Abstract
Microcephaly and macrocephaly are overrepresented in individuals with autism and are thought to be disease-related risk factors or endophenotypes. Analysis of DNA microarray results from a family with a low functioning autistic child determined that the proband and two additional unaffected family members who carry a rare inherited 760 kb duplication of unknown clinical significance at 19p13.12 are macrocephalic. Consideration alongside overlapping deletion and duplication events in the literature provides support for a strong relationship between gene dosage at this locus and head size, with losses and gains associated with microcephaly (p=1.11x10-11) and macrocephaly (p=2.47x10-11), respectively. Data support A kinase anchor protein 8 and 8-like (AKAP8 and AKAP8L) as candidate genes involved in regulation of head growth, an interesting finding given previous work implicating the AKAP gene family in autism. Towards determination of which of AKAP8 and AKAP8L may be involved in the modulation of head size and risk for disease, we analyzed exome sequencing data for 693 autism families (2591 individuals) where head circumference data were available. No predicted loss of function variants were observed, precluding insights into relationship to head size, but highlighting strong evolutionary conservation. Taken together, findings support the idea that gene dosage at 19p13.12, and AKAP8 and/or AKAP8L in particular, play an important role in modulation of head size and may contribute to autism risk. Exome sequencing of the family also identified a rare inherited variant predicted to disrupt splicing of TPTE / PTEN2, a PTEN homologue, which may likewise contribute to both macrocephaly and autism risk.
Introduction
The autism spectrum disorders are defined by impairments in social communication and restricted or stereotypical interests or behaviors. According to the Center for Disease Control and Prevention, these disorders are common, with a reported prevalence of 1/68 [1]. Genetic analyses have identified a large and growing number of genes and loci that are known to be risk associated [2,3], however, it is now well recognized that the relationship between genetic variation and clinical diagnosis is inexact. In fact, many variants first identified in autism have been observed subsequently in individuals with intellectual disability (ID) and schizophrenia [4].
Many have sought to leverage this apparent blurring of disease boundaries by seeking out loci that map better onto disease-related traits. This has been met with success in studies of social cognition [5,6] and language performance [7–9]. Another such autism-related trait is head size. Larger than average heads were observed by Leo Kanner in his initial description of children with autism [10], and subsequent work has found additional support for involvement [11–13]. Reported rates of macrocephaly in the autism population are 14–24% [11,14–16], while reported rates of microcephaly vary from 3–15% [11,14,15]. Although the link between extreme head size and autism is clearly established, how macrocephaly and microcephaly may contribute to autism risk remains unclear. Some believe that people with autism and macrocephaly or autism and microcephaly represent different autism subgroups [11,14,17]. Furthermore, it is hypothesized that macrocephaly and microcephaly may arise from defects in neural progenitor proliferation and/or synaptic pruning, which in turn can affect brain growth and brain structure [18,19].
Many genes have been shown to be associated with extreme head size and autism with one of the most well established genes being PTEN. Mutations in PTEN have been associated with autism and macrocephaly [20], and it is currently estimated that ~5% of individuals with autism have a mutation in PTEN [21]. Rare mutations in other autism related genes such as CNTNAP2 and CHD8 are also associated with both autism and macrocephaly [22,23],while mutations in the risk genes AUTS2 and DHCR7 give rise to autism and microcephaly [24–26]. Another established neurodevelopmental disease risk loci, 16p11.2, causes a mirror effect with regards to gene dosage and head size. Patients with a 16p11.2 deletion often have macrocephaly while duplication patients often have microcephaly [27]. A similar phenomenon was seen within the 1q21.1 risk locus, whereby deletions and duplications give rise to microcephaly and macrocephaly, respectively [28]. Furthermore, a relationship between dosage and head size is observed in individuals with autism associated structural variants at 17q12 and 22q13.3 (Phelan-McDermid Syndrome), where deletions at both loci are associated with macrocephaly [29,30].
We present here a patient with autism and ID with an inherited duplication at 19p13.12 and discuss the relationship between altered gene dosage at this locus, involving AKAP8 and/or AKAP8L in particular, and each of head size and autism. Additionally, we identified a rare maternally inherited variant in TPTE / PTEN2, a PTEN homologue, via exome sequencing of the proband and his family and propose that disruption of this gene may also contribute to regulation of head size and perhaps autism risk.
Materials and Methods
Ethics Statement
The Tg64 family was enrolled with written informed consent through the Department of Genetics at Albert Einstein College of Medicine, Bronx, NY (Internal Review Board approved program, protocol #20110456). For minors enrolled in this study, informed written consent was obtained via a parent. All consent forms are stored in a locked filing cabinet and uploaded online to a HIPPA compliant password protected database. This consent procedure was executed as written in our Internal Review Board approved program. The individuals in this manuscript have given written informed consent (as outlined in PLOS consent form) to publish these case details.
Array CGH and FISH Confirmation
Genomic DNA was isolated from blood using the Puregene Genomic DNA Purification kit (Gentra, Minneapolis, MN) and aCGH performed in the context of a clinical evaluation using a custom 44k Agilent array (Santa Clara, CA). Probes were placed at 5–10 kb intervals in subtelomeric, pericentromeric, and known microdeletion/microduplication regions, providing ~50 kb resolution. Additional probes were placed every 50–100 kb across the remaining euchromatic genome resulting in a ~500 kb resolution. Samples were labeled and hybridized, washed, and scanned (CGH Analytic 3.5.14 software) according to the manufacturer’s protocol. FISH confirmation of the duplication was done by Quest Diagnostics (Madison, NJ) using the DNA probe RP11-637P24.
