Abstract
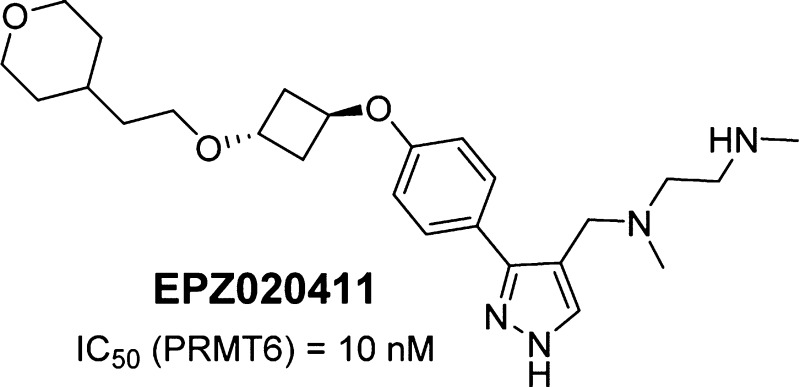
A novel aryl pyrazole series of arginine methyltransferase inhibitors has been identified. Synthesis of analogues within this series yielded the first potent, selective, small molecule PRMT6 inhibitor tool compound, EPZ020411. PRMT6 overexpression has been reported in several cancer types suggesting that inhibition of PRMT6 activity may have therapeutic utility. Identification of EPZ020411 provides the field with the first small molecule tool compound for target validation studies. EPZ020411 shows good bioavailability following subcutaneous dosing in rats making it a suitable tool for in vivo studies.
Keywords: PRMT6, protein methyltransferase, oncology, tool compound
PRMT6 is a member of the protein arginine methyltransferase (RMT) family, which comprises 45 enzymes, nine of which are known to catalyze protein arginine N-methylation reactions. These post-translational modifications are important regulators of RNA processing, transcriptional regulation, signal transduction, and other cellular processes.1,2
PRMT6 is a nuclear-localized RMT capable of creating omega-N(G)-monomethylarginine and asymmetric omega-N(G),N(G)-dimethylarginine derivatives on histone and other protein substrates containing a GAR motif;3 it is the only RMT known to methylate the H3R2 mark.4,5 This mark can act in opposition to the activating H3K4me3 mark, effectively acting as a transcriptional repressor.6
PRMT6 has been reported to play a role in a variety of cellular processes including maintenance of stem cell pluripotency,7 regulation of cell cycle,8 DNA repair,9 regulation of nuclear receptor-mediated transcription,10 and viral transactivation.11 PRMT6 overexpression has been reported in several cancer types including melanoma12 and bladder, lung,13 and prostate14 carcinoma, suggesting that inhibition of PRMT6 may have therapeutic utility and supporting development of small molecule inhibitors for use as tool compounds for in vitro and in vivo target validation studies.
An aryl pyrazole bearing a diamine side-chain, 1, was found to have potent PRMT1, PRMT6, and PRMT8 inhibitory activity through screening of the Epizyme internal library.
A 2.4 Å resolution crystal structure of a ternary complex of 1, SAH, and PRMT6 was obtained and is shown in Figure 1a,b (4Y2H). The diamine side-chain occupies the putative site of the substrate arginine side-chain. The terminal nitrogen atom is 3.4 Å away from the sulfur atom of SAH. The terminal NH2 group makes multiple direct hydrogen bonds to the Glu155 side-chain and backbone carbonyl and water-mediated hydrogen bond interactions with the backbone carbonyl of Trp156 and the side-chain of Glu164. In addition, the His317 side-chain makes a hydrogen bond to the tertiary amine of the diamine side-chain. The pyrazole makes a hydrogen bond to the side chain of Glu59. The aryl ring of 1 makes π–π interactions with the side chains of Glu64 and Tyr159. This extensive set of interactions, in addition to numerous van der Waals interactions between the protein and 1, is consistent with low nanomolar potency for this compound. In addition, as shown in Figure 1b, there is more space in the pocket for expansion of the ligand beyond the para-position of the aryl group; hence, this region was further explored.
