PCR product
For sequencing of PCR products, approximately 500 ng of each PCR products obtained above were used and cleaned using the following reaction. 1 U Shrimp Alkaline Phospatase (Fermentas) and 10 U of Exonuclease I (Fermentas) in a final volume of 10 μl at 37°C for 15 min followed by enzyme inactivation at 85°C for 15 min.
One μl (~50 ng) of each of the above cleaned samples was directly used as template for sequencing. The reaction was set-up using 10 pmole primer and 0.5 μl Big-dye chemistry v3.1 (ABI) in a final volume of 10 μl. The sequence of cycles was set-up with the following profile at a ramp rate of 3.35°C/second: denaturation at 96°C for 10s, primer annealing at 50°C for 5 s and extension at 60°C for 4 min for a total of 30 cycles. Gene Amp PCR system 9700 (Applied Biosystem) was used for PCR amplification. The fluorescently labelled PCR products were analysed using an ABI 3730xl sequencer.
Sequence alignment and SNP detection
Sequence alignment and SNP detection were performed using software CLC genomics/DNA workbench (http://www.clcbio.com). In order to identify quality SNPs, specific criteria based on the read depth, minor allele frequency and the quality of flanking regions were used. Each high quality SNP was identified in a segment of appropriate size, where all bases matched except the SNP identified, so that a 15-bp flanking region on each side of an identified SNP had no extra SNPs or indels [28, 29]. Only SNPs with minor allele frequency of no less than 5% in the population were declared as quality SNPs.
Marker-trait association
Descriptive statistics for all nine traits including TGW were obtained using SPSS. Association analysis was conducted using General Linear Model (GLM) with 1000 permutations with the help of software TASSEL (http://www.maizegenetics.net). Significance of the association was determined by p-value (<0.05). Mann-Witney (non-parametric test) was applied to test the significance of difference for TGW between the two allele classes of each SNP locus using SPSS. Analysis of variance (ANOVA) was conducted by PROC GLM in the Statistical Analysis System (SAS Institute, 1997) to test the significant differences of TGW among different haplotypes.
RNA extraction and qRT PCR
Total RNA was extracted from immature seed (15 DAP = days after pollination) from 10 genotypes (5 genotypes with SNP-494_A and 5 genotypes with SNP-494_G) using Sigma Aldrich’s Spectrum Plant Total RNA kit. Quantitative Real-time PCR (qRT-PCR) was used to analyze the transcript level of TaGW2-6A (primer sequences: TaGW2-6A_For: AAGCATGGGTGCTGCGGAA, TaGW2-6A_Rev: GTCAGCAAAAGGCAACGGTA [30]). qRT-PCR was performed with Thermo Scientific’s DyNAmo Flash SYBR Green qPCR kit, using Applied Biosystem’s 7500 Fast RT-PCR System according to the manufacturer’s instructions. qRT-PCR reaction was set up with the following thermal profile using a ramp at the rate of 3.5°C/second: 95°C for 15 min (initial denaturation), followed by 40 cycles with 95°C for 10 s (denaturation) and 60°C for 30 s (annealing/extension). The relative transcript level of TaGW2-6A was calculated using 2− ΔΔCT method [31]. TaActine gene (primer sequences TaActine_For: CACTGGAATGGTCAAGGCTG, TaActine_Rev: CTCCATGTC ATCCCAGTTG) was used as internal control and HI 1500 genotype (with minimum expression level) was used as a reference. For expression analysis, two biological replications for each genotype were performed and three technical replications were analyzed for each biological replication.
Motif search in amplified promoter (regulatory) sequence
For motif search, promoter region involving ~1-kb segment upstream of the TaGW2-6A gene was examined using PLACE database (http://www.dna.affrc.go.jp/htdocs/PLACE/) [32].
Development of functional marker
Phenotyping and genotypic data were used to identify the causal SNP (at -494bp). The causal SNP was then converted into a CAPS (cleaved amplified polymorphism sequence) marker. Restriction site was identified using dCAPS Finder 2.0 program. Promoter region of TaGW2-6A was first amplified using Hap-6A-P1_For and Hap-6A-P1_Rev, followed by a second PCR (primer pair: Hap-6A-P2_For and Hap-6A-P2_Rev [22]) to get smaller specific fragment. The amplified product (1μg DNA) of second PCR was then digested with FauI (New England Biolabs) using 1 unit enzyme at 55°C for one h. The fragments resulting due to digestion were separated on 2% agarose gel.