Additional Clinical Genetic Testing
Standard karyotyping (550–650 band resolution) was carried out by G banding by Quest Diagnostics on fixed cells. A subtelomeric FISH assay was also performed by Quest Diagnostics with ToTelVysion sub-telomeric probes (Abbot Molecular, Abbot Park, IL). Fragile X testing was performed by Quest Diagnostics via Southern blot hybridization. MELAS (mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) and NARP (neuropathy, ataxia, and retinitis pigmentosa) testing were performed at Columbia University (NY, NY) by PCR and RFLP analysis of the mitochondrial genes MTTL1 (NC_012920.1:m.3243A>G, dbSNP:rs199474657) and MTATP6 (NC_012920.1:m.[8993T>G; 8993T>C], dbSNP:rs199476133) for MELAS and NARP, respectively. JNCL (juvenile neuronal ceroid lipofuscinosis) testing was performed at IBR-Specialty Clinical Laboratories (Staten Island, NY) via PCR testing for a 1.02 kb deletion within the CLN3 gene (NC_000016.10:g.28485965_28486930del966).
Affy6.0 Arrays
Genomic DNA was isolated from blood using the Puregene Genomic DNA Purification kit as described above. Affymetrix 6.0 microarrays (946 000 CNV and 906 600 SNP probes) were processed according to manufacturer protocols (www.affymetrix.com). Arrays were washed on an Affymetrix fluidics station and scanned using a GeneChip Scanner 3000 7G; image intensities were extracted as. CEL files. Quality control, genotype calling, and copy number analysis were done using the Affymetrix Genotyping Console (v4.1.4). The Birdseed algorithm (v2) was used for genotype calling and the CN5 algorithm was used for copy number analysis. Data was visualized using Affymetrix’s Chromosome Analysis Suite (ChAS) Software (v2.1).
Exome Sequencing
Genomic DNA was isolated from blood using the Puregene Genomic DNA Purification kit as described above. Libraries were then generated according to the manufacturer’s instructions using the TruSeq DNA Sample Preparation kit (Illumina, San Diego, CA). Capture was performed using the NimbleGen SeqCap EZ Human Exome v2.0 kit (Roche, Basel, Switzerland), which targets 34 Mb corresponding to ~180 000 coding exons / 20 000 genes. Paired-end sequencing was performed using an Illumina HiSeq2000 instrument to generate 76 base pair reads. Sequence analysis was performed in the Albert Einstein Computational Core Facility. Briefly, raw sequence data were aligned to the human reference genome (Hg19) using BWA (v0.5.9) [31,32], and PCR duplicates were eliminated with Picard MarkDuplicates (v1.45) (http://broadinstitute.github.io/picard/). Local re-alignment and base quality recalibration was then carried out using GATK (v1.5) [33,34]. BAM files were merged by Picard and GATK was used for SNP/INDEL detection. This was followed by the determination of minor allele frequencies via dB SNP (v131) and 1000 Genomes Project (v2010.12 release) and prediction of the functional impact of nonsynonymous variants by BLOSUM62, Polyphen2, and SIFT [35–38]. Variants with a read depth ≥10 were considered for review. Results from a meta-analysis of separate exome sequencing data for 2591 individuals from the Simons Simplex Collection [39] (693 carefully phenotyped families with a single autistic child in each) were also reviewed for predicted loss of function variants within AKAP8 and AKAP8L. For this dataset, only variants with a read depth ≥20 were considered for review.
Validation of Exome Sequencing Variants
Validation of variants identified in exome sequencing was done by performing Sanger sequencing using an ABI 3730 on PCR products generated using the following primers:
ARGHAP11A (chr15:32920998; hg19):
F- TGTGAAGTTGTAATTGCTTATGCC
R- TTGAACTATTTTCACACGCTTA
ARHGAP11A (chr15:32927988; hg19):
F- TCCCTTTTTAAATCAGCTAAAGATT
R- TCTGGGCTAAAAAGCAAACC
TPTE / PTEN2 (chr21:10910401–10910402; hg19):
F-TTTTTTAGCATCTTGACTTTGTG
R-GTCTCAGAAAACAAAAAGCAAATGT
Variant Submission
Copy number and sequence level variants identified and confirmed in the Tg64 family have been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/); Accession numbers SCV000212157, SCV000212158, and SCV000212159 correspond to variants in ARHGAP11A (NP_055598.1:p.[Arg311Ile];[Ser452Cys], TPTE / PTEN2 (NM_199261.3:c.1357-3_1357-2delTA), and the 19p13.12 duplication, respectively.
Normalization of Head Circumference
Published reference data were used to convert head circumference (measured in centimeters) into standardized scores. Age and gender based distributions were used for individuals under 18 years of age [40] and height and gender based norms used for individuals over 18 years of age [41].
Subject Photographs
A 3dMDface System (Atlanta, Georgia) was used to capture subject photographs as described elsewhere [42,43]. Images were viewed in the 3dMDVultusViewer.
Literature and DECIPHER Database Search
Additional overlapping cases were found in the literature using the search term “19p13.12”. Overlapping cases in DECIPHER were found using the search term “chr19:15051982–15809751”. Only cases where head circumference data were available or mention of clinical evaluation of head size were included in our analyses.
Statistical Analysis
To determine the likelihood of seeing frequencies of microcephaly and macrocephaly in individuals with structural variants at 19p13.12 by chance alone, we compared observed counts to the expected (one-tailed) binomial distributions. Calculations were based on the assumption that head circumference is normally distributed within a population.
