Abstract
Assisted reproductive technology (ART) is being increasingly applied to overcome infertility. However, the in vitro production process, the main procedure of ART, can lead to aberrant embryonic development and health-related problems in offspring. Understanding the mechanisms underlying the ART-induced side effects is important to improve the ART process. In this study, we carried out comparative transcriptome profiling between in vivo- (IVO) and in vitro- produced (IVP) mouse blastocysts. Our results suggested that aberrant actin organization might be a major factor contributing to the impaired development of IVP embryos. To test this, we examined the effect of actin disorganization on the development of IVP preimplantation embryos. Specific disruption of actin organization by cytochalasin B (CB) indicated that well-organized actin is essential for in vitro embryonic development. Supplementing the culture medium with 10–9 M melatonin, a cytoskeletal modulator in adult somatic cells, significantly reversed the disrupted expression patterns of genes related to actin organization, including Arhgef2, Bcl2, Coro2b, Flnc, and Palld. Immunofluorescence analysis showed that melatonin treatment of IVP embryos significantly improved the distribution and organization of actin filaments (F-actin) from the 8-cell stage onwards. More importantly, we found that melatonin alleviated the CB-mediated aberrant F-actin distribution and organization and rescued CB-induced impaired embryonic development. This is the first study to indicate that actin disorganization is implicated in impaired development of IVP embryos during the preimplantation stage. We also demonstrated that improving actin organization is a promising strategy to optimize existing IVP systems.
Introduction
The number of patients receiving assisted reproductive technology (ART) to treat infertility has increased annually. It is estimated that approximately 1.5 million ART cycles are performed annually worldwide, resulting in 5,000,000 live births [1]. However, ART is associated with risks, such as aberrant embryonic development and health-related problems in the offspring [2]. Studies have shown that in vitro fertilization (IVF) and in vitro culture (IVC), the main procedures of ART, have a profound effect on gene/protein expression patterns and phenotypes in pre- and post-implantation mouse embryos [3, 4]. Moreover, ART is associated with long-term effects on the offspring’s health, such as fetal complications [5], and an increased risk of diseases in childhood and adulthood [6]. Using high-throughput methods, such as microarrays and RNA sequencing (RNA-seq), many studies have reported that a series of biological processes, including energy metabolism [5], genetic information processing [7], and epigenetic modifications [8, 9], are disrupted in IVP embryos. However, these deductions were generally based on the functional analysis of high-throughput data, which need to be further confirmed. Moreover, in-depth understanding of mechanisms of these effects would be beneficial to optimize the IVP processes.
Thus far, improvements of in vitro embryonic development have primarily depended on modifying the in vitro culture system. Some improvements have shown beneficial effects on in vitro embryonic development, and optimizations have included the use of chemically defined culture media [10], two-step sequential culture system (e.g., G1/G2 [11]), and supplementation of medium with specific physiological factors (e.g., glutathione [12], cysteine [13] and melatonin [14]). Melatonin (N-acetyl-5-methoxytryptamine), a natural molecule mainly secreted by the pineal gland in mammals, apparently improves in vitro embryonic development in mice [14], cattle [15], sheep [16], pigs [17] and humans [18]. Previously, the pervasive antioxidant and apoptosis inhibition abilities [15, 17, 19] of melatonin were found thought to be responsible for the improvement of in vitro embryonic development. However, melatonin has multiple functions. It participates in many cellular functions, including prompting mitochondrial biogenesis [20], regulating nuclear transcriptional activity [21] and facilitating gap-junction commutation [22].
In this study, we carried out comparative transcriptome profiling between in vivo- (IVO) and in vitro- (IVP) produced mouse blastocysts. Functional clustering showed that aberrant actin organization might be a major factor contributing to the impaired development of IVP preimplantation embryos. The results of CB-induced actin disorganization further suggested that well-organized actin organization is essential for in vitro embryonic development. These results also implied that actin organization could be a target for improving in vitro embryonic development. In previous studies, melatonin was shown to act as a cytoskeletal modulator in somatic cells and cancer cells [23]. Therefore, we hypothesized that melatonin might also participate in actin organization during in vitro embryonic development, which contributes to the beneficial effect it exerts on in vitro embryonic development. Our results showed that melatonin could reverse the dysregulated expression of genes associated with actin organization, and alleviated the actin disorganization in IVP preimplantation embryos. Moreover, melatonin could rescue the cytochalasin B (CB)-induced actin disorganization and impaired preimplantation embryonic development. Our study, indicated, for the first time, that actin disorganization is a major factor in the impaired development of IVP embryos during the preimplantation stage. More importantly, we also demonstrated that improving actin organization is a promising strategy to optimize existing IVP systems.
Materials and Methods
Ethics Statement
The protocols for the animal studies were approved by, and performed in accordance with, the requirements of the Institutional Animal Care and Use Committee of China Agricultural University.
Animals
We used female Institute for Cancer Research (ICR) mice aged 7 to 8- weeks and male ICR mice aged 12 to 14 weeks. The mice were fed ad libitum and housed under controlled lighting conditions (12:12-h light:dark photoperiod).
Chemicals
All chemicals used were purchased from the Sigma-Aldrich Chemical Company (St. Louis, MO, USA) unless otherwise specified.
Embryo Preparation
The female mice were superovulated by intraperitoneal injection of 5 IU pregnant mare serum gonadotropin (PMSG) and 5 IU of human chorionic gonadotropin (hCG) after 47 h. In the IVO group, the superovulated female mice were mated individually with male mice. The following morning, successful mating was confirmed by detecting the presence of a vaginal plug.
In the IVP group, IVF was performed as previously described [4]. Briefly, cumulus-enclosed oocyte complexes (COCs) were collected from the ampullae 14 h after the hCG injection, and the cumulus cells were removed by digesting the COCs with hyaluronidase for 3−5 min. The oocytes were first rinsed in human tubal fluid (HTF) medium (Sage, Bedminster, NJ, USA), and placed in 60-μL drops of HTF medium, covered with paraffin oil and equilibrated overnight at 37°C and 5% CO2. Sperm was obtained from the cauda epididymis and capacitated for 1 h in HTF medium at 37°C and 5% CO2. Sperm insemination was carried out 15 h after hCG injection. After 4 h in the incubator, the oocytes/embryos were washed several times in potassium simplex optimization medium containing amino acids (KSOM + AA; Millipore, Billerica, MA, USA) and transferred to 60-μL drops of KSOM + AA medium covered with paraffin oil. The embryos were cultured at 37°C with 5% CO2.