Results
Variation for TGW and eight other agronomic traits in 207 Indian wheat genotypes
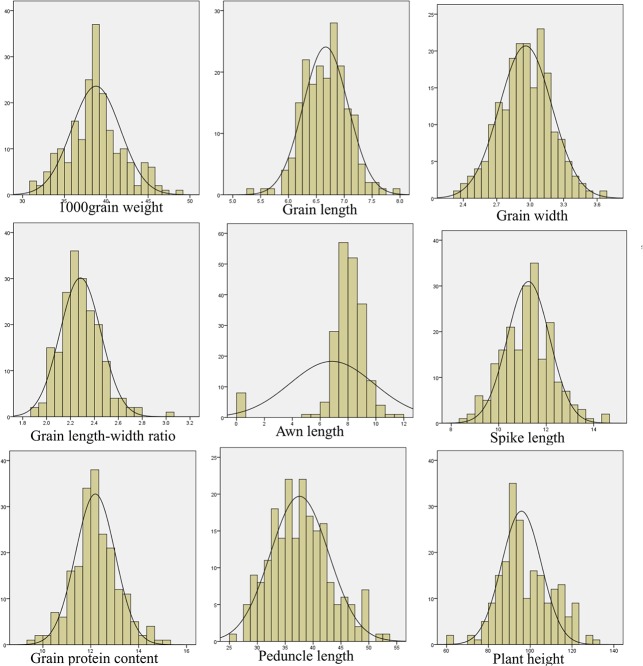
TGW in 207 Indian wheat genotypes ranged from 31.1 to 48.5 g with a mean of 38.7 g. The data gave a good fit to normal distribution with a standard deviation of 3.26 and coefficient of variation (CV) of 8.41%. Descriptive statistics for the remaining eight agronomic traits are presented in Table 1. Frequency distributions of genotypes with different class intervals of nine agronomic traits including TGW is presented in Fig 1.
Table 1. Descriptive statistics for 1000 grain weight and other agronomic traits.
| Traits | Minimum | Maximum | Mean | Std. Deviation | CV (%) |
|---|---|---|---|---|---|
| 1000 grain weight (g) | 31.10 | 48.50 | 38.75 | 3.26 | 8.41 |
| Grain length (mm) | 5.33 | 7.97 | 6.67 | 0.40 | 6.05 |
| Grain width (mm) | 2.32 | 3.67 | 2.96 | 0.23 | 7.94 |
| Length-width ratio | 1.91 | 3.03 | 2.28 | 0.17 | 7.54 |
| Awn length (cm) | 0.00 | 11.59 | 7.83 | 1.82 | 23.19 |
| Spike length (cm) | 8.64 | 14.55 | 11.21 | 1.06 | 9.48 |
| Grain protein content (%) | 9.57 | 15.32 | 12.19 | 0.95 | 7.81 |
| Peduncle length (cm) | 25.93 | 53.10 | 37.69 | 5.35 | 14.20 |
| Plant height (cm) | 62.00 | 131.00 | 97.87 | 12.77 | 13.05 |
Fig 1. Frequency distribution curve for nine agronomic traits including 1000 grain weight in 207 Indian wheat genotypes used in association mapping study.
Identification of two novel SNPs in the promoter region of TaGW2-6A in Indian wheat genotypes
PCR amplification (Fig 2) and sequencing of the amplified promoter region of gene TaGW2-6A in 207 Indian wheat genotypes allowed identification of four SNPs, at positions -988bp, -739bp, -593bp and -494bp (S1 Fig) with minor allele frequencies of 7.2%, 15.0%, 14.0% and 6.3%, respectively. The details of the four SNPs are presented in Table 2. All the four SNPs were biallelic and all were transitions with G-A/A-G substitutions. Alignment of the amplified sequences of 207 Indian wheat genotypes and those reported by Su et al. [22] was done using multiple sequence alignment online software ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2). The sequence alignment revealed that out of the four SNPs, two SNPs (at -988bp and at -494bp) were novel and were reported for the first time, while the remaining two SNPs at -739bp and -593bp positions were also reported earlier by Su et al. [22].
Fig 2. Representative gel picture of PCR amplification of promoter region of TaGW2-6A gene in 15 Indian wheat genotypes.
Table 2. Summary of four SNPs (identified by sequence alignment) with their position, variation, frequency and count.
| Consensus position | Consensus | Allele variations | Frequencies | Counts |
|---|---|---|---|---|
| -988 | G | G/A | 92.8/7.2 | 192/15 |
| -739 | A | A/G | 85.0/15.0 | 175/31 |
| -593 | G | G/A | 86.0/14.0 | 178/29 |
| -494 | G | G/A | 93.7/6.3 | 194/13 |
Marker-trait association using individual SNP
Association mapping using general linear model (GLM) and Mann-Whitney U test revealed that out of the four SNPs identified during the present study, only one SNP (G/A at -494bp) was associated with TGW (Table 3); 13 genotypes with SNP allele having A at position 494 exhibited significantly higher TGW (41.1 g as against a mean TGW of 38.6 g in genotypes with SNP allele having G at position 494). None of the two SNPs earlier reported by Su et al. [22] showed association with TGW. SNP-494 was also found to be associated with grain length-width ratio and five other agronomic traits (awn length, spike length, grain protein content, peduncle length and plant height; Table 4), but not with grain length and grain width.
