Abstract
Since its identification in 2000, sulfotransferase (SULT) 4A1 has presented an enigma to the field of cytosolic SULT biology. SULT4A1 is exclusively expressed in neural tissue, is highly conserved, and has been identified in every vertebrate studied to date. Despite this singular level of conservation, no substrate or function for SULT4A1 has been identified. Previous studies demonstrated that SULT4A1 does not bind the obligate sulfate donor, 3′-phosphoadenosine-5′-phosphosulfate, yet SULT4A1 is classified as a SULT superfamily member based on sequence and structural similarities to the other SULTs. In this study, transcription activator-like effector nucleases were used to generate heritable mutations in the SULT4A1 gene of zebrafish. The mutation (SULT4A1Δ8) consists of an 8-nucleotide deletion within the second exon of the gene, resulting in a frameshift mutation and premature stop codon after 132 AA. During early adulthood, casual observations were made that mutant zebrafish were exhibiting excessively sedentary behavior during the day. These observations were inconsistent with published reports on activity in zebrafish that are largely diurnal organisms and are highly active during the day. Thus, a decrease in activity during the day represents an abnormal behavior and warranted further systematic analysis. EthoVision video tracking software was used to monitor activity levels in wild-type (WT) and SULT4A1Δ8/Δ8 fish over 48 hours of a normal light/dark cycle. SULT4A1Δ8/Δ8 fish were shown to exhibit increased inactivity bout length and frequency as well as a general decrease in daytime activity levels when compared with their WT counterparts.
Introduction
The cytosolic sulfotransferases (SULTs) comprise a superfamily of enzymes that catalyze a metabolic reaction wherein a sulfonate moiety is transferred from the obligate donor, 3′-phosphoadenosine-5′-phosphosulfate (PAPS), onto the substrate for conjugation