Treatment with melatonin or CB
The treatment of embryos with CB was performed as previously described [24], with minor modifications. In brief, CB was dissolved as a stock solution (20 mg/ml) in dimethyl sulfoxide (DMSO) and stored at −20°C. The stock solution was later diluted to the working concentrations (5μg/ml, 10 μg/ml and 20 μg/ml) in KSOM + AA medium, before use. We set two controls: untreated control (IVP group) and vehicle control (supplementation with DMSO; 0.1% v/v, the same amount of DMSO as the embryos receiving 20 μg/ml CB treatment).
Melatonin was dissolved as a stock solution (15mM) in DMSO and stored at −80°C. The stock solution was later diluted to the working concentrations (10−7~10−10 M) in KSOM + AA medium before use. For melatonin treatment, the concentration of DMSO supplemented into the KSOM + AA medium (< 0.001% v/v, the same amount of DMSO as the embryos receiving melatonin treatment in 10−7 M group) was very low compared with the CB-vehicle control (0.1% v/v); therefore, we did not perform this vehicle control for melatonin treatment.
After IVF, zygotes were further cultured in KSOM + AA medium supplemented with melatonin or/ and CB. For CB treatment, the zygotes were treated for 3 h, and then removed from the medium containing CB. After washing at least three times in KSOM + AA medium, embryos were further cultured in CB-free KSOM + AA medium. For melatonin treatment, the IVF zygotes were cultured in KSOM + AA medium containing melatonin until the embryos were collected at different stages.
Embryo Collection
Embryos were collected according to a previously described method [25]. Briefly, IVO embryos at the 4-cell, 8-cell and morula stages were obtained from the recipient females by flushing the Fallopian tube with M2 medium (at 54–56, 68–70, and 76–78 h post-hCG, respectively), while embryos at the blastocyst stage were obtained from recipient females by flushing the uterus with M2 medium (96−100 h post-hCG).
The IVP embryos were collected at different stages on the basis of their developmental progress and embryo morphology, as described in previous reports [3, 4, 26].
High-throughput RNA Sequencing
Total RNAs were extracted from blastocysts using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Polyadenylated RNAs were isolated using an Oligotex mRNA midi kit (Qiagen, Valencia, CA, USA). RNA-seq libraries were constructed using a SOLiD whole transcriptome analysis kit (Applied Biosystems, Foster City, CA, USA) according to the standard protocol, and sequenced on an Applied Biosystems SOLiD platform to generate high-quality single-end reads. The raw reads were aligned to genome sequences, after trimming off a nucleotide each from the 5ʹ- and 3ʹ-ends and allowing up to two mismatches. Reads mapped to multiple locations were discarded and only uniquely mapped reads were used for subsequent analysis. The gene expression levels were measured as reads per kilobase of exon model per million mapped reads (RPKM). Biological processes in response to different physiological conditions were annotated using the database for annotation, visualization, and integrated discovery (DAVID v6.7; http://david.abcc.ncifcrf.gov). Differentially expressed proteins (DEPs) were run through the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string.embl.de/) to build a protein-protein interaction (PPI) network, and for comparison. Phenotype annotations of the DEPs were analyzed using the Mouse Genome Informatics database (MGI; http://www.informatics.jax.org/phenotypes.shtml).
Immunofluorescence and Immunofluorescence Quantification
To detect the abundance and distribution of actin filaments, fluorescein isothiocyanate labeled phalloidin (FITC- phalloidin) was used. FITC- phalloidin was prepared in accordance with the manufacturer's instructions. Briefly, FITC- phalloidin was dissolved as a stock solution (0.1 mg/ml) in DMSO and stored at −20°C. The stock solution was later diluted to the working concentrations (5 μg/ml) in PBST-PVA (0.2% Triton-X100 and 0.1% polyvinyl alcohol [PVA] in PBS) before use. Embryos were fixed with 3.7% formaldehyde for 1 h at 4°C, and then permeabilized in PBST-PVA for 20 min at room temperature. After washing three times with 0.1% PBS/PVA at 37°C for 5 min, the embryos were incubated with FITC- phalloidin overnight at 4°C. After washing three times with 0.1% PBS/PVA at 37°C for 5 min, the embryos were counterstained with 4’,-6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA) for 10 min and mounted on glass-bottomed culture dishes (Nest) with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA).
Fluorescence signals were observed using a confocal scanning laser microscope (Digital Eclipse C1, Nikon, Japan). As described previously [27], we generated a Z-stack using the acquisition function: choosing the upper and lower limits of the Z-stack, and then Z-stack images with 14 consequential sections for each embryo were taken. The full project image was generated from the Z-stack files. For all embryos identical image acquisition (laser power, light path, objective, gains and offsets) and Z-stack settings were used to minimize potential differences in signal strength from embryo to embryo. Moreover, to minimize bleaching, the embryos were not exposed to light emitted from mercury lamps and were only briefly exposed to laser light to set the Z-positions and during image acquisition.
Quantitative analysis of F-actin was carried out as described previously [28], with minor modifications. Fluorescence intensity was assessed using Image J software (NIH). Within the region of interest (ROI) in the immunofluorescence images, the average fluorescence intensity per unit area was analyzed. Three independent quantifications were performed on each embryo, and average values of all measurements were used to determine the final average intensities.
Real-time Quantitative PCR (qPCR) Analysis
qPCR was carried out as described previously [17]. Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, embryos were extracted with TRIzol reagent by centrifugation, washed with 1 mL of 75% ethanol, and dissolved in 20 μL of RNase-free water. For reverse transcription, a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Hanover, MD, USA) was used to generate cDNA, according to the manufacturer’s protocol. The real-time PCR amplification mixture consisted of 1 μL of cDNA, 5 μL of SsoFast EvaGreen Supermix (Bio-Rad), and 0.5 μL of both forward and reverse primers (Table 1), in a total volume of 10 μL. The PCR procedure was performed according to the manufacturer’s protocol. The PCR parameters were as follows: incubation at 95°C for 30 s; followed by 45 cycles of amplification at 95°C for 5 s, at gene-specific annealing temperature (Table 1) for 5 s, and a melting curve program at 65–95°C (starting fluorescence acquisition at 65°C, with measurements obtained at 5-s intervals until the temperature reached 95°C). Three replicates were performed, and the mRNA level of each group was normalized to the Gapdh mRNA level.