Results
Clinical Report
A 3.5-year-old male of African and European descent (Tg64.001), the second child born in an unremarkable pregnancy to healthy non-consanguineous parents, was seen at the Children’s Hospital at Montefiore. He presented with autism, developmental delay, and macrocephaly. Supraventricular tachycardia was observed at birth but later resolved. At age 11, the proband was additionally given a diagnosis of ID. At 15 years old, an MRI revealed maldevelopment of the posterior cerebral white matter and a CT head scan revealed optic atrophy.
Molecular Findings
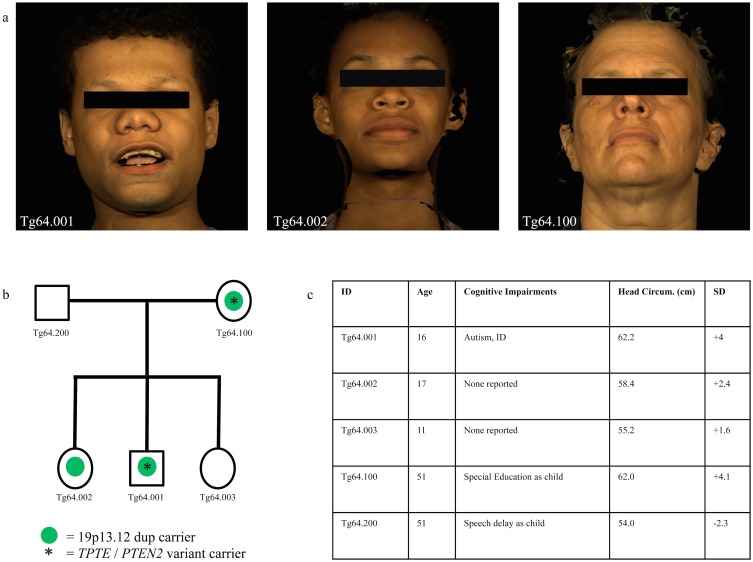
Standard karyotyping (550 band resolution), subtelomeric FISH, and testing for Fragile X, MELAS, NARP, and JNCL syndromes revealed no abnormalities in the proband. A custom 44k Agilent array, however, identified a 15 gene 645 kb duplication of unknown clinical significance at 19p13.12 (Fig 1). The duplication was confirmed by FISH and familial testing identified the identical duplication in the proband’s mother (Tg64.100), and in one of his two sisters (Tg64.002). Breakpoints were subsequently refined using an Affy 6.0 SNP array (Fig 1) to a 19 gene 760 kb region (chr19:15051982–15809751; hg19). Reexamination of available clinical data revealed macrocephaly not only in the carrier proband, but also in the two additional familial carriers, consistent with a relationship between macrocephaly and duplication status (Fig 2). Although the proband’s carrier mother was enrolled in special education classes during her schooling, his carrier sister (now in college) did not encounter such academic difficulties.
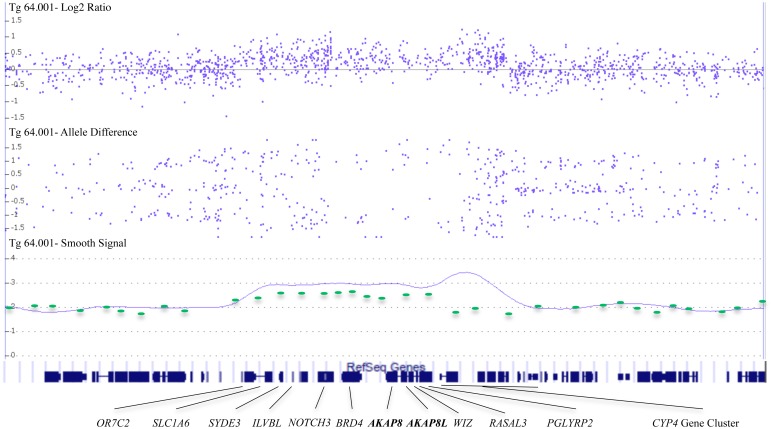
Fig 1. Chr19p13.12 duplication breakpoints.
Results from an Affy 6.0 SNP array of Tg64.001 reveal a 760 kb duplication at 19p13.12. The top panel displays the log2 ratio of normalized intensity. The middle panel shows the difference of A allele signal and B allele signal. The bottom panel displays a Gaussian smoothed copy number estimate. Intensity values for probes included on the 44k Agilent array used in initial clinical evaluation are overlaid in green ovals.
Fig 2. Characterization the of Tg64 family.
(a) 3dMD photos of all Tg64 duplication carriers: proband (left), carrier sister (middle), carrier mother (right). (b) Carrier status of Tg64 family members for the gain observed at 19p13.12 (chr19:15051982–15809751; hg19) and sequence variant at TPTE / PTEN2 (NM_199261.3:c.1357-3_1357-2delTA). (c) Summary of head circumference measurements and developmental concerns for Tg64 family members.
Beyond the carriers described here, examination of the literature [44–50] and the DECIPHER database [51] identified an additional 16 individuals harboring overlapping copy number variants. As summarized in Table 1, review of clinical data for all available cases suggests a gene dosage effect whereby 8 of 11 individuals with losses are microcephalic (<2 SD) (p = 1.11x10-11) and 7 of 8 individuals harboring gains are macrocephalic (>2 SD) (p = 2.47x10-11). Two non-microcephalic deletion carriers are in the 10th centile or lower for head size [46,49]. No relationship to carrier status, however, was observed for either DECIPHER ID 257523 (duplication) or 265764 (deletion) where microcephaly and normal head size were observed, respectively.