Table 3. Summary of marker-trait association using single SNP for 1000 grain weight.
Significantly associated SNP is marked with *.
Table 4. Marker-trait association of SNP-494 with 1000 grain weight and other eight agronomic traits.
| Traits | General Linear Model | Mann-Whitney U test | |||
|---|---|---|---|---|---|
| p-value | R2-value | G allele effect | A allele effect | p-value | |
| 1000 grain weight | 0.006** | 0.036 | -2.54 | 0.00 | 0.001** |
| Grain length | 0.306 | - | - | - | 0.328 |
| Grain width | 0.208 | - | - | - | 0.218 |
| Length-width ratio | 0.026* | 0.026 | 0.12 | 0.00 | 0.016* |
| Awn length | 0.048* | 0.019 | -1.03 | 0.00 | 0.003** |
| Spike length | 0.005** | 0.037 | 0.84 | 0.00 | 0.007** |
| Grain protein content (%) | 0.036* | 0.003 | -0.59 | 0.00 | 0.021* |
| Peduncle length | 0.000** | 0.082 | -6.31 | 0.00 | 0.000** |
| Plant height | 0.000** | 0.064 | -13.31 | 0.00 | 0.000** |
*, ** significant at 0.05, and 0.01 levels, respectively.
Haplotype analysis and their association with TGW
Using four SNPs, following five haplotypes could be constituted [Fig 3: Hap1 (G_G_G_G), Hap2 (A_G_A_G), Hap3 (G_G_A_G), Hap4 (G_A_G_G_) and Hap5 (G_A_G_A)]. Out of 207 genotypes, Hap1 occured in 2 (0.97%) genotypes with a mean TGW of 36.33 g, Hap2 occurred in 15 (7.25%) genotypes with a mean TGW of 39.16 g, Hap3 occurred in 14 (6.76%) genotypes with a mean TGW of 38.7 g, Hap 4 occurred in 163 (78.74%) genotypes with a mean TGW of 38.7 g and Hap 5 occurred in 13 (6.28%) genotypes with a mean TGW of 41.13 g.
Fig 3. Five haplotypes with single nucleotide polymorphisms (SNPs) in the promoter regions of TaGW2-6A.

Frequency of each haplotype is given in parentheses. SNPs are highlighted with yellow (G allele) and red (A allele) colours. CGCG motif is shown in a box. Significant difference of mean TGW (g) is represented by bars. Haplotypes covered by a single bar represent no significant difference for TGW and vice-versa.
Analysis of variance (ANOVA) showed significant difference for TGW among 5 haplotypes (p < 0.01; Table 5), and also between Hap5 and the remaining four haplotypes (Hap5 vs others). A comparisons between pairs also showed that the mean TGW of Hap5 was significantly higher than the mean TGW of Hap1, Hap3 and Hap4; however no significant difference for TGW was observed between Hap5 and Hap2 (Fig 3).
Table 5. Analysis of variance for TGW using five different haplotypes.
| Source | DF* | Mean Square | F Value | p-value | R2-value |
|---|---|---|---|---|---|
| Haplotype | 4 | 31.094 | 3.04 | 0.0183 | 0.056806 |
| Hap 5 vs. Haps 1, 2, 3, 4 | 1 | 92.842 | 9.08 | 0.0029 | |
| Error | 202 | 10.223 |
*DF = Degree of freedom
SNPs and motifs in the promoter region of TaGW2-6A
We also analysed if any of the SNPs detected in the ~1 Kbp promoter region of TaGW2-6A during the present study had association with any specific motif. The analysis led to the identification of several putative binding sites within the above region of promoter that was analysed during the present study (see S2 Fig). Out of the four SNPs, SNP -494 showing significant association with TGW was located in the ‘CGCG’ motif (see Fig 3).