Table 1. Primer sequences used for real-time PCR.
| Gene | Primer sequence (5'-3') | Tm(°C) | Product size (bp) |
|---|---|---|---|
| Arhgef2 | F-GCTGCTGATGACAGATGT; R-AGGCGGTCCAGAACTAAT | 60 | 160 |
| β-actin | F-AGGTCATCACTATTGGCAAC; R-ACTCATCGTACTCCTGCTTG | 60 | 357 |
| Bcl2 | F-GATGGTGTGGTTGCCTTA; R-GGTATATCCGCTACAAGTTAC | 60 | 232 |
| Ccdc88a | F-TCCAGACACTAATGCTACAG; R-ATCCAGTTGCCTCTCCTT | 60 | 196 |
| Coro2b | F-GAGCAGACAGGCAGAATC; R-GAGCAAGAGGCGATGATG | 60 | 110 |
| Elmo1 | F-GCCAACTCATTCCTCATCT; R-TCTCCTGTCTCCTCTCATC | 60 | 108 |
| Enah | F-GCTGTGATGGTCTATGATGA; R-AATGTGTTGTTGCCTGTATG | 60 | 104 |
| Flnc | F-AATTGTCACCAAGGATGCT; R-CACGATGATGCTGTAGTCT | 60 | 148 |
| Gapdh | F-TGCCCCCATGTTTGTGATG; R-TGTGGTCATGAGCCCTTCC | 60 | 151 |
| Palld | F-TTCAGGAGCGATTCTTCAG; R-GGACCAGCATCTTGTGAG | 60 | 174 |
| Tns4 | F-CGAGAGCAAGCAATCAATC; R-CAGGAAGTGACGGATAAGG | 60 | 171 |
| Vil1 | F-GGTGGTTAGAGAAGTTGCTA; R-TGGAAGAGTTGTTGGAAGAT | 60 | 246 |
Statistical Analysis
All data are presented as the means ± standard deviation (SD). One-way analysis of variance was used to compare differences among groups, followed by post-hoc comparisons between groups, using IBM SPSS statistics software version 20.
Results
Actin Disorganization Contributes to the Impaired Development of IVP Embryos
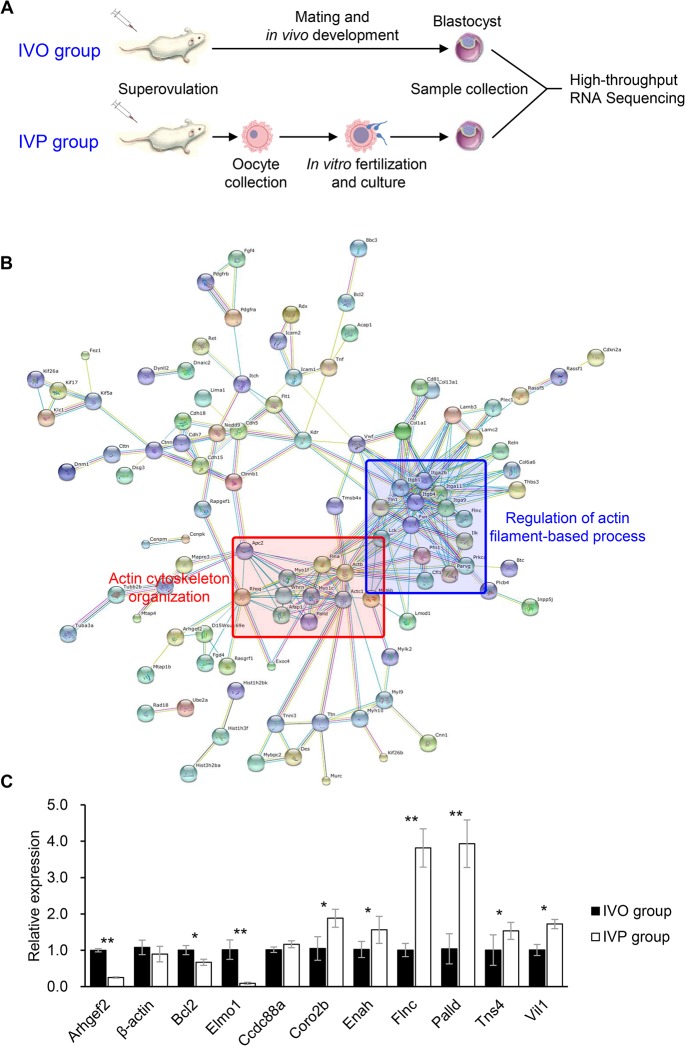
To investigate the potential effects of the IVP process on the global gene expression patterns in preimplantation embryos, we sampled 8,400 blastocysts for high-throughput mRNA sequencing (Fig 1A). The results of gene ontology (GO) classification showed that many differentially expressed genes (DEGs) were functionally associated with the “actin binding” and “cytoskeletal protein binding” clusters (Table 2). Using the DEGs that were functionally associated with cytoskeletal organization as seed nodes, a detailed PPI network was constructed (Fig 1B). Many genes that encoded actin-binding proteins or were responsible for actin organization were clustered tightly in the network. Clusters involved in actin cytoskeleton organization and the regulation of actin filament-based processes were located at the center of the network. These results indicated that cytoskeletal organization, especially actin organization, was disrupted in IVP blastocysts. We verified this observation by evaluating the expressions of some of the DEGs involved in actin organization using qPCR (Fig 1C).
Fig 1. In vitro-produced (IVP) blastocysts are characterized by abnormal actin cytoskeleton organization.
(A) Overview of the experimental design. Blastocysts were collected after either mating and in vivo development (IVO group) or in vitro fertilization and culture (IVP group). (B) Protein-protein interaction (PPI) network. Differentially expressed genes (DEGs) in the IVO and IVP blastocysts showed a tightly interconnected network, as observed from a web-based search of the STRING database. (C) Expression of actin-associated genes in the IVP and IVO blastocysts detected by quantitative real-time PCR analysis. The asterisks (* or **) in each column represent statistically significant differences (P < 0.05 and P < 0.01, respectively).
