Abstract
How an amino acid sequence encodes the information necessary for a protein to adopt a unique tertiary structure remains unresolved. We are addressing this problem by designing "from scratch" protein molecules that will adopt predetermined three-dimensional structures. Based on this strategy, two identical four-stranded beta-sheets were designed to dimerize and form a beta-sandwich protein, called betadoublet. A synthetic gene encoding half the beta-sandwich protein was expressed in Escherichia coli, and the protein was purified to homogeneity. Biophysical characterization of betadoublet in aqueous solution demonstrated that the disulfide formed between the two sheets and that the dimer was a compact unaggregated globular protein, consisting predominantly of beta-sheet and stable to thermal denaturation. It has some backbone amide protons whose exchange is slow enough to be measured by NMR but binds more of the dye 1-anilinonaphthalene-8-sulfonate than a well-folded protein.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allured V. S., Collier R. J., Carroll S. F., McKay D. B. Structure of exotoxin A of Pseudomonas aeruginosa at 3.0-Angstrom resolution. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1320–1324. doi: 10.1073/pnas.83.5.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becktel W. J., Schellman J. A. Protein stability curves. Biopolymers. 1987 Nov;26(11):1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Beta-turns in proteins. J Mol Biol. 1977 Sep 15;115(2):135–175. doi: 10.1016/0022-2836(77)90094-8. [DOI] [PubMed] [Google Scholar]
- DeGrado W. F., Wasserman Z. R., Lear J. D. Protein design, a minimalist approach. Science. 1989 Feb 3;243(4891):622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Wilcox W., Eshita S. M., Pryciak P. M., Ho S. P., DeGrado W. F. The design, synthesis, and crystallization of an alpha-helical peptide. Proteins. 1986 Sep;1(1):16–22. doi: 10.1002/prot.340010105. [DOI] [PubMed] [Google Scholar]