Relationship among SNP-494, TaGW2-6A expression and TGW
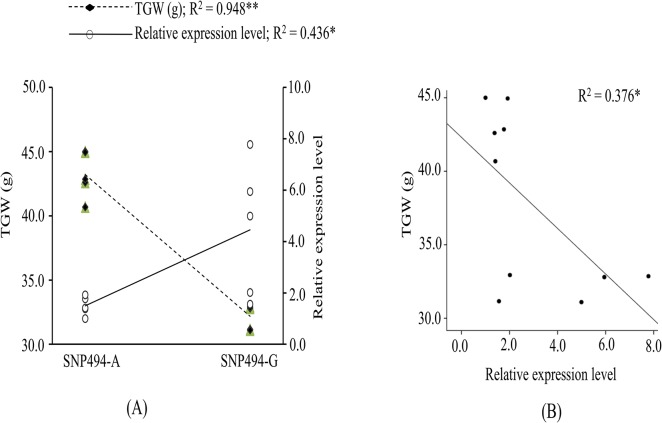
Association of SNP-494 with expression of TaGW2-6A was also examined using five genotypes each with alleles A and G of this SNP. Genotypes with SNP-494_A had expression level, which was 1.0 to 1.9 fold (average = 1.49 fold), and the genotypes with SNP-494_G had expression level, which was 1.5 to 7.7 fold (average = 4.45 fold) relative to expression in HI 1500, used as a reference (see Fig 4). There was not much variation in the expression level among the five genotypes with SNP-494_A, although the expression level in the five genotypes with SNP-494_G differed markedly. Regression of the expression level of the gene TaGW2-6A and TGW on SNP-494 genotypes was significant, with A allele having significantly lower expression and higher TGW relative to that in genotypes with G allele (Fig 5A). TGW also exhibited a significant regression on the expression of the gene TaGW2-6A, suggesting that the expression of the gene TaGW2-6A has negative association with TGW (Fig 5B).
Fig 4. Bar diagrams showing (A) relative expression level of TaGW2-6A in immature seeds at 15dpf.
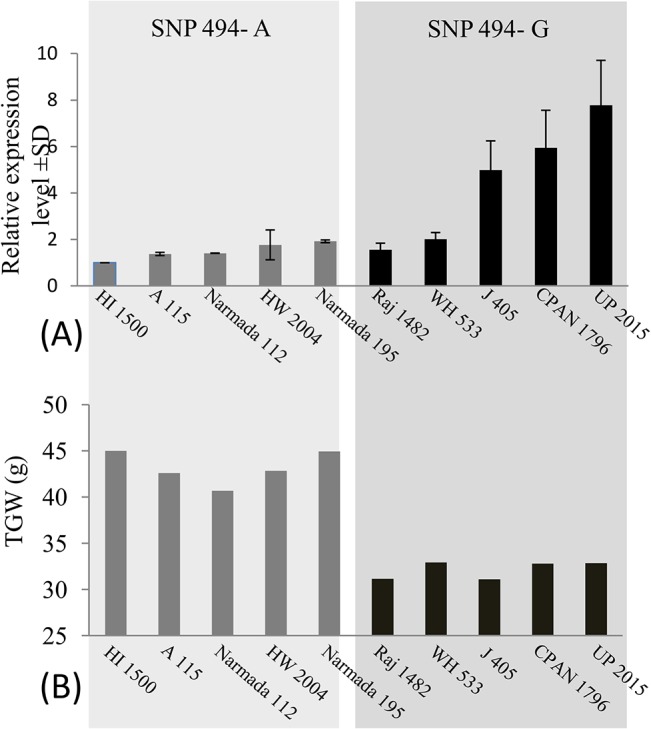
Actin gene was used as the endogenous control and variety HI 1500 used as reference; (B) TGW of varieties having SNP-494_A on the left panel and those with SNP-494_G in the right panel.
Fig 5. Plot showing significant regression of (A) TaGW2-6A expression level and thousand grain weight (TGW) in genotypes with two different alleles (A and G) of SNP-494 identified in promoter region, and (B) TGW with relative expression level in same genotypes.
* and ** indicate significance at 0.05 and 0.01 levels, respectively.
Development of functional marker for utilization of TaGW2-6A for MAS
The causal SNP (SNP-494) was converted into a CAPS (cleaved amplified polymorphism sequence) marker to distinguish the alleles of TaGW2-6A. After digestion of the PCR product by FauI, a length polymorphism (363-bp vs 418-bp) was observed in the cleavage products, which could be easily distinguished on agarose gels (Fig 6).
Fig 6. Validation of CAPS in genotypes with causal SNP_G and SNP_A on 2% agarose gel.

G and A represent undigested PCR products of genotypes with SNP_G and SNP_A; G1–G4 and A1–A4 are genotypes with SNP_G and SNP_A after digestion with FauI, M = marker.
Discussion
Two hundred seven (207) Indian wheat genotypes used in the present study were released over a period of ~100 (1910–2006) years and captured almost the entire genetic variation in TGW among Indian wheat genotypes. TGW in these 207 genotypes was normally distributed suggesting suitability of the mapping panel for conducting candidate gene-based association analysis. Similar candidate gene-based association mapping studies involving TGW and grain length were earlier conducted in China mainly using Chinese wheat germpalsm [33–35].
In the present study, we focused on sequence polymorphism in the promoter region of the gene TaGW2-6A, and detected two novel SNPs in this region. However, an insertion of a single base (T) was also earlier reported in the coding region, generating a pre-mature stop codon [23]. Thus, altogether, four SNPs in the promoter region and one insertion in the coding region of TaGW2-6A, are now known (present study and two earlier studies [22–23]). This supports the prevalent view that more SNPs occur in the promoter region than in the coding region of individual genes [22]. In future, more SNPs, indels and desirable haplotypes are likely to be identified, if screening of world wheat collection is undertaken. This variability in TaGW2-6A and similar other genes involved in grain weight may prove useful for the improvement of grain weight and related traits in bread wheat.