Table 2. Representative gene ontology (GO) terms associated with actin cytoskeleton organization based on the differentially expressed genes (DEGs, fold changes > 2) between IVP and IVO blastocysts.
| Annotation Cluster 1 | Enrichment Score: 4.54 | Count | P_Value | Benjamini |
| GOTERM_CC_FAT | extracellular region | 203 | 0.000091 | 0.021 |
| Annotation Cluster 2 | Enrichment Score: 4.38 | Count | P_Value | Benjamini |
| GOTERM_BP_FAT | chemical homeostasis | 65 | 3.8E-07 | 0.0012 |
| GOTERM_BP_FAT | ion homeostasis | 54 | 0.0000014 | 0.0023 |
| GOTERM_BP_FAT | homeostatic process | 89 | 0.0000026 | 0.0028 |
| GOTERM_BP_FAT | cation homeostasis | 38 | 0.0000046 | 0.0036 |
| GOTERM_BP_FAT | cellular chemical homeostasis | 49 | 0.0000058 | 0.0037 |
| GOTERM_BP_FAT | cellular ion homeostasis | 48 | 0.0000063 | 0.0033 |
| GOTERM_BP_FAT | cellular cation homeostasis | 32 | 0.00002 | 0.0078 |
| GOTERM_BP_FAT | cellular homeostasis | 56 | 0.000036 | 0.013 |
| GOTERM_BP_FAT | di-, tri-valent inorganic cation homeostasis | 30 | 0.00006 | 0.017 |
| GOTERM_BP_FAT | cellular di-, tri-valent inorganic cation homeostasis | 28 | 0.000083 | 0.022 |
| GOTERM_BP_FAT | metal ion homeostasis | 23 | 0.00023 | 0.039 |
| GOTERM_BP_FAT | cellular metal ion homeostasis | 22 | 0.00027 | 0.04 |
| GOTERM_BP_FAT | calcium ion homeostasis | 20 | 0.00098 | 0.11 |
| GOTERM_BP_FAT | cellular calcium ion homeostasis | 19 | 0.0015 | 0.13 |
| GOTERM_BP_FAT | regulation of membrane potential | 19 | 0.025 | 0.5 |
| Annotation Cluster 3 | Enrichment Score: 3.5 | Count | P_Value | Benjamini |
| GOTERM_CC_FAT | sarcomere | 20 | 0.00012 | 0.019 |
| GOTERM_CC_FAT | Z disc | 14 | 0.00023 | 0.021 |
| GOTERM_CC_FAT | I band | 15 | 0.00029 | 0.019 |
| GOTERM_CC_FAT | contractile fiber part | 20 | 0.00034 | 0.02 |
| GOTERM_CC_FAT | contractile fiber | 21 | 0.00047 | 0.022 |
| GOTERM_CC_FAT | myofibril | 20 | 0.00072 | 0.027 |
| Annotation Cluster 4 | Enrichment Score: 3.39 | Count | P_Value | Benjamini |
| GOTERM_MF_FAT | actin binding | 47 | 0.00043 | 0.087 |
| GOTERM_MF_FAT | cytoskeletal protein binding | 62 | 0.00052 | 0.087 |
| Annotation Cluster 5 | Enrichment Score: 2.75 | Count | P_Value | Benjamini |
| GOTERM_CC_FAT | extracellular matrix | 48 | 0.00068 | 0.028 |
| GOTERM_CC_FAT | proteinaceous extracellular matrix | 46 | 0.00095 | 0.034 |
| GOTERM_CC_FAT | extracellular region part | 95 | 0.0058 | 0.14 |
| Annotation Cluster 6 | Enrichment Score: 2.72 | Count | P_Value | Benjamini |
| GOTERM_MF_FAT | ion binding | 456 | 0.0000011 | 0.00056 |
| GOTERM_MF_FAT | cation binding | 448 | 0.0000027 | 0.00095 |
| GOTERM_MF_FAT | metal ion binding | 442 | 0.0000056 | 0.0015 |
| GOTERM_MF_FAT | transition metal ion binding | 275 | 0.058 | 0.72 |
| GOTERM_MF_FAT | zinc ion binding | 222 | 0.085 | 0.78 |
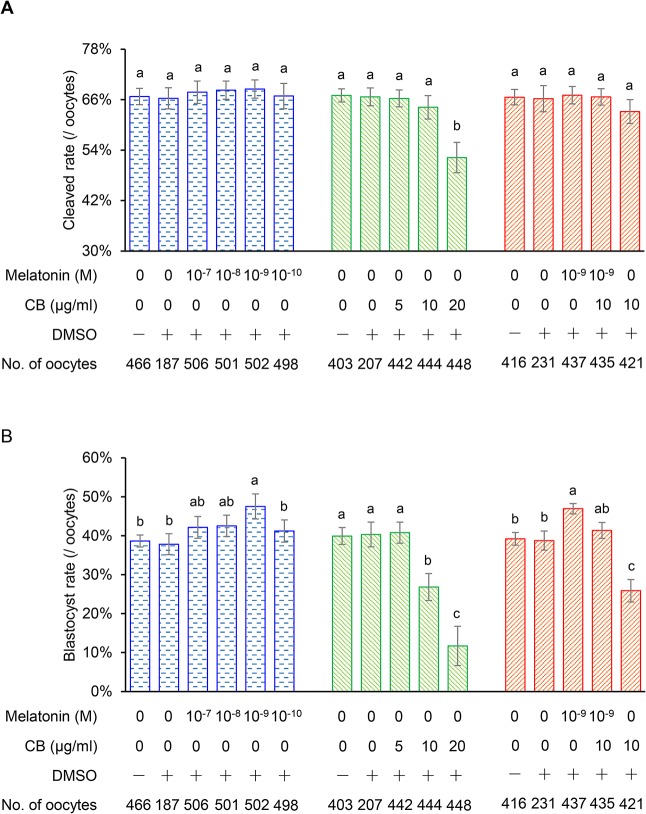
To confirm the critical role of actin organization in early embryonic development, we supplemented the culture medium with CB, which inhibits actin filament polymerization and specifically disrupts actin organization [24]. As shown in Fig 2, 10 μg/mL of CB for 3 h mildly reduced the blastocyst rate to a relatively low level, while higher concentrations (20 μg/mL) of CB led to a severe decrease in the blastocyst rate. This implied that well-organized actin is essential for in vitro embryonic development.
Fig 2. Effects of different concentrations of melatonin and/or CB supplementation on the embryonic development.