Figure 1.
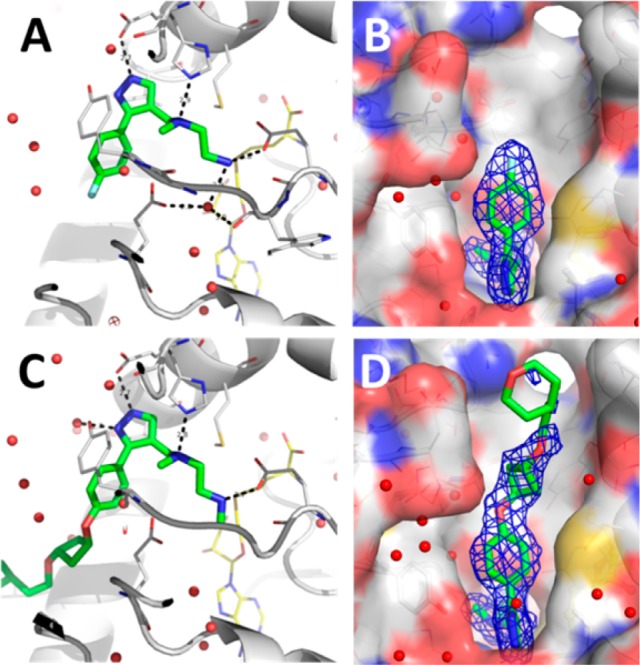
Crystal structures of 1 (A) and EPZ020411 (C) (green) showing SAH (yellow), and 2Fo – Fc electron density maps for ligands 1 (B) and EPZ020411 (D) at 1σ (blue mesh).
A series of analogues of compound 1 was synthesized,15 which retained balanced PRMT1, PRMT6, and PRMT8 activity (compounds 1–11, Table 1).
Table 1. Compounds with Balanced PRMT6/8/1 (1–11) or Decreased PRMT6 (12–15) Activity.
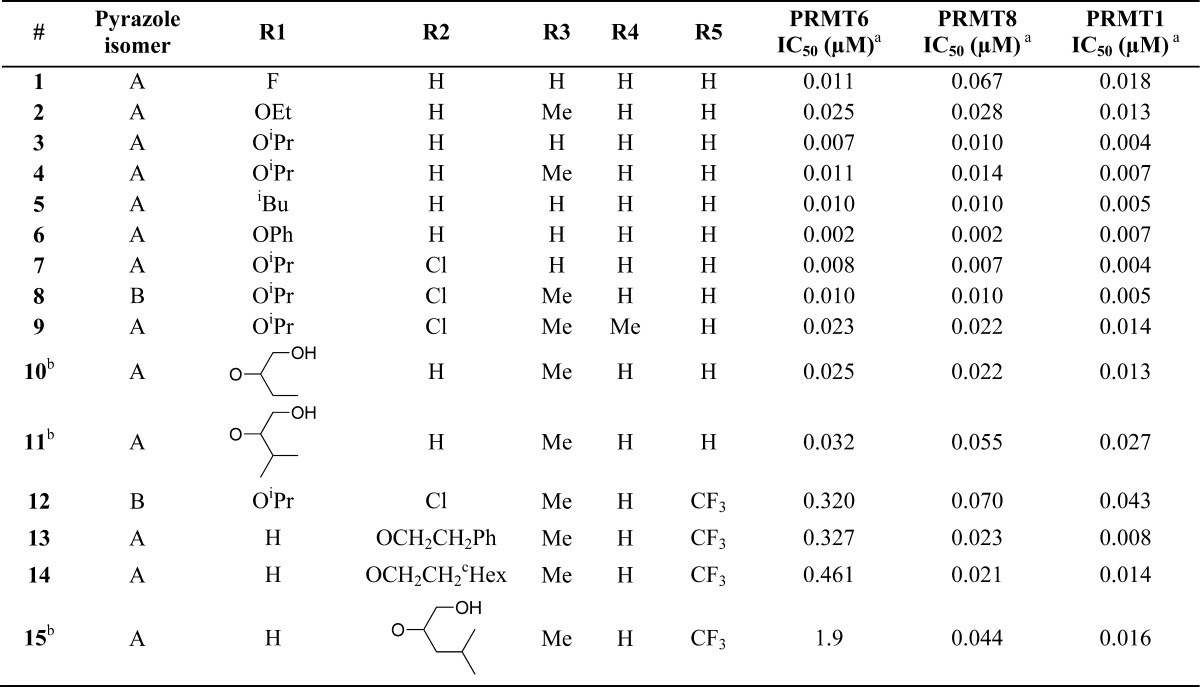
IC50s were determined from at least two independent experiments, see Supporting Information for details.