- Fedorov A. N., Dolgikh D. A., Chemeris V. V., Chernov B. K., Finkelstein A. V., Schulga A. A., Alakhov YuB, Kirpichnikov M. P., Ptitsyn O. B. De novo design, synthesis and study of albebetin, a polypeptide with a predetermined three-dimensional structure. Probing the structure at the nanogram level. J Mol Biol. 1992 Jun 20;225(4):927–931. doi: 10.1016/0022-2836(92)90092-x. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Hahn K. W., Klis W. A., Stewart J. M. Design and synthesis of a peptide having chymotrypsin-like esterase activity. Science. 1990 Jun 22;248(4962):1544–1547. doi: 10.1126/science.2360048. [DOI] [PubMed] [Google Scholar]
- Handel T. M., Williams S. A., DeGrado W. F. Metal ion-dependent modulation of the dynamics of a designed protein. Science. 1993 Aug 13;261(5123):879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Richardson J. S., Richardson D. C., Ogden R. C. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990 Aug 24;249(4971):884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Jr Protein secondary structure and circular dichroism: a practical guide. Proteins. 1990;7(3):205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- Kamtekar S., Schiffer J. M., Xiong H., Babik J. M., Hecht M. H. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993 Dec 10;262(5140):1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- Mutter M. Nature's rules and chemist's tools: a way for creating novel proteins. Trends Biochem Sci. 1988 Jul;13(7):260–265. doi: 10.1016/0968-0004(88)90159-4. [DOI] [PubMed] [Google Scholar]
- Pace C. N. Conformational stability of globular proteins. Trends Biochem Sci. 1990 Jan;15(1):14–17. doi: 10.1016/0968-0004(90)90124-t. [DOI] [PubMed] [Google Scholar]
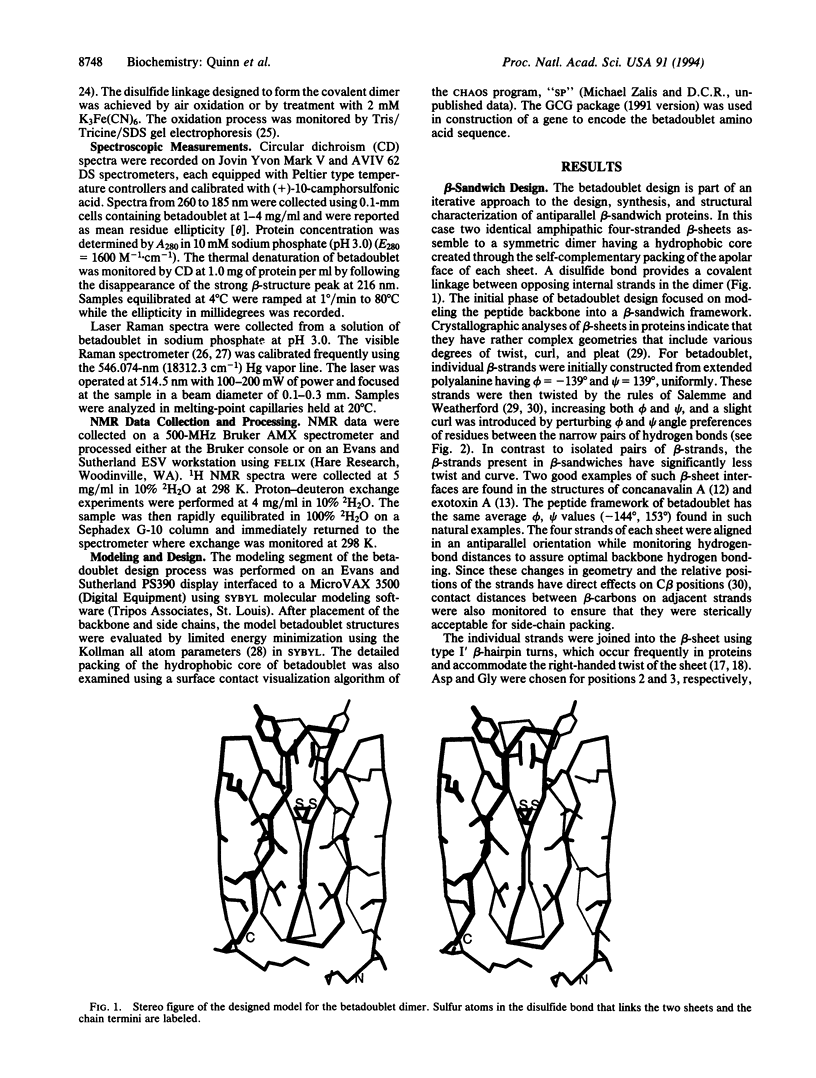
- Poljak R. J., Amzel L. M., Chen B. L., Phizackerley R. P., Saul F. The three-dimensional structure of the fab' fragment of a human myeloma immunoglobulin at 2.0-angstrom resolution. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3440–3444. doi: 10.1073/pnas.71.9.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Reeke G. N., Jr, Becker J. W., Edelman G. M. The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure. J Biol Chem. 1975 Feb 25;250(4):1525–1547. [PubMed] [Google Scholar]