Table 1. Summary of cytogenetic and clinical findings in cases with overlapping events at 19p13.12.
| Case ID | Event Type | Coordinates (Hg19) | Size (Mb) | Inheritance | Gender | Head Size | Developmental Status |
|---|---|---|---|---|---|---|---|
| DECIPHER 257523 | Gain | 12.84–15.93 | 3.09 | de novo | M | Micro | Intellectual Disability |
| Gallant et al., 2011 | Loss | 13.90–16.52 | 2.62 | de novo | F | Micro | Unavailable |
| DECIPHER 285763 | Loss | 13.93–16.32 | 2.39 | de novo | F | Micro | Intellectual Disability |
| DECIPHER 283124 | Loss | 13.93–16.32 | 2.39 | de novo | F | Micro | Intellectual Disability |
| DECIPHER 284366 | Gain | 13.99–24.30 | 10.31 | Unknown | F | Macro | Global Developmental Delay |
| Engels et al., 2007/Bonaglia et al. 2010 | Loss | 14.10–16.67 | 2.57 | Unknown | F | Micro | Intellectual Disability |
| Bonaglia et al., 2010 | Loss | 14.27–16.19 | 1.92 | Mat. Inherited | M | 5–10 centile | Intellectual Disability |
| Van der Aa et al., 2010/DECIPHER 255743 | Loss | 14.38–15.49 | 1.11 | de novo | M | Micro at birth | Intellectual Disability |
| DECIPHER 249355 | Loss | 14.66–15.66 | 1.00 | Unknown | F | Micro | Unavailable |
| DECIPHER 249428 | Gain | 15.05–16.03 | 0.98 | Inherited-parental origin unknown | M | Macro | Intellectual Disability |
| Sanders et al., 2011 | Gain | 15.05–15.89 | 0.84 | de novo | M | Macro | Unavailable |
| Tg64.100 (Carrier Mother) | Gain | 15.05–15.81 | 0.76 | Unknown | F | Macro | Special Education as Child |
| Tg64.002 (Carrier Sib) | Gain | 15.05–15.81 | 0.76 | Mat. Inherited | F | Macro | Typically Developing |
| Tg64.001 (index) | Gain | 15.05–15.81 | 0.76 | Mat. Inherited | M | Macro | Intellectual Disability |
| DECIPHER 265764 | Loss | 15.05–16.24 | 1.19 | de novo | M | Normal | Intellectual Disability |
| Jelsig et al., 2012 | Loss | 15.18–16.62 | 1.44 | de novo | M | Micro | Intellectual Disability |
| DECIPHER 250827 | Gain | 15.18–16.46 | 1.28 | de novo | F | Macro | Intellectual Disability |
| DECIPHER 255839 | Loss | 15.19–16.62 | 1.43 | de novo | M | Micro | Intellectual Disability |
| Kosaki et al., 2011 | Loss | 15.44–16.20 | 0.76 | de novo | F | 10th centile | Mild Developmental Delay |
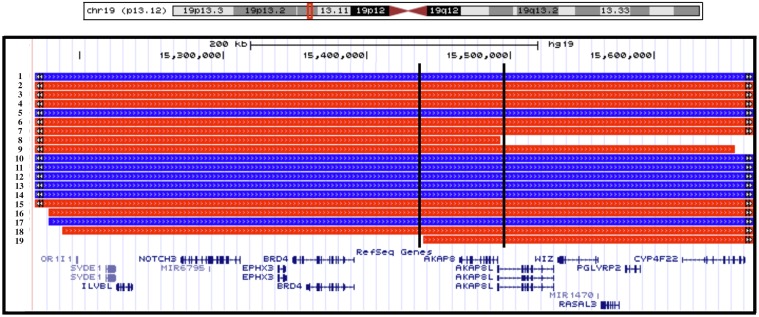
Comparison of breakpoints in carrier individuals point to a 54 kb critical region (chr19:15439339–15492848; hg19) consisting of two genes: AKAP8 and AKAP8L (Fig 3). Although the number of informative cases is small, this relationship of gene dosage and head size was observed in individuals of different genders and ethnicities. Almost all cases with deletions or duplications that span the locus are reported to have some degree of developmental delay (15/16 informative cases), with many having ID (12/16 informative cases), although neither was observed in Tg64.002.
Fig 3. Summary of cases with copy number variants at 19p13.12 in relation to RefSeq genes.
Black bars represent the critical interval breakpoints. Deletions and duplications are presented in red and blue, respectively. Cases are encoded as follows: 1- DECIPHER 257523; 2- Gallant et al., 2011; 3- DECIPHER 285763; 4- DECIPHER 283124; 5- DECIPHER 284366; 6- Engels et al., 2007/Bonaglia et al., 2010; 7- Bonaglia et al., 2010; 8- Van der Aa et al., 2010/DECIPHER 255743; 9- DECIPHER 249355; 10- DECIPHER 249428; 11- Sanders et al., 2011; 12- Tg64.100; 13- Tg64.002; 14- Tg64.001; 15- DECIPHER 265764; 16- Jelsig et al., 2012; 17- DECIPHER 250827; 18- DECIPHER 255839; 19- Kosaki et al., 2011.
In an attempt to explore the possible role of AKAP8 and AKAP8L in the modulation of head size, we looked for stop gains and splice site mutations in exome sequencing data from 2591 individuals where head size was available. These analyses were non informative, as no events likely to be deleterious were observed in either gene. The absence of any disruptive variants in this large number of individuals is, however, consistent with strong evolutionary pressure on both genes. Additionally, interrogation of the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/homev.20141016release) [52] failed to identify any exonic duplications overlapping AKAP8 or 8L, but did show three deletions, a 1900 bp event across AKAP8, a 183 kb event spanning AKAP8 and AKAP8L, and a 1.8 Mb even spanning AKAP8, AKAP8L, and 4 other genes in the region. Although head size and other phenotypic information for individuals in which these events were identified is unknown, these data show that structural variants at the locus are rare in the general population. Lastly, GWASs for brain and intracranial volume on data from more than 15,000 people failed to see any common variant signal within this region [53,54].