Compound is an undetermined mixture of isomers at the chiral center.
Replacement of the R1 fluorine in compound 1 with oxygen gave a synthetic handle with which to explore the unoccupied space extending off the para-position of the aryl group. Activity against PRMT1, PRMT6, and PRMT8 was retained as the fluorine was replaced with ethers of increasing size; ethoxy (2), isopropoxy (3), and phenoxy (6).
Exploration of SAR around the isopropoxy analogue 3 showed that similar activity was retained when chlorine was introduced into the ortho-position (7) and with the regioisomeric pyrazole core (8). The pyrazole cores could also be N-methylated in the 2-position (9) with minimal change in activities.
Polarity is tolerated in the para-position as illustrated by compounds 10 and 11. The oxygen linker was not necessary for activity and could be converted to carbon as in compound 5. The terminal primary amine of the original hit 1 was subject to N-acetylation in vivo, which could be blocked by N-methylation with retention of activity as in compound pairs 3 → 4 and 7 → 8.16
PRMT6 activity could be reduced through di-meta substitution of the aryl ring, in particular by introduction of a trifluoromethyl group in one of the meta-positions (compounds 12–15, Table 1). For example, introduction of a meta-trifluoromethyl group to the isopropoxy chloro analogue 8 gave decreased activity against PRMT6 compared to PRMT1 and PRMT8, compound 12.
Removal of the substituent in the para-position, 13 and 14, gave a slight potency improvement against PRMT1 and PRMT8 while retaining decreased PRMT6 activity. Introduction of polarity into the meta-side chain further decreased PRMT6 activity (15).
Analogues with selectivity for PRMT6 over both PRMT1 and PRMT8 were obtained by extending further off the para-position of the aryl group (Table 2). Increased potency for PRMT6 compared to PRMT1 and PRMT8 was achieved with oxygen- and nitrogen-linked alkyl compounds EPZ020411 and 17/18. Single digit nanomolar inhibitory activity at PRMT6 with selectivity over PRMT1 and PRMT8 was obtained with phenoxy analogues 19–21.
Table 2. PRMT6 Selective Compounds.
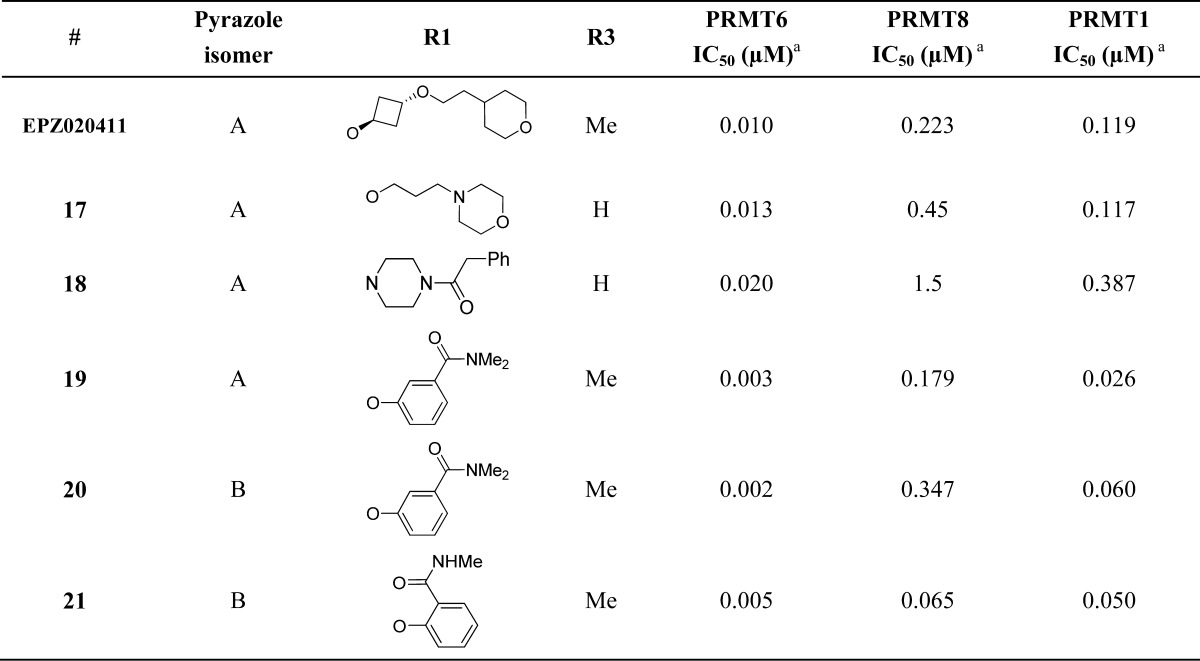