- Regan L., DeGrado W. F. Characterization of a helical protein designed from first principles. Science. 1988 Aug 19;241(4868):976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C., Tweedy N. B., Gernert K. M., Quinn T. P., Hecht M. H., Erickson B. W., Yan Y., McClain R. D., Donlan M. E. Looking at proteins: representations, folding, packing, and design. Biophysical Society National Lecture, 1992. Biophys J. 1992 Nov;63(5):1185–1209. [PMC free article] [PubMed] [Google Scholar]
- Richardson J., Thomas K. A., Rubin B. H., Richardson D. C. Crystal structure of bovine Cu,Zn superoxide dismutase at 3 A resolution: chain tracing and metal ligands. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1349–1353. doi: 10.1073/pnas.72.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Ellman's reagent: 5,5'-dithiobis(2-nitrobenzoic acid)--a reexamination. Anal Biochem. 1979 Apr 1;94(1):75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- Salemme F. R. Structural properties of protein beta-sheets. Prog Biophys Mol Biol. 1983;42(2-3):95–133. doi: 10.1016/0079-6107(83)90005-6. [DOI] [PubMed] [Google Scholar]
- Salemme F. R., Weatherford D. W. Conformational and geometrical properties of beta-sheets in proteins. II. Antiparallel and mixed beta-sheets. J Mol Biol. 1981 Feb 15;146(1):119–141. doi: 10.1016/0022-2836(81)90369-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
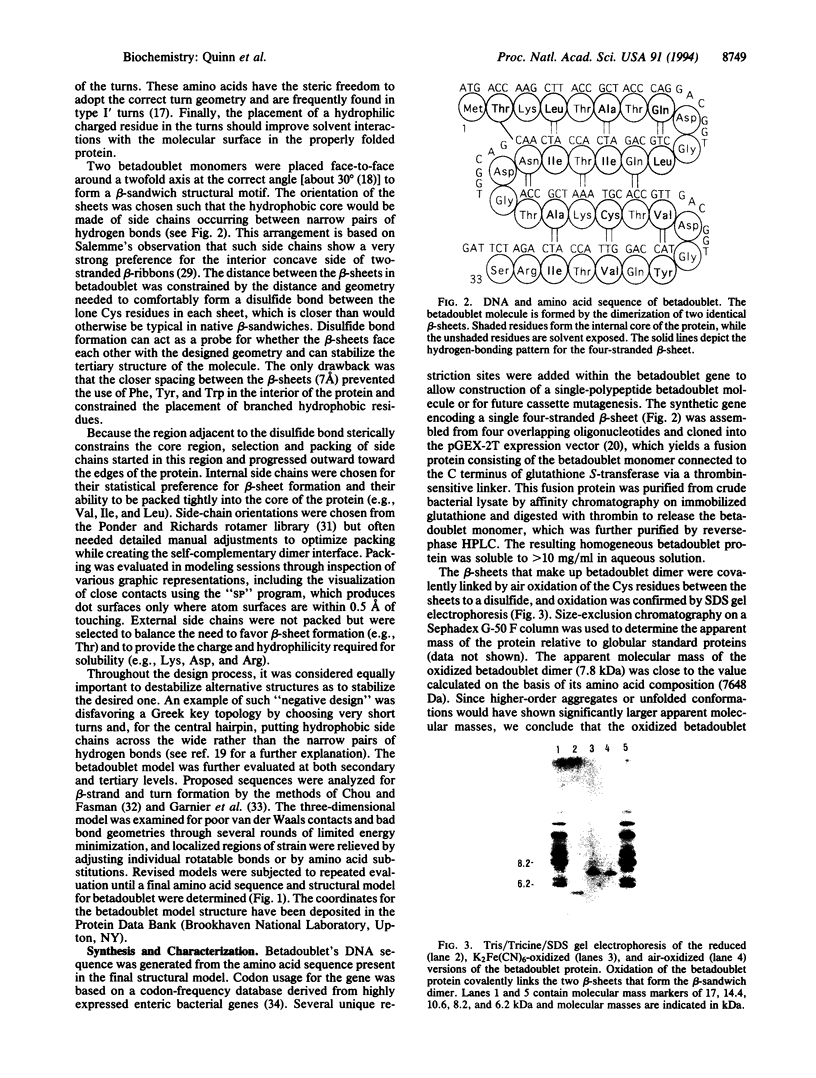
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas' A. F., Gilmanshin R. I. Study of the "molten globule" intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991 Jan;31(1):119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- Sibanda B. L., Thornton J. M. Beta-hairpin families in globular proteins. Nature. 1985 Jul 11;316(6024):170–174. doi: 10.1038/316170a0. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Williams R. W., Beeler T. J. Secondary structure of calsequestrin in solutions and in crystals as determined by Raman spectroscopy. J Biol Chem. 1986 Sep 15;261(26):12408–12413. [PubMed] [Google Scholar]
- Williams R. W. Protein secondary structure analysis using Raman amide I and amide III spectra. Methods Enzymol. 1986;130:311–331. doi: 10.1016/0076-6879(86)30016-8. [DOI] [PubMed] [Google Scholar]