(A) Cleavage rate. (B) Blastocyst rate. The vehicle control was also tested (0.1% v/v, the highest concentration of DMSO supplementation). All experiments were replicated at least three times independently. Different superscript letters (a, b, c) indicate significant differences (P < 0.05).
Melatonin Improves in vitro Embryonic Development
As the optimal melatonin dose for in vitro development depends on the culture system [29] and species [15], we preselected the optimal concentration of melatonin treatment to suit the experimental conditions used in our study. There were no differences in the cleavage rates between the treatment and control groups (P > 0.05; Fig 2A). However, the blastocyst rate was significantly higher in the group treated with 10−9 M of melatonin than that in the control group (47.53 ± 3.22% vs. 38.67 ± 1.54%, P < 0.05) (Fig 2B). Therefore, 10−9 M was selected as the optimal concentration of melatonin to test the effect of melatonin treatment on the actin organization in IVP preimplantation embryos.
Melatonin Modulates the Impaired actin Organization of IVP Embryos
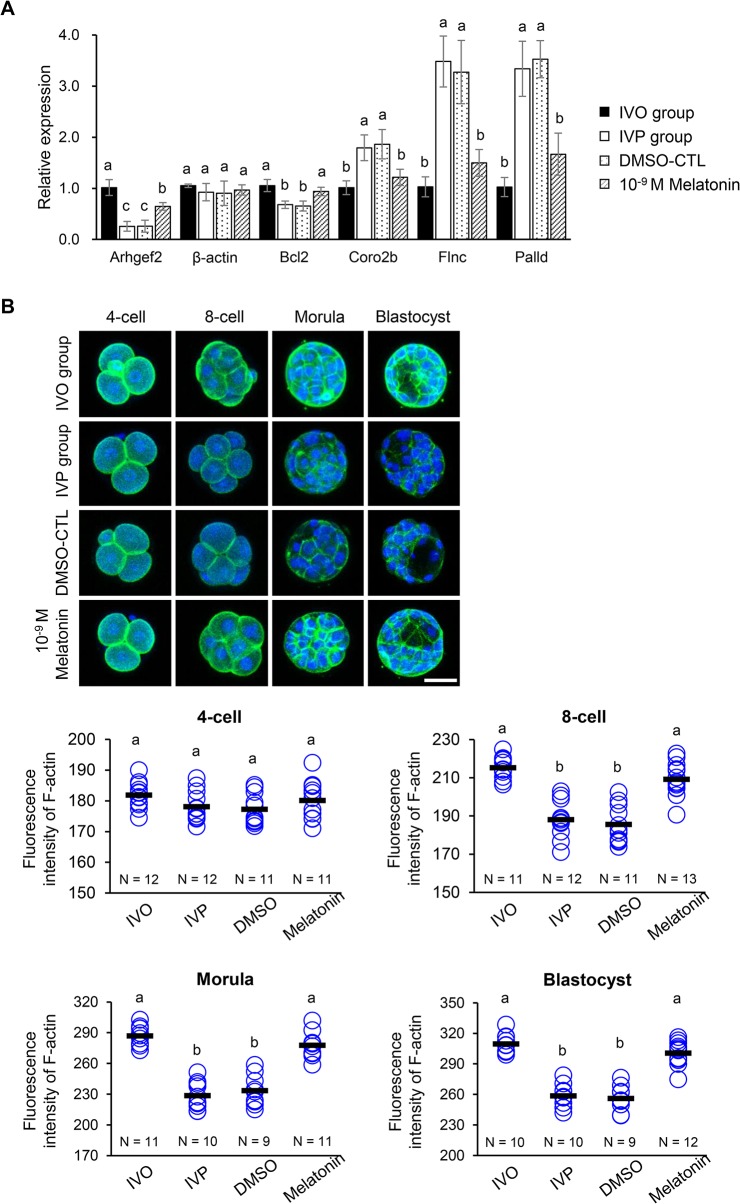
Melatonin treatment was found to alleviate or significantly reverse the disturbed expression patterns of certain genes, including Arhgef2, Bcl2, Coro2b, Flnc, and Palld (P < 0.05; Fig 3A). Among these genes, the expression levels of Arhgef2 and Bcl2 were significantly elevated (by 2.5-fold and 1.4-fold) relative to the corresponding expression levels in the control group, while the expression levels of Coro2b, Flnc, and Palld were significantly decreased (by 1.5-, 2.3-, and 2.0-fold, respectively) compared to the corresponding values in the control group (IVP group).
Fig 3. Effect of melatonin on the distribution and organization of F-actin in preimplantation embryos.
(A) Comparative expression of the genes involved in actin organization in the IVP, IVO, and melatonin-treated blastocysts detected by qPCR analysis. DMSO-CTL indicates the vehicle control, in which the concentration of DMSO was 0.1% (v/v). (B) FITC-phalloidin immunofluorescence analysis to detect F-actin (green) in IVO, IVP, DMSO-CTL and 10-9M melatonin treated embryos at different stages during preimplantation development (4-cell, 8-cell, morula, and blastocyst). Nuclei (blue) are stained with 4′,6-diamidino-2-phenylindole (DAPI). Bar, 50 μm. Each circle represents the fluorescence intensity in each embryo. Horizontal lines represent the mean values. The number of embryos in each group is indicated. Different superscript letters (a, b, c) indicate significant differences (P < 0.05).
The differential expression patterns of the genes involved in actin organization led us to hypothesize that IVP embryos have faulty actin organization. Thus, we used FITC-phalloidin immunofluorescence analysis to study the distribution of F-actin, filamentous actin [30], in preimplantation IVO and IVP embryos at different stages (4-cell, 8-cell, morula, and blastocyst stages; Fig 3B). The results showed that IVP embryos had a lower abundance of F-actin (measured by fluorescence intensity of FITC-phalloidin staining) from the 8-cell stage onwards, and some cells in a proportion of the IVP morulae and blastocyst had disrupted F-actin organization. There was no obvious difference in distribution between the IVP and IVO embryos at the 4-cell stage. We quantified the average fluorescence intensities of the pixels, using previously described method [28]. The fluorescence intensity of F-actin significantly decreased in the IVP embryos from the 8-cell stage onwards compared with IVO embryos (P < 0.05). Furthermore, melatonin treatment reversed the adverse effect of the IVP process on F-actin organization (indicated by the fluorescence distribution and intensity) from the 8-cell stage onwards (average fluorescence intensity in melatonin-treated IVP embryos vs. control IVP embryos: 8-cell, 209.29 vs. 188.14; morula, 277.79 vs. 228.55; blastocyst, 300.66 vs. 258.72; P < 0.05).