Towards isolation of one or more additional variants that could better explain the clinical presentation of Tg64.001, exome sequencing was performed on the proband, mother, and father. Consideration of rare alleles (MAF < 1% based on a combination of European, African, and Asian ancestry data from the 1000 Genomes Project) predicted to be disruptive by at least two in silico prediction tools as well as splicing, frameshift, and non-frameshift insertion/deletion variants identified thirteen variants in twelve genes (S1 Table). Among these were two putatively deleterious variants—one inherited from each parent—in ARHGAP11A (NP_055598.1:p.[Arg311Ile];[Ser452Cys], dbSNP: rs372419991 and rs146176251, respectively) on 15q13.3, a gene within the RhoGAP family. Other RhoGAP family members have been implicated in autism and ID [55–58], making it an excellent candidate gene. Sanger sequencing of all family members confirmed the presence of both variants in the proband, but also found each to be present in his younger unaffected sibling (Tg64.003) ruling out any straightforward relationship to disease.
Also identified through exome sequencing was an apparent de novo variant in TPTE / PTEN2 (NM_199261.3:c.1357-3_1357-2delTA). Interestingly, TPTE / PTEN2 is a PTEN homologue, a gene in which mutations give rise to autism with macrocephaly [21,59,60]. Attempts at validation by Sanger sequencing determined that this rare variant in TPTE / PTEN2 was not in fact de novo, but rather maternally inherited (Fig 2b). However, consistent with functionality, the observed allele is predicted to disrupt a canonical splice acceptor site and result in the elimination of 31 amino acids from a conserved C2 domain (pfam10409). Moreover, no variant at this position in TPTE / PTEN2 was observed in the Exac database [61], a compendium of exome sequence data for more than 60,000 individuals, although the same allele we observed was identified in the 1000 Genomes Project at a frequency of 1/500 (rs149363218) (S2 Table). Consistent with a possible role for variation in TPTE / PTEN2 in regulation of head size is the fact that the proband and his mother who each carry the gain encompassing AKAP8 / AKAP8L and the TPTE / PTEN2 variant predicted to disrupt splicing have heads 1.4 SD and 1.5 SD greater than the macrocephalic sibling who carries only the gain. Exome sequencing provided no other obvious candidates to account for the proband’s clinical presentation.
Discussion
We describe a male with autism, ID, and macrocephaly. He and two non-autistic family members have a 760 kb duplication on 19p13.12 and macrocephaly. Results presented here suggest that gene dosage at this locus, and the AKAP8 and/or AKAP8L genes in particular, is strongly associated with head size. A clear positive relationship between 19p13.12 dosage and head size is observed in 17/19 informative cases. While no such relationship was seen in two carriers with overlapping copy number variants, a number of genes are thought to play a role in head size [62–64], and so it is likely there are additional variants in these individuals acting as modifiers.
AKAP8 and AKAP8L are anchoring proteins known to regulate chromatin condensation during mitosis [65,66]. AKAP8 has been shown to interact with various cyclins that regulate G1 phase and G1/S transition [67,68], and disruption of AKAP8 in vitro inhibits DNA replication initiation [69]. Knockdown of AKAP8 and AKAP8L together causes a G2/M arrest and aberrant chromosomal morphology [70]. Also relevant is that both AKAP8 and AKAP8L are highly expressed in the human fetal brain, most prominently in transient forebrain structures. Like other microcephaly genes [64], AKAP8 is present at high levels in the ventricular and subventricular zones, areas where the birth and division of progenitor cells occur [71]. These data suggest that altered AKAP8 and/or AKAP8L dosage may impact head size through regulation of cellular proliferation in the brain. Consistent with involvement in disease, AKAP8 has been shown to interact physically with CC2D1A, an ID causing gene [72]. Moreover, a recent meta-analysis of autism GWAS data implicates the AKAP gene family in disease and suggests that many family members are targets of autism-implicated miRNAs [73].
The identification of a rare maternally inherited variant in TPTE / PTEN in the proband is intriguing given the well-established role for PTEN in autism. Previous work suggests that ~5% of children with autism carry germline mutations in PTEN [21], with all of these patients showing macrocephaly. TPTE / PTEN2 expression was initially thought to be testis specific [74], however, more recent analyses shows expression in multiple structures in human fetal brain [71]. Although little is known about TPTE / PTEN2 function, in vitro enzymatic analyses show that this protein acts as a phosphatase to remove the 3-phosphate from substrates overlapping with those modified by PTEN [75]. Moreover, the specific variant identified here is predicted to disrupt a conserved C2 domain (pfam10409) that in PTEN is involved in the interaction with phospholipid membranes and the suppression of glioblastoma cell growth [76]. If this C2 domain acts in a similar fashion in TPTE / PTEN2, disruption may serve to accelerate cellular proliferation in brain development. Although phenotypic characterization of additional individuals who harbor rare variants in TPTE / PTEN2 is of course required, the proband and his mother who carry both the duplication encompassing AKAP8 / AKAP8L and the TPTE/ PTEN2 predicted splice variant have heads much larger (1.4 and 1.5 SD, respectively) than the macrocephalic sibling who carries only the duplication. This suggests rare variation in TPTE / PTEN2 may be involved in biological processes underlying head growth and perhaps autism pathogenesis.