SNPs and haplotype associated with grain weight
During the present study, association of TGW with only one novel SNP that occurred in the promoter region of TaGW2-6A, was detected. This SNP was available at -494bp position within the promoter; the other SNP that occurred at -593bp position was also reported earlier by Su et al. [22], but was not found to be associated with TGW during the present study. The association of SNP-494 suggested an involvement of this particular SNP in regulation of the expression of gene TaGW2-6A, as also indicated by the results of expression analysis conducted during the present study. Association of an insertion in the coding region of TaGW2-6A with grain weight was also reported in an earlier study [23], but could not be confirmed during the present study, which focused on the promoter region only.
Several earlier reports are available on candidate gene-based sssociation studies in wheat involving a variety of traits including TGW and grain length [33–35]; In an earlier study, association of two SNPs in the promoter region of the gene TaGW2-6A with TGW was reported in a Chinese wheat collection [22]. During the present study, similar information on this gene in Indian wheat germplasm was collected, which led to the identification of four SNPs including a novel SNP in the promoter region showing association with grain size including TGW and length-width ratio. Simultaneous association of this novel SNP with five other agronomic traits suggested that TaGW2-6A was also involved in controlling agronomic traits other than grain size. A user-friendly CAPS marker for the causal SNP was also developed for exploitation of the variation in TaGW2-6A gene for improvement in TGW and other associated agronomic traits through marker-assisted selection (MAS) in wheat. During the present study, only five of the 16 possible haplotypes (involving four SNPs) were available. A failure to detect all the possible haplotypes may be attributed to small population size as well as strong LD. Using the above five haplotypes, we conducted haplotype-based marker-trait association analysis to study intragenic interaction. Of all the five haplotypes, Hap5 (G_A_G_A) had significantly higher TGW than other haplotypes except Hap2, which did not show any significant difference from Hap5 (Fig 3). This suggested presence of some intragenic interaction among at least some of the SNPs.
SNP in CGCG motif of promoter region and the putative pathway
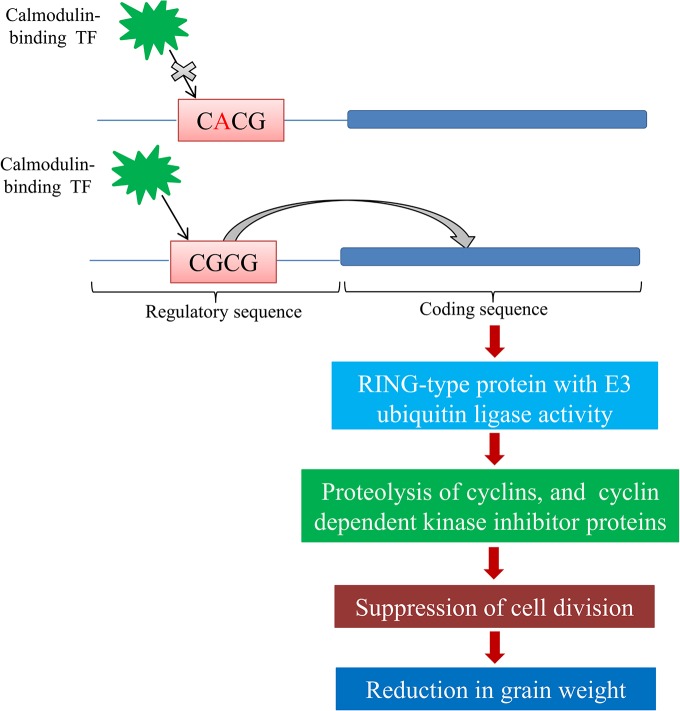
The CGCG motif is an important motif, which occurs as a cis-regulatory element within the promoter of many genes that are under Ca2+/calmodulin (CAM) regulation [36–38], and provides a site for the binding of a calmodulin-binding transcription factor [36, 38]. We detected two “CCGCGG” motifs in the promoter region of TaGW2-6A, one at -810 bp and other at -495bp. The presence of more than one Ca2+/calmodulin-responsive cis regulatory elements in the promoter region of TaGW2-6A favours the possibility of a Ca2+-mediated regulation of TaGW2-6A gene expression in a manner similar to that of calcium-dependent protein kinases (CDPKs). In rice, a CDPK with calmodulin like domain (SPK) has been shown to be involved in accumulation of storage products during seed development [39]. It is possible that a similar mechanism is involved in the regulation of TaGW2-6A in wheat also. Interestingly, the SNP-494 was located in one of the two “CGCG” motifs (at -495bp). Expression analysis also revealed that SNP-494 was involved in the regulation of the expression of TaGW2-6A. In view of the above, a hypothetical pathway of CGCG mediated regulation of TaGW2-6A gene is proposed (Fig 7). The pathway indicate that a calmodulin-binding transcription factor binds to the cis regulatory CGCG motif allowing enhanced expression of TaGW2-6A, which encodes a ring type protein with E3 ubiquitin ligase activity. The ring type protein with E3 ubiquitin ligase activity bind with substrates like cyclins, and cyclin dependent kinase inhibitor proteins, allowing the proteolysis of these important proteins, which have a key role in the progression of the cell division cycle [40]. This leads to suppression of cell division and consequent reduction in TGW. Reverse is the case if CGCG box got mutated into CACG, which leads to reduced expression of TaGW2-6A, thus leading to higher grain weight.