Melatonin rescues preimplantation development of the impaired IVP embryos via improved distribution and organization of F-actin
To prove that the melatonin-induced improvement of in vitro embryonic development at least partly depends on its actin organization-modulating role, we supplemented the culture medium with CB and melatonin. As shown in Fig 2, supplementation with 10 μg/mL of CB for 3 h mildly decreased the blastocyst rate. However, 10−9 M melatonin in the medium significantly alleviated the impaired embryonic development (Fig 2). Similarly, melatonin could overcome the adverse effects of treatment with 10 μg/mL CB on the distribution and organization of F-actin (Fig 4).
Fig 4. Effect of CB and/or melatonin on the distribution and organization of F-actin.
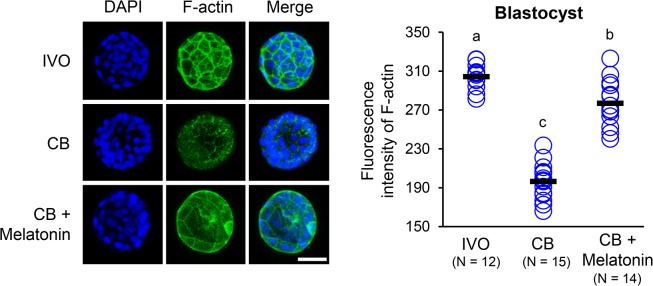
Immunofluorescence analysis to detect F-actin (green) in the blastocysts of CB (10 μg/mL) and/or melatonin-treated (10−9 M) groups. Nuclei (blue) are stained with DAPI. Bar, 50 μm. Each circle represents the fluorescence intensity in each embryo. Horizontal lines represent the mean values. The number of embryos in each group is indicated. Different superscript letters (a, b, c) indicate significant differences (P < 0.05).
Discussion
Our high-throughput RNA sequencing data suggested that an aberrant actin cytoskeleton organization might be a major factor contributing to the observed compromised in vitro embryonic development. Several previous studies have shown that IVP embryos undergo aberrant cytoskeletal organization. Long et al. showed that porcine IVP embryos were bundled or clumped, and had unevenly shaped cells [31]. Tremoleda et al. observed irregularly distributed microfilaments in horse IVP embryos [32]. Wang et al. also suggested that abnormal actin filament distribution might be a possible reason for the abnormal preimplantation development in IVP porcine embryos [33]. Functional analysis of DEGs using DAVID and STRING protein-PPI network clustering both revealed that many DEGs in the “actin cytoskeleton organization” and “regulation of actin filament-based processes” clusters were enriched. Furthermore, MGI analysis indicated that deficiency or dysregulation of these genes might lead to impaired embryonic development and mortality. For example, Bcl2 enhances actin polymerization [34], homozygous Bcl2-null mutants show oocyte maturation arrest and apoptosis induction during early embryonic development [35]. Palld is involved in modulating the actin cytoskeleton and in nervous system development [36], and its dysregulation leads to ventral closure defects and complete lethality throughout fetal growth and development [37]. Enah encodes an actin regulatory protein involved in the control of cell motility and adhesion. Deficiency of this gene has been shown to cause defects in the major axonal projection pathways of the brain [38]. Taken together, these results indicated that deficiency or dysregulation of genes involved in actin organization might be a major factor leading to impaired early embryonic development.
Our immunofluorescence analysis focused on the aberrant actin organization in IVP embryos. We found that F-actin was diffusely distributed in IVP embryos from the 8-cell stage onwards. Actin is a multifunctional protein that participates in many important cellular processes, such as cell division and cytokinesis, cell signaling, and the establishment and maintenance of cell junctions and cell shape [39–41]. Many of these processes are mediated by extensive and intimate interactions of actin with cellular membranes [42]. This diffuse distribution pattern of F-actin indicated that actin might not function properly in IVP embryos. Furthermore, quantification of the fluorescence intensity showed a significantly reduced abundance of F-actin in IVP embryos compared with IVO embryos. Studies have shown that deficiency or dysregulation of F-actin leads to a series of abnormal phenotypes, such as retarded embryogenesis, decreased body growth/size, and mortality/aging [43, 44]. Actin plays a crucial role in various dynamic events during oogenesis, fertilization, and embryonic development [45]; therefore, the decreased abundance as well as the suboptimal distribution, might adversely affect in vitro embryonic development. The detrimental effect of actin organization on early in vitro development was further confirmed by CB-induced actin disorganization, which dramatically decreased the developmental rate of IVP embryos.
Previous studies indicated that melatonin could improve the efficiency and quality of in vitro preimplantation development [15] and could increase the implantation rate after embryo transfer [46]. These beneficial effects of melatonin are frequently associated with its free radical scavenging and antiapoptotic abilities [19]. Considering the function of melatonin in actin organization, we questioned whether the beneficial effect of melatonin on in vitro embryonic development is dependent on the improved actin organization.
Our data demonstrated that melatonin could modulate actin organization in preimplantation embryos, which in turn might improve in vitro embryonic development. The results of the immunofluorescence analysis revealed that melatonin treatment improved the organization and distribution of F-actin, which mirrored the increased blastocyst rate. Moreover, the melatonin-mediated improvement of F-actin organization could be explained by the reversion to normal expression levels of dysregulated genes involved in actin organization (Arhgef2, Bcl2, Coro2b, Flnc, and Palld). It should also be mentioned that the effects of melatonin on actin organization are not mediated by membrane receptors. In a previous study, melatonin-induced microfilament rearrangement and microtubule enlargement was found to occur even in the presence of large concentrations of the MT1 and MT2 melatonin membrane receptor antagonist luzindole [47]. In addition, considering the fact that preimplantation embryos do not express these two melatonin membrane receptors [17, 48], we hypothesized that melatonin’s involvement in the modulation of actin organization is receptor independent.
To further confirm that actin disorganization is an essential factor that impairs the development of IVP embryos, we tested whether melatonin could alleviate CB-induced actin disorganization and rescue the impaired embryonic development. CB is a well-known specific inhibitor of actin filament polymerization that reduces the actin polymerization rate and inhibits the interaction of actin filaments in solution; however, CB has little effect on the rate of monomer addition and the rate of filament annealing [49]. When the in vitro culture medium was supplemented with CB alone, the blastocyst rate of IVP embryos significantly decreased, and the distribution of F-actin was severely disrupted. In contrast, the aberrant actin organization in IVP embryos after combined treatment with CB and melatonin appeared to be rescued, with a developmental rate comparable to IVP embryos. These results suggested that melatonin could restore the impaired embryonic development caused by actin disorganization, probably by improving actin organization.