Our report, together with data from others, provides strong support for a positive relationship between gene dosage at 19p13.12 and head size where gene dosage gives rise to mirror phenotypes. Effects may be mediated by AKAP8 and/or AKAP8L. Additionally, there may be a contributory role for gene dosage at this region to autism. Our results are consistent with the idea that genetic variation at 19p13.12 can play an important role in modulation of disease without being directly associated with diagnosis. Our data also suggest that TPTE / PTEN2 may be involved in regulation of head size and perhaps autism risk.
Supporting Information
Exome sequencing was performed on samples from the proband (Tg64.001), his mother (Tg64.100), and his father (Tg64.200). Variants with a read depth ≥10, a minor allele frequency <1% (based a combination of European, African, and Asian ancestry data from the 1000 Genomes Project), and with potential for functional impact are listed.
(XLSX)
(XLSX)
Acknowledgments
This study makes use of data generated by the DECIPHER Consortium funded by the Wellcome Trust. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. We also thank the Molecular Cytogenetics Core at Einstein for DNA extraction from blood, the Genomics Core at Einstein for running the Affy 6.0 SNP arrays, the Epigenomics Core at Einstein for performing exome sequencing, and the Computational Genetics Core at Einstein for analysis of exome sequence data. Special thanks also to Deqiong Ma, Rizwan Naeem, Joy Samanich, and Bernice E. Morrow (Einstein / Montefiore), Stephan J. Sanders and Matt W. State (UCSF), Jesse Knowles (3dMD), and the Tg64 family who made this work possible.
Data Availability
All relevant data are within the paper and its Supporting Information files. Confirmed copy number and sequence level variants have been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
Funding Statement
This work was supported through an Albert Einstein New Investigator Development Award, an IDDRC pilot award (P30HD071593; National Institute of Child Health Development, http://www.nichd.nih.gov/Pages/index.aspx), and an Autism Center of Excellence Network Award (9R01 MH100027; National Institute of Mental Health; http://www.nimh.nih.gov/index.shtml) to BSA. Support for RAN was provided through the Training Program in Cellular and Molecular Biology and Genetics (T32 GM007491; National Institute of General Medical Sciences; http://www.nigms.nih.gov/Pages/default.aspx). Clinical recruitment efforts were supported by CTSA Grants (UL1RR025750, KL2RR025749, and TL1RR025748; National Center for Research Resources; http://www.nih.gov/about/almanac/organization/NCRR.htm and 8UL1 TR000086; National Center for Advancing Translational Sciences; http://www.ncats.nih.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators, Centers for Disease Control and Prevention (CDC). Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014. March 28;63(2):1–21. [PubMed] [Google Scholar]
- 2. Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism. 2013;4(1):36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014. February;15(2):133–41. 10.1038/nrg3585 [DOI] [PubMed] [Google Scholar]
- 4. Guilmatre A, Dubourg C, Mosca A-L, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009. September;66(9):947–56. 10.1001/archgenpsychiatry.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu AT-H, Yoon J, Geschwind DH, Cantor RM. QTL replication and targeted association highlight the nerve growth factor gene for nonverbal communication deficits in autism spectrum disorders. Molecular Psychiatry. 2013. February;18(2):226–35. 10.1038/mp.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skuse DH, Lori A, Cubells JF, Lee I, Conneely KN, Puura K, et al. Common polymorphism in the oxytocin receptor gene (OXTR) is associated with human social recognition skills. Proc Natl Acad Sci USA. 2014. February 4;111(5):1987–92. 10.1073/pnas.1302985111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitehouse AJO, Bishop DVM, Ang QW, Pennell CE, Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes, Brain and Behavior. 2011. June;10(4):451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008. November 27;359(22):2337–45. 10.1056/NEJMoa0802828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, Association, and Gene-Expression Analyses Identify CNTNAP2 as an Autism-Susceptibility Gene. The American Journal of Human Genetics. 2008. January;82(1):150–9. 10.1016/j.ajhg.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanner L, Asperger H, Eisenberg L, Mosse HL, Bender L, Eveloff HH, et al. Library of the History of Autism Research, Behaviorism & Psychiatry. Nervous Child. 1943;2:217–50. [Google Scholar]
- 11. Deutsch CK, Joseph RM. Brief report: cognitive correlates of enlarged head circumference in children with autism. J Autism Dev Disord. 2003. April;33(2):209–15. [DOI] [PubMed] [Google Scholar]
- 12. Chaste P, Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, et al. Adjusting head circumference for covariates in autism: clinical correlates of a highly heritable continuous trait. Biological Psychiatry. 2013. October 15;74(8):576–84. 10.1016/j.biopsych.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stevenson RE, Schroer RJ, Skinner C, Fender D, Simensen RJ. Autism and macrocephaly. Lancet. 1997. June 14;349(9067):1744–5. [DOI] [PubMed] [Google Scholar]
- 14. Miles JH, Hadden LL, Takahashi TN, Hillman RE. Head circumference is an independent clinical finding associated with autism. Am J Med Genet. 2000. December 11;95(4):339–50. [PubMed] [Google Scholar]
- 15. Fombonne E, Rogé B, Claverie J, Courty S, Frémolle J. Microcephaly and macrocephaly in autism. J Autism Dev Disord. 1999. April;29(2):113–9. [DOI] [PubMed] [Google Scholar]