Fig 7. A hypothetical pathway of CGCG mediated regulation of TaGW2-6A.
Of the 207 wheat genotypes examined during the present study, as many as 194 genotypes carried CGCG motif with G allele and the remaining set of 13 genotypes had CACG motif with ‘A’ allele at the SNP-494 locus. This suggested that the motif CGCG (carrying SNP allele G) is the predominant wild type and the other motif CACG carrying the allele A evolved during the course of evolution. A perusal of TGW values of these two sets of haplotypes revealed that haplotype with CACG motif had significantly higher mean TGW than the haplotype with CGCG; this suggested that CACG motif might have evolved later due to selection for higher grain weight.
TaGW2-6A as a negative regulator of grain size
TaGW2-6A in wheat and its homologues in rice and maize are constitutively expressed [22, 41–42]. In rice, the gene OsGW2 for grain size encodes a RING-type protein with E3 ubiquitin ligase that negatively regulates grain width through control of cell division in the spikelet hull. Loss-of-function mutations in the coding sequence, or interference with the expression level of OsGW2, resulted in enhanced grain width, grain weight and grain yield [41]. In wheat, two earlier studies involving TaGW2-6A concluded that like rice gene OsGW2, its wheat otrthologue TaGW2-6A is also a negative regulator of grain-width and grain-weight [22, 23]. The present study also suggested negative regulation of grain size in wheat by TaGW2-6A. However, Bednarek et al. [24] reported that RNAi-based down-regulation of TaGW2 expression resulted in a significant reduction in final grain weight and size. Following may be the possible reasons for these apparently opposite results: (1) the gene TaGW2-6A may have different genetic backgrounds in the genotypes used in different studies; (2) there may be other genes, which may be silenced during the study conducted by Bednarek et al. [24], who used full-length sequence of ~1275bp to construct the RNAi cassette which might have resulted in off-target effects to silence other genes; (3) three homoeologues of TaGW2 may have different effects on grain weight, so that silencing of all the three genes might result in reduction in grain weight: this contention received support from a recent study [30], where it has been reported that transcript abundance of TaGW2-6A is negatively associated with the grain width, but the transcript levels of TaGW-2B and TaGW-2D were positively associated with the grain width in the same bread wheat accessions. This suggested that triplicate homoeologues of TaGW2 might have different functions in grain development, and that there is a balance among three genes finally determining the grain size in bread wheat.
TaGW2-6A with other yield related genes in wheat
Beside TaGW2, three other genes, namely TaGASR7-A1, TaGS-D1 and 6-SFT-A2 which control grain weight and/or length have been recently reported [33–35]. However, there must be a number of other yield-related important genes controlling grain weight in wheat. A number of such genes (e.g., GS3, GW5, GW8, TGW6, Ghd7 and GIF1) have actually been isolated and cloned in rice [43–48] and there is no reason why orthologues of these genes may not be available in wheat. The availability of draft genome sequence in wheat should facilitate prediction and cloning of a number of these yield-related genes, so that it will be possible to identify favourable alleles and develop functional markers for these genes. This knowledge about yield related genes including TaGW2 used for the present study may prove useful for development of high yielding wheat cultivars through marker-assisted selection.
Conclusions
A novel SNP (SNP-494) was identified in the promoter region of the gene TaGW2-6A, which significantly affects TGW, grain length-width ratio and five other agronomic traits in wheat. This SNP was also a part of a haplotype and was located in an important motif (CGCG), which may possibly be a site for one or more calmodulin-binding transcription factors and eventually may be involved in regulation of the expression of the TaGW2-6A gene. This SNP was found to regulate the expression of the gene TaGW2-6A. The findings of the present study provide an initial step toward dissecting the molecular mechanism underlying seed development and TGW in wheat. The functional CAPS marker developed for causal SNP during the present study is recommended for use in marker-assisted selection for improvement of TGW along with other agronomic traits in wheat.
Supporting Information
SNPs are highlighted with yellow (G allele) and red (A allele). SNP involving CGCG motif is represented with box.