Conclusions
Our study used high-throughput data, immunofluorescence and in vitro experiments using mouse embryos cultured with CB to study the effect of actin organization on IVP preimplantation embryos. Our results revealed that aberrant actin organization might contribute considerably to the impaired development of IVP preimplantation embryos. Furthermore, melatonin treatment improved actin organization and showed an ability to rescue this impaired development. Our results demonstrated that actin disorganization would be a reasonable target to optimize existing IVP systems. This study identified a promising strategy to improve the clinical ART process.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31472092), the Earmarked Fund for the Innovative Teams of Beijing Swine Industrialization Research Program, and the National High-Tech R&D Program (Nos. 2011AA100303, 2013AA102506).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (No. 31472092), the Earmarked Fund for the Innovative Teams of Beijing Swine Industrialization Research Program, and the National High-Tech R&D Program (Nos. 2011AA100303, 2013AA102506). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. European Society of Human Reproduction and Embryology, ART fact sheet Beigem, Belgium: European Society of Human Reproduction and Embryology; June 2014. Available: http://www.eshre.eu/Guidelines-and-Legal/ART-fact-sheet.aspx. Accessed 2015 Jan 12. [Google Scholar]
- 2. Delle Piane L, Lin W, Liu X, Donjacour A, Minasi P, Revelli A, et al. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod. 2010;25(8):2039–2046. 10.1093/humrep/deq165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giritharan G, Talbi S, Donjacour A, Di Sebastiano F, Dobson AT, Rinaudo PF. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction. 2007;134(1):63–72. [DOI] [PubMed] [Google Scholar]
- 4. Nie J, An L, Miao K, Hou Z, Yu Y, Tan K, et al. Comparative analysis of dynamic proteomic profiles between in vivo and in vitro produced mouse embryos during postimplantation period. J Proteome Res. 2013;12(9):3843–3856. 10.1021/pr301044b [DOI] [PubMed] [Google Scholar]
- 5. Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346(10):731–737. [DOI] [PubMed] [Google Scholar]
- 6. Rosenwaks Z, Bendikson K. Further evidence of the safety of assisted reproductive technologies. Proc Natl Acad Sci U S A. 2007;104(14):5709–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Driver AM, Penagaricano F, Huang W, Ahmad KR, Hackbart KS, Wiltbank MC, et al. RNA-Seq analysis uncovers transcriptomic variations between morphologically similar in vivo- and in vitro-derived bovine blastocysts. BMC Genomics. 2012;13:118 10.1186/1471-2164-13-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oliver VF, Miles HL, Cutfield WS, Hofman PL, Ludgate JL, Morison IM. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril. 2012;97(1):147–153 e147. 10.1016/j.fertnstert.2011.10.027 [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Cui J, Zhao C, Zhou L, Guo X, Shen R, et al. Differential expression of microRNAs in 2-cell and 4-cell mouse embryos. Zygote. 2013:1–7. [DOI] [PubMed]
- 10. Pinyopummintr T, Bavister BD. In vitro-matured/in vitro-fertilized bovine oocytes can develop into morulae/blastocysts in chemically defined, protein-free culture media. Biol Reprod. 1991;45(5):736–742. [DOI] [PubMed] [Google Scholar]
- 11. Biggers JD, McGinnis LK, Lawitts JA. One-step versus two-step culture of mouse preimplantation embryos: is there a difference? Hum Reprod. 2005;20(12):3376–3384. [DOI] [PubMed] [Google Scholar]
- 12. Ali AA, Bilodeau JF, Sirard MA. Antioxidant requirements for bovine oocytes varies during in vitro maturation, fertilization and development. Theriogenology. 2003;59(3–4):939–949. [DOI] [PubMed] [Google Scholar]
- 13. Abeydeera LR. In vitro production of embryos in swine. Theriogenology. 2002;57(1):256–273. [DOI] [PubMed] [Google Scholar]
- 14. Wang F, Tian X, Zhang L, Tan D, Reiter RJ, Liu G. Melatonin promotes the in vitro development of pronuclear embryos and increases the efficiency of blastocyst implantation in murine. J Pineal Res. 2013;55(3):267–274. 10.1111/jpi.12069 [DOI] [PubMed] [Google Scholar]
- 15. Wang F, Tian X, Zhang L, Gao C, He C, Fu Y, et al. Beneficial effects of melatonin on in vitro bovine embryonic development are mediated by melatonin receptor 1. J Pineal Res. 2014;56(3):333–342. 10.1111/jpi.12126 [DOI] [PubMed] [Google Scholar]
- 16. Abecia JA, Forcada F, Zuniga O. The effect of melatonin on the secretion of progesterone in sheep and on the development of ovine embryos in vitro. Vet Res Commun. 2002;26(2):151–158. [DOI] [PubMed] [Google Scholar]
- 17. Pang YW, An L, Wang P, Yu Y, Yin QD, Wang XH, et al. Treatment of porcine donor cells and reconstructed embryos with the antioxidant melatonin enhances cloning efficiency. J Pineal Res. 2013;54(4):389–397. 10.1111/jpi.12024 [DOI] [PubMed] [Google Scholar]
- 18. Tamura H, Takasaki A, Miwa I, Taniguchi K, Maekawa R, Asada H, et al. Oxidative stress impairs oocyte quality and melatonin protects oocytes from free radical damage and improves fertilization rate. J Pineal Res. 2008;44(3):280–287. 10.1111/j.1600-079X.2007.00524.x [DOI] [PubMed] [Google Scholar]
- 19. Galano A, Tan DX, Reiter RJ. On the free radical scavenging activities of melatonin's metabolites, AFMK and AMK. J Pineal Res. 2013;54(3):245–257. 10.1111/jpi.12010 [DOI] [PubMed] [Google Scholar]
- 20.Teodoro BG, Baraldi FG, Sampaio IH, Bomfim LH, Queiroz AL, Passos MA, et al. Melatonin prevents mitochondrial dysfunction and insulin resistance in rat skeletal muscle. J Pineal Res. 2014. [DOI] [PubMed]