IC50s were determined from at least two independent experiments; see Supporting Information for details.
Further characterization, including PK, of selected analogues identified EPZ020411 to have a profile suitable for use as a PRMT6 inhibitor tool compound. In biochemical assays EPZ020411 was over 100-fold selective for PRMT6/8/1 compared to other histone methyltransferases including four arginine methyltransferases (PRMT3, PRMT4, PRMT5, and PRMT7).
A 2.1 Å resolution crystal structure of the tool compound, EPZ020411, with SAH and PRMT6 is shown in Figure 1c,d (4Y30). The interactions between EPZ020411 and PRMT6 are quite similar to those observed for compound 1 with the majority of the interactions occurring with the diamine side-chain and the pyrazole core. Since the R3 substituent is a methyl instead of a hydrogen in this case, the hydrogen bonds to both the Glu155 backbone carbonyl and water are lost, but the methyl picks up additional van der Waals interactions that are not present when R3 is a hydrogen atom. The entirety of the oxygen linked alkyl side chain is not well ordered in the structure with electron density not observed for the terminal tetrahydropyran group.
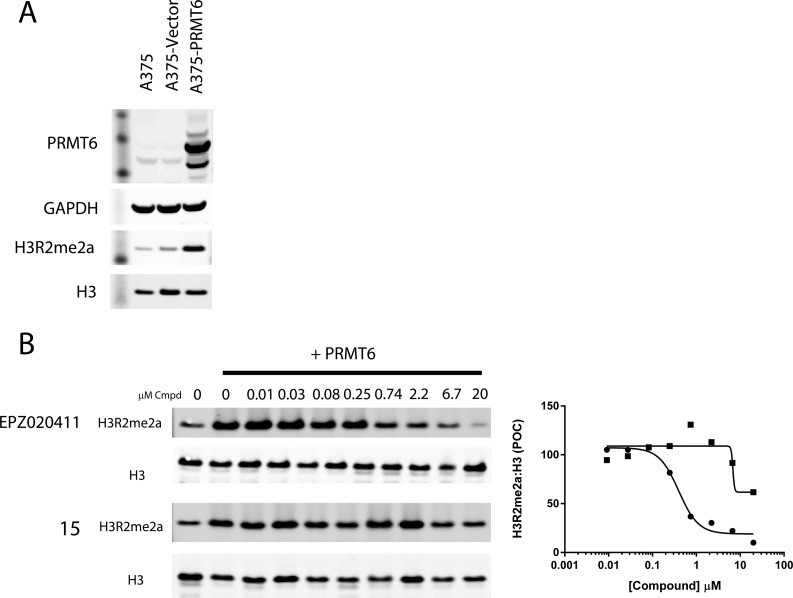
The PRMT6 cellular activity of compounds 15 and EPZ020411 was tested in an engineered model in which PRMT6 was transiently expressed in an A375 cell line background. Notably, compound treatment did not affect levels of PRMT6 expression (Supplemental Figure 3). Selective methylation of the PRMT6 substrate H3R2 was robustly induced upon 48 h of PRMT6 expression as shown in Figure 2A. Treatment with EPZ020411 resulted in a dose-dependent decrease in H3R2 methylation (IC50 = 0.637 ± 0.241 μM), while treatment with the PRMT6-inactive compound 15 did not generate an IC50 at concentrations up to 20 μM (Figure 2). In order to determine the cellular activity of EPZ020411 against PRMT1, monomethyl R*GG was quantified, which has previously been demonstrated to be selectively modulated by PRMT1 and not PRMT6.17 As seen in Supplemental Figure 4, EPZ020411 had a >10-fold less potent effect on this PRMT1-specific methylmark than was seen on the PRMT6-mediated H3R2 methylmark, consistent with the biochemical potencies of EPZ020411 on these two enzymes.