(DOCX)
(DOCX)
Acknowledgments
We thank Indian Institute of Wheat and Barley Research (IIWBR), Karnal for providing seed material of 207 Indian wheat genotypes. Thanks are due to the Head, Department of Genetics and Plant Breeding, Ch. Charan Singh University, Meerut, India for providing facilities to carry out this study. We also acknowledge the use of bioinformatics facilities available in the BIF laboratory supported by the Department of Biotechnology, New Delhi. PKG also thanks National Academy of Sciences India for the award of a Platinum Jubilee Senior Scientist position, during the tenure of which this work was undertaken.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Thanks are due to the Department of Biotechnology, New Delhi, Department of Science Technology (Funds for Improvement of Science and Technology infrastructure program), New Delhi and University Grants Commission (Special Assistance Program-Departmental Research Support Program), New Delhi for support which facilitated this study. PKG was awarded the position of NASI-Senior Scientist, VG was awarded SRF under the same program. CSIR awarded SRF to VJ.
References
- 1. Marshall DR, Mares DJ, Moss HJ, Ellison FW (1986) Effects of grain shape and size on milling yields in wheat. II. Experimental studies. Aust J Agr Res 37: 331–342. [Google Scholar]
- 2. Ramya P, Chaubal A, Kulkarni K, Gupta L, Kadoo N, Dhaliwal HS, et al. (2010) QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J Appl Genet 51: 421–429. [DOI] [PubMed] [Google Scholar]
- 3. Botwright TL, Condon AG, Rebetzke GJ, Richards RA (2002) Field evaluation of early vigour for genetic improvement of grain yield in wheat. Aust J Agric Res 53: 1137–1146. [Google Scholar]
- 4. Peng J, Ronin Y, Fahima T, Roder MS, Li Y, Nevo E, et al. (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci USA 100: 2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kato K, Miura H, Sawada S (2000) Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor Appl Genet 101: 1114–1121. [Google Scholar]
- 6. Varshney RK, Prasad M, Roy JK, Kumar N, Harjit-Singh, Dhaliwal HS, et al. (2000) Identification of eight chromosomes and a microsatellite marker on 1AS associated with QTL for grain weight in bread wheat. Theor Appl Genet 100: 1290–1294. [Google Scholar]
- 7. Dholakia BB, Ammiraju JSS, Singh H, Lagu MD, Roder MS, Rao VS, et al. (2003) Molecular marker analysis of kernel size and shape in bread wheat. Plant Breed 122: 392–395. [Google Scholar]
- 8. Groos C, Robert N, Bervas E, Charmet G (2003) Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor Appl Genet 106: 1032–1040. [DOI] [PubMed] [Google Scholar]
- 9. Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, et al. (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110: 865–880. [DOI] [PubMed] [Google Scholar]
- 10. McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N, Noll J, et al. (2005) Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452×AC domain. Genome 48: 870–883. [DOI] [PubMed] [Google Scholar]
- 11. Marza F, Bai GH, Carver BF, Zhou WC (2005) Quantitative trait loci for yield and related traits in the wheat population Ning7840 × Clark. Theor Appl Genet 21: 1–11. [DOI] [PubMed] [Google Scholar]
- 12. Huang X- Q, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, et al. (2006) Molecular detection of QTLs for agronomic and quality traits in a double haploid population derived from two Canadian wheats (Triticum aestivium L.). Theor Appl Genet 113: 753–766. [DOI] [PubMed] [Google Scholar]
- 13. Kumar N, Kulwal PL, Gaur A, Tyagi AK, Khurana JP, Khurana P, et al. (2006) QTL analysis for kernel weight in common wheat. Euphytica 151: 135–144. [Google Scholar]
- 14. Kunert A, Naz AA, Dedeck O, Pillen K, Léon J (2007) AB-QTL analysis in winter wheat: I. Synthetic hexaploid wheat (T. turgidum ssp. dicoccoides × T. tauschii) as a source of favourable alleles for milling and baking quality traits. Theor Appl Genet 115: 683–695. [DOI] [PubMed] [Google Scholar]
- 15. Li S, Jia J, Wei X, Zhang X, Li L, Chen H, et al. (2007) An intervarietal genetic map and QTL analysis for yield traits in wheat. Mol Breed 20: 167–178. [Google Scholar]
- 16. Roder MS, Huang XQ, Borner A (2008) Fine mapping of the region on wheat chromosome 7D controlling grain weight. Funct Integr Genomics 8: 79–86. [DOI] [PubMed] [Google Scholar]
- 17. Sun XY, Wu K, Zhao Y, Kong FM, Han GZ, Jiang HM, et al. (2009) QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 165: 615–624. [Google Scholar]
- 18. Mir RR, Kumar N, Jaiswal V, Girdharwal N, Prasad M, Balyan HS, et al. (2012) Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol Breed 29: 963–972. [Google Scholar]
- 19. Maphosa L, Langridge P, Taylor H, Parent B, Emebiri LC, Kuchel H, et al. (2014) Genetic control of grain yield and grain physical characteristics in a bread wheat population grown under a range of environmental conditions. Theor Appl Genet 127:1607–1624. 10.1007/s00122-014-2322-y [DOI] [PubMed] [Google Scholar]