- 21. Natarajan M, Reiter RJ, Meltz ML, Herman TS. Effect of melatonin on cell growth, metabolic activity, and cell cycle distribution. J Pineal Res. 2001;31(3):228–233. [DOI] [PubMed] [Google Scholar]
- 22. Sekaran S, Foster RG, Lucas RJ, Hankins MW. Calcium imaging reveals a network of intrinsically light-sensitive inner-retinal neurons. Curr Biol. 2003;13(15):1290–1298. [DOI] [PubMed] [Google Scholar]
- 23. Benitez-King G, Soto-Vega E, Ramirez-Rodriguez G. Melatonin modulates microfilament phenotypes in epithelial cells: implications for adhesion and inhibition of cancer cell migration. Histol Histopathol. 2009;24(6):789–799. [DOI] [PubMed] [Google Scholar]
- 24. Zhu ZY, Chen DY, Li JS, Lian L, Lei L, Han ZM, et al. Rotation of meiotic spindle is controlled by microfilaments in mouse oocytes. Biol Reprod. 2003;68(3):943–946. [DOI] [PubMed] [Google Scholar]
- 25. Nagy A. Manipulating the mouse embryo: a laboratory manual 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 26. Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17(1):1–14. [DOI] [PubMed] [Google Scholar]
- 27. Kuijk EW, van Tol LT, Van de Velde H, Wubbolts R, Welling M, Geijsen N, et al. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139(5):871–882. 10.1242/dev.071688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan X, Dai XX, Wang T, Liu HL, Sun SC. Melamine negatively affects oocyte architecture, oocyte development and fertility in mice. Hum Reprod. 2015. 10.1093/humrep/dev091 [DOI] [PubMed]
- 29. Ishizuka B, Kuribayashi Y, Murai K, Amemiya A, Itoh MT. The effect of melatonin on in vitro fertilization and embryo development in mice. J Pineal Res. 2000;28(1):48–51. [DOI] [PubMed] [Google Scholar]
- 30. Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. [DOI] [PubMed] [Google Scholar]
- 31. Long CR, Dobrinsky JR, Garrett WM, Johnson LA. Dual labeling of the cytoskeleton and DNA strand breaks in porcine embryos produced in vivo and in vitro. Mol Reprod Dev. 1998;51(1):59–65. [DOI] [PubMed] [Google Scholar]
- 32. Tremoleda JL, Stout TA, Lagutina I, Lazzari G, Bevers MM, Colenbrander B, et al. Effects of in vitro production on horse embryo morphology, cytoskeletal characteristics, and blastocyst capsule formation. Biol Reprod. 2003;69(6):1895–1906. [DOI] [PubMed] [Google Scholar]
- 33. Wang WH, Abeydeera LR, Han YM, Prather RS, Day BN. Morphologic evaluation and actin filament distribution in porcine embryos produced in vitro and in vivo. Biol Reprod. 1999;60(4):1020–1028. [DOI] [PubMed] [Google Scholar]
- 34. Ke H, Parron VI, Reece J, Zhang JY, Akiyama SK, French JE. BCL2 inhibits cell adhesion, spreading, and motility by enhancing actin polymerization. Cell Res. 2010;20(4):458–469. 10.1038/cr.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boumela I, Assou S, Aouacheria A, Haouzi D, Dechaud H, De Vos J, et al. Involvement of BCL2 family members in the regulation of human oocyte and early embryo survival and death: gene expression and beyond. Reproduction. 2011;141(5):549–561. 10.1530/REP-10-0504 [DOI] [PubMed] [Google Scholar]
- 36.Niedenberger BA, Chappell VA, Otey CA, Geyer C. Actin dynamics regulate subcellular localization of PALLD in mouse Sertoli cells. Reproduction. 2014. [DOI] [PubMed]
- 37. Luo H, Liu X, Wang F, Huang Q, Shen S, Wang L, et al. Disruption of palladin results in neural tube closure defects in mice. Mol Cell Neurosci. 2005;29(4):507–515. [DOI] [PubMed] [Google Scholar]
- 38. Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, et al. Mena is required for neurulation and commissure formation. Neuron. 1999;22(2):313–325. [DOI] [PubMed] [Google Scholar]
- 39. Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–288. [DOI] [PubMed] [Google Scholar]
- 41. Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, et al. Cellular control of cortical actin nucleation. Curr Biol. 2014;24(14):1628–1635. 10.1016/j.cub.2014.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. 10.1146/annurev.biophys.37.032807.125912 [DOI] [PubMed] [Google Scholar]
- 43. Shawlot W, Deng JM, Fohn LE, Behringer RR. Restricted beta-galactosidase expression of a hygromycin-lacZ gene targeted to the beta-actin locus and embryonic lethality of beta-actin mutant mice. Transgenic Res. 1998;7(2):95–103. [DOI] [PubMed] [Google Scholar]
- 44. Okamoto H, Nakae J, Kitamura T, Park BC, Dragatsis I, Accili D. Transgenic rescue of insulin receptor-deficient mice. J Clin Invest. 2004;114(2):214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun QY, Schatten H. Regulation of dynamic events by microfilaments during oocyte maturation and fertilization. Reproduction. 2006;131(2):193–205. [DOI] [PubMed] [Google Scholar]
- 46. Tian XZ, Wen Q, Shi JM, Liang W, Zeng SM, Tian JH, et al. Effects of melatonin on in vitro development of mouse two-cell embryos cultured in HTF medium. Endocr Res. 2010;35(1):17–23. 10.3109/07435800903539607 [DOI] [PubMed] [Google Scholar]
- 47. Mathes AM, Wolf B, Rensing H. Melatonin receptor antagonist luzindole is a powerful radical scavenger in vitro. J Pineal Res. 2008;45(3):337–338. 10.1111/j.1600-079X.2008.00583.x [DOI] [PubMed] [Google Scholar]
- 48. Gao C, Han HB, Tian XZ, Tan DX, Wang L, Zhou GB, et al. Melatonin promotes embryonic development and reduces reactive oxygen species in vitrified mouse 2-cell embryos. J Pineal Res. 2012;52(3):305–311. 10.1111/j.1600-079X.2011.00944.x [DOI] [PubMed] [Google Scholar]
- 49. MacLean-Fletcher S, Pollard TD. Mechanism of action of cytochalasin B on actin. Cell. 1980;20(2):329–341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.