Figure 2.
(A) H3R2 methylation is induced by 48 h overexpression of PRMT6, but not an empty vector, in A375 cells. (B) PRMT6-induced H3R2 methylation is inhibited by 48 h exposure to EPZ020411 (circles, IC50 = 0.637 ± 0.241 μM) but not by 15 (squares, IC50 > 20 μM).
EPZ020411 had a free fraction of 0.51 ± 0.01 and 0.52 ± 0.02 in rat and human plasma, respectively. The compound showed poor permeability in the parallel artificial membrane permeation assay (PAMPA; 0.1 × 10–6 cm/s), in line with the observation of low bioavailability after oral dosing in rats (<5%, data not shown). Pharmacokinetic (PK) studies in rats were also performed by intravenous (i.v.) bolus and subcutaneous (s.c.) administration. Blood PK parameters derived from noncompartmental analysis are displayed in Table 3. Male Sprague–Dawley rats administered a single dose of EPZ020411 at 1 mg/kg by i.v. bolus showed a moderate clearance (CL) of 19.7 ± 1.0 mL/min/kg, with a volume of distribution at steady state (Vss) of 11.1 ± 1.6 L/kg, translating to a mean terminal half-life (t1/2) of 8.54 ± 1.43 h. Following 5 mg/kg s.c. dosing, a good bioavailability of 65.6 ± 4.3% was observed, leading to EPZ020411 unbound blood concentration remaining above the PRMT6 biochemical IC50 value for more than 12 h.
Table 3. Pharmacokinetic Parameters for EPZ020411 Following i.v. and s.c. Bolus Administration to Sprague-Dawley Rats; Expressed as Mean ± SD, n = 3.
| parameter | 1 mg/kg i.v. | 5 mg/kg s.c. |
|---|---|---|
| CL (mL/min/kg) | 19.7 ± 1.0 | |
| Vss (L/kg) | 11.1 ± 1.6 | |
| t1/2 (h) | 8.54 ± 1.43 | 9.19 ± 1.60 |
| tmax (h) | 0.444 | |
| Cmax (ng/mL) | 844 ± 306 | |
| AUC0-τ (h·ng/mL) | 745 ± 34 | 2456 ± 135 |
| AUC0-inf (h·ng/mL) | 846 ± 45 | 2775 ± 181 |
| F (%) | 65.6 ± 4.3 |
In conclusion, we report here the first potent, selective, small molecule PRMT6 inhibitors (EPZ020411 and 17–21). Further characterization of EPZ020411 showed it to have good bioavailability following subcutaneous dosing in rats making it a suitable tool for potential in vivo target validation studies.
Glossary
ABBREVIATIONS
- CL
clearance
- DMSO
dimethyl sulfoxide
- i.v.
intraveneous
- Ni-NTA
nickel-nitrilotriacetic acid
- PAMPA
parallel artificial membrane permeation assay
- PK
pharmacokinetic
- PRMT
protein arginine methyltransferase
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- s.c.
subcutaneous
- Vss
volume of distribution at steady state
Supporting Information Available
ADME/PK methods, H3R2 cell assay conditions, biochemical assay conditions, crystallography methods and data, and synthesis and data for EPZ020411. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): All authors are stockholders of Epizyme, Inc.