- 16. Lainhart JE, Bigler ED, Bocian M, Coon H, Dinh E, Dawson G, et al. Head circumference and height in autism: a study by the Collaborative Program of Excellence in Autism. Am J Med Genet. 2006. November 1;140(21):2257–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sacco R, Militerni R, Frolli A, Bravaccio C, Gritti A, Elia M, et al. Clinical, morphological, and biochemical correlates of head circumference in autism. Biological Psychiatry. 2007. November 1;62(9):1038–47. [DOI] [PubMed] [Google Scholar]
- 18. McCaffery P, Deutsch CK. Macrocephaly and the control of brain growth in autistic disorders. Progress in Neurobiology. 2005. September;77(1–2):38–56. [DOI] [PubMed] [Google Scholar]
- 19. Wollnik B. News and views. Nature Publishing Group. Nature Publishing Group; 2010. November 1;42(11):923–4. 10.1038/ng1110-923 [DOI] [PubMed] [Google Scholar]
- 20. Butler MG. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. Journal of Medical Genetics. 2005. April 1;42(4):318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genetics in medicine: official journal of the American College of Medical Genetics. 2013. pp. 399–407. 10.1038/gim.2013.32 [DOI] [PubMed] [Google Scholar]
- 22. Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006. March 30;354(13):1370–7. [DOI] [PubMed] [Google Scholar]
- 23. Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell. Elsevier Inc; 2014. July 17;158(2):263–76. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagamani SCS, Erez A, Ben-Zeev B, Frydman M, Winter S, Zeller R, et al. Detection of copy-number variation in AUTS2gene by targeted exonic array CGH in patients with developmental delay and autistic spectrum disorders. Nature Publishing Group; 2012. August 8;21(3):343–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and Regulation of AUTS2, a Gene Implicated in Autism and Human Evolution. Noonan J, editor. PLoS Genet. 2013. January 17;9(1):e1003221 10.1371/journal.pgen.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryan AK, Bartlett K, Clayton P, Eaton S, Mills L, Donnai D, et al. Smith-Lemli-Opitz syndrome: a variable clinical and biochemical phenotype. Journal of Medical Genetics. 1998. July;35(7):558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shinawi M, Liu P, Kang SHL, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. Journal of Medical Genetics. 2010. May 7;47(5):332–41. 10.1136/jmg.2009.073015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008. November 23;40(12):1466–71. 10.1038/ng.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, et al. AR TICLEDeletion 17q12 Is a Recurrent Copy Number Variant that Confers High Risk of Autism and Schizophrenia. The American Journal of Human Genetics. The American Society of Human Genetics; 2010. November 12;87(5):618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarasua SM, Dwivedi A, Boccuto L, Chen C-F, Sharp JL, Rollins JD, et al. 22q13.2q13.32 genomic regions associated with severity of speech delay, developmental delay, and physical features in Phelan–McDermid syndrome. Genet Med. 2013. October 17;16(4):318–28. 10.1038/gim.2013.144 [DOI] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009. July 15;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010. March 1;26(5):589–95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010. September;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Publishing Group. 2011. May;43(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE. A method and server for predicting damaging missense mutations. Nature. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eddy SR. Where did the BLOSUM62 alignment score matrix come from? Nat Biotechnol. 2004. August;22(8):1035–6. [DOI] [PubMed] [Google Scholar]
- 37. Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002. March;12(3):436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sunyaev S, Ramensky V, Koch I. Prediction of deleterious human alleles. Human molecular …. 2001. [DOI] [PubMed] [Google Scholar]
- 39. He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9(8):e1003671 10.1371/journal.pgen.1003671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roche AF, Mukherjee D, Guo S, Moore WM. Head circumference reference data: birth to 18 years. 1987. [PubMed] [Google Scholar]
- 41. Bushby KM, Cole T, Matthews JN, Goodship JA. Centiles for adult head circumference. Archives of Disease in Childhood. 1992. October 1;67(10):1286–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Littlefield TR, Kelly KM, Cherney JC, Beals SP, Pomatto JK. Development of a new three-dimensional cranial imaging system. J Craniofac Surg. 2004. January;15(1):175–81. [DOI] [PubMed] [Google Scholar]
- 43. Hutton TJ, Buxton BF, Hammond P, Potts HWW. Estimating average growth trajectories in shape-space using kernel smoothing. IEEE Trans Med Imaging. 2003. June;22(6):747–53. [DOI] [PubMed] [Google Scholar]
- 44. Gallant NM, Baldwin E, Salamon N, Dipple KM, Quintero-Rivera F. Pontocerebellar hypoplasia in association with de novo 19p13.11p13.12 microdeletion. Am J Med Genet. 2011. October 12;155(11):2871–8. 10.1002/ajmg.a.34286 [DOI] [PubMed] [Google Scholar]
- 45. Engels H, Brockschmidt A, Hoischen A, Landwehr C, Bosse K, Walldorf C, et al. DNA microarray analysis identifies candidate regions and genes in unexplained mental retardation. Neurology. 2007. March 6;68(10):743–50. [DOI] [PubMed] [Google Scholar]
- 46. Bonaglia MC, Marelli S, Novara F, Commodaro S, Borgatti R, Minardo G, et al. Genotype-phenotype relationship in three cases with overlapping 19p13.12 microdeletions. Eur J Hum Genet. 2010. December;18(12):1302–9. 10.1038/ejhg.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van der Aa N, Vandeweyer G, Kooy RF. A boy with mental retardation, obesity and hypertrichosis caused by a microdeletion of 19p13.12. European Journal of Medical Genetics. Elsevier Masson SAS; 2010. September 10;53(5):291–3. [DOI] [PubMed] [Google Scholar]