- 20. Wei L, Bai S, Li J, Hou X, Wang X, Li H, et al. (2014) QTL positioning of thousand wheat grain weight in Qaidam basin. Open J of Genet 4: 239–244. [Google Scholar]
- 21. Wu QH, Chen YX, Zhou S- H, Fu L, Chen JJ, Xiao Y, et al. (2015) High-density genetic linkage map construction and QTL mapping of grain shape and size in the wheat population Yanda1817 × Beinong6. PLoS ONE 10: e0118144 10.1371/journal.pone.0118144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Su Z, Hao C, Wang L, Dong Y, Zhang X (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl Genet 122: 211–223. 10.1007/s00122-010-1437-z [DOI] [PubMed] [Google Scholar]
- 23. Yang Z, Bai Z, Li X, Wang P, Wu Q, Yang L, et al. (2012) SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor Appl Genet 125: 1057–1068. 10.1007/s00122-012-1895-6 [DOI] [PubMed] [Google Scholar]
- 24. Bednarek J, Boulaflous A, Girousse C, Ravel C, Tassy C, Barret P, et al. (2012) Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J Exp Bot 63: 5945–5955. 10.1093/jxb/ers249 [DOI] [PubMed] [Google Scholar]
- 25.Kundu S, Shoran J, Mishra B, Gupta RK (2006) Indian wheat varieties at a glance. Directorate of Wheat Research, Karnal-132001, India. Research Bulletin No. 21, p: 447
- 26. Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard W (1984) Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci 91: 5466–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 28. Liu S, Zhou Z, Lu J, Sun F, Wang S, Liu H, et al. (2011) Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12: 53 10.1186/1471-2164-12-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao Z, Luo W, Liu H, Zeng C, Liu X, Yi S, et al. (2012) Transcriptome analysis and SSR/SNP markers information of the blunt snout bream (Megalobrama amblycephala). PLoS ONE 7: e42637 10.1371/journal.pone.0042637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hong Y, Chen L, Du L, Su Z, Wang J, Ye X, et al. (2014) Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct Integr Genomics 14:341–349. 10.1007/s10142-014-0380-5 [DOI] [PubMed] [Google Scholar]
- 31. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 32. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Liu J, Xia X, He Z (2014) TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed 34: 1097–1107. [Google Scholar]
- 34. Dong L, Wang F, Liu T, Dong Z, Li A, Jing R, et al. (2014) Natural variation of TaGASR7-1 A1 affects grain length in common wheat under multiple cultivation conditions. Mol Breed 34: 937–947. [Google Scholar]
- 35. Yue A, Li A, Mao X, Chang X, Li R, Jing R (2015) Identification and development of a functional marker from 6-SFT-A2 associated with grain weight in wheat. Mol Breed 35: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang T, Poovaiah BW (2002) A Calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chemis 277: 45049–45058. [DOI] [PubMed] [Google Scholar]
- 37. Campos-Soriano L, Gómez-Ariza J, Bonfante P, Segundo BS (2011) A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biol 11: 90 10.1186/1471-2229-11-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D (2012) SR1, a calmodulin-binding transcription factor, modulates plant defence and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158: 1847–1859. 10.1104/pp.111.192310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Asano T, Kunieda N, Omura Y, Ibe H, Kawasaki T, Takano M, et al. (2002) Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell 14: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teixeira LK, Reed SI (2013). Ubiquitin ligases and cell cycle control. Ann Rev Biochem 82: 387–414. 10.1146/annurev-biochem-060410-105307 [DOI] [PubMed] [Google Scholar]
- 41. Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630. [DOI] [PubMed] [Google Scholar]
- 42. Li Q, Li L, Yang X, Waburton ML, Bai G, Dai J, et al. (2010) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol 10: 143 10.1186/1471-2229-10-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, et al. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- 44. Weng JF, Gu SH, Wan XY, Gao H, Guo T, Su N, et al. (2008) Isolation and initial characterization of GW5, a major QTL associated with rice kernel width and weight. Cell Res 18: 1199–1209. 10.1038/cr.2008.307 [DOI] [PubMed] [Google Scholar]
- 45. Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950–954. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- 46. Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45: 707–711. 10.1038/ng.2612 [DOI] [PubMed] [Google Scholar]
- 47. Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- 48. Wang ET, Wang JJ, Zhu XD, Hao W, Wang LY, Li Q, et al. (2008) Control of rice kernel filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs are highlighted with yellow (G allele) and red (A allele). SNP involving CGCG motif is represented with box.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.