Supplementary Material
References
- Bedford M. T.; Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18 (3), 263–72. [DOI] [PubMed] [Google Scholar]
- Richon V. M.; Johnston D.; Sneeringer C. J.; Jin L.; Majer C. R.; Elliston K.; Jerva L. F.; Scott M. P.; Copeland R. A. Chemogenetic analysis of human protein methyltransferases. Chem. Biol. Drug Des 2011, 78 (2), 199–210. [DOI] [PubMed] [Google Scholar]
- Frankel A.; Yadav N.; Lee J.; Branscombe T. L.; Clarke S.; Bedford M. T. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002, 277 (5), 3537–43. [DOI] [PubMed] [Google Scholar]
- Hyllus D.; Stein C.; Schnabel K.; Schiltz E.; Imhof A.; Dou Y.; Hsieh J.; Bauer U. M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007, 21 (24), 3369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neault M.; Mallette F. A.; Vogel G.; Michaud-Levesque J.; Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012, 40 (19), 9513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E.; Bassi C.; Casadio F.; Martinato F.; Cesaroni M.; Schuchlautz H.; Lüscher B.; Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449 (7164), 933–7. [DOI] [PubMed] [Google Scholar]
- Lee Y. H.; Ma H.; Tan T. Z.; Ng S. S.; Soong R.; Mori S.; Fu X. Y.; Zernicka-Goetz M.; Wu Q. Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells Dev. 2012, 21 (14), 2613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt M. A.; de Graaf P.; van Teeffelen H. A.; Timmers H. T. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One 2012, 7 (8), e41446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Andaloussi N.; Valovka T.; Toueille M.; Steinacher R.; Focke F.; Gehrig P.; Covic M.; Hassa P. O.; Schär P.; Hübscher U.; Hottiger M. O. Arginine methylation regulates DNA polymerase beta. Mol. Cell 2006, 22 (1), 51–62. [DOI] [PubMed] [Google Scholar]
- Harrison M. J.; Tang Y. H.; Dowhan D. H. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 2010, 38 (7), 2201–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger M. C.; Liang C.; Russell R. S.; Lin R.; Bedford M. T.; Wainberg M. A.; Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 2005, 79 (1), 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limm K.; Ott C.; Wallner S.; Mueller D. W.; Oefner P.; Hellerbrand C.; Bosserhoff A. K. Deregulation of protein methylation in melanoma. Eur. J. Cancer 2013, 49 (6), 1305–13. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu M.; Toyokawa G.; Hayami S.; Unoki M.; Tsunoda T.; Field H. I.; Kelly J. D.; Neal D. E.; Maehara Y.; Ponder B. A.; Nakamura Y.; Hamamoto R. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 2011, 128 (3), 562–73. [DOI] [PubMed] [Google Scholar]
- Vieira F. Q.; Costa-Pinheiro P.; Ramalho-Carvalho J.; Pereira A.; Menezes F. D.; Antunes L.; Carneiro I.; Oliveira J.; Henrique R.; Jerónimo C. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr. Relat. Cancer 2014, 21 (1), 51–61. [DOI] [PubMed] [Google Scholar]
- Compounds were synthesized using the methods described in Chesworth R.; Mitchell L. H.; Shapiro G.; Boriack-Sjodin A.; Moradei O. M.; Lei J.; Duncan K. W.. Preparation of substituted pyrazoles as arginine methyltransferase inhibitors and uses therof. WO 2014178954. The synthesis of EPZ020411 is described in the Supporting Information.
- Rioux N.; et al. Unpublished results.
- Dhar S.; Vemulapalli V.; Patananan A. N.; Huang G. L.; Di Lorenzo A.; Richard S.; Comb M. J.; Guo A.; Clarke S. G.; Bedford M. T. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013, 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.