- 48. Jelsig AM, Brasch-Andersen C, Kibæk M, Fagerberg CR. A case of microdeletion of 19p13 with intellectual disability, hypertrichosis, synophrys, and protruding front teeth. European Journal of Medical Genetics. 2012. October;55(10):564–7. 10.1016/j.ejmg.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 49. Kosaki K, Saito H, Kosaki R, Torii C, Kishi K, Takahashi T. Branchial arch defects and 19p13.12 microdeletion: Defining the critical region into a 0.8 M base interval. Am J Med Genet. 2011. August 3;155(9):2212–4. 10.1002/ajmg.a.33908 [DOI] [PubMed] [Google Scholar]
- 50. Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. Elsevier Inc; 2011. June 9;70(5):863–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. The American Journal of Human Genetics. 2009. April;84(4):524–33. 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Research. 2014. January;42(Database issue):D986–92. 10.1093/nar/gkt958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. Nature Publishing Group; 2012. April 15;44(5):552–61. 10.1038/ng.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ikram MA, Fornage M, Smith AV, Seshadri S, Schmidt R, Debette S, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012. April 15;44(5):539–44. 10.1038/ng.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Billuart P, Bienvenu T, Ronce N, Portes des V, Vinet MC, Zemni R, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998. April 30;392(6679):923–6. [DOI] [PubMed] [Google Scholar]
- 56. Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, et al. The novel Rho-GTPase activating gene MEGAP/srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci USA. National Acad Sciences; 2002;99(18):11754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010. July 15;466(7304):368–72. 10.1038/nature09146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sakai Y, Shaw CA, Dawson BC, Dugas DV, Al-Mohtaseb Z, Hill DE, et al. Protein Interactome Reveals Converging Molecular Pathways Among Autism Disorders. Science Translational Medicine. 2011. June 8;3(86):86ra49–9. 10.1126/scitranslmed.3002166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008. May;9(5):341–55. 10.1038/nrg2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abrahams BS, Geschwind DH. Genetics of autism. Springer; 2010;:699–714. [Google Scholar]
- 61.Exome Aggregation Consortium (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org) [9 December 2014].
- 62. Mochida GH, Walsh CA. Molecular genetics of human microcephaly. Current Opinion in Neurology. 2001. April 1;14(2):151 [DOI] [PubMed] [Google Scholar]
- 63. Williams CA, Dagli A, Battaglia A. Genetic disorders associated with macrocephaly. Journal of Medical Genetics. 2008. [DOI] [PubMed] [Google Scholar]
- 64. Chae TH, Walsh CA. Genes that control the size of the cerebral cortex. Cortical development: genes and genetic …. 2007. [DOI] [PubMed] [Google Scholar]
- 65. Collas P, Le Guellec K, Tasken K. The A-kinase-anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. The Journal of Cell Biology. 1999. December 13;147(6):1167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martins SB, Eide T, Steen RL, Jahnsen T, Skålhegg BS, Collas P. HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. Journal of Cell Science. 2000. November;113 Pt 21:3703–13. [DOI] [PubMed] [Google Scholar]
- 67. Arsenijevic T, Degraef C, Dumont JE, Roger PP, Pirson I. A novel partner for D-type cyclins: protein kinase A-anchoring protein AKAP95. Biochem J. 2004. March 1;378(Pt 2):673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arsenijevic T, Degraef C, Dumont JE, Roger PP, Pirson I. G1/S Cyclins interact with regulatory subunit of PKA via A-kinase anchoring protein, AKAP95. Cell Cycle. 2006. June;5(11):1217–22. [DOI] [PubMed] [Google Scholar]
- 69. Eide T, Tasken KA, Carlson C, Williams G, Jahnsen T, Tasken K, et al. Protein Kinase A-anchoring Protein AKAP95 Interacts with MCM2, a Regulator of DNA Replication. Journal of Biological Chemistry. 2003. July 18;278(29):26750–6. [DOI] [PubMed] [Google Scholar]
- 70. Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes & Development. 2006. September 15;20(18):2566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.BrainSpan: Atlas of the Developing Human Brain [Internet]. Funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01, and 1RC2MH089929-01. © 2011. Available from: http://developinghumanbrain.org.
- 72. Al-Tawashi A, Jung SY, Liu D, Su B, Qin J. Protein implicated in nonsyndromic mental retardation regulates protein kinase A (PKA) activity. Journal of Biological Chemistry. 2012. April 27;287(18):14644–58. 10.1074/jbc.M111.261875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Poelmans G, Franke B, Pauls DL, Glennon JC, Buitelaar JK. AKAPs integrate genetic findings for autism spectrum disorders. Transl Psychiatry. 2013;3:e270 10.1038/tp.2013.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen H, Rossier C, Morris MA, Scott HS, Gos A, Bairoch A, et al. A testis-specific gene, TPTE, encodes a putative transmembrane tyrosine phosphatase and maps to the pericentromeric region of human chromosomes 21 and 13, and to chromosomes 15, 22, and Y. Hum Genet. 1999. November;105(5):399–409. [DOI] [PubMed] [Google Scholar]
- 75. Wu Y, Dowbenko D, Pisabarro MT, Dillard-Telm L, Koeppen H, Lasky LA. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J Biol Chem. 2001. June 15;276(24):21745–53. [DOI] [PubMed] [Google Scholar]
- 76. Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999. October 29;99(3):323–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exome sequencing was performed on samples from the proband (Tg64.001), his mother (Tg64.100), and his father (Tg64.200). Variants with a read depth ≥10, a minor allele frequency <1% (based a combination of European, African, and Asian ancestry data from the 1000 Genomes Project), and with potential for functional impact are listed.
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Confirmed copy number and sequence level variants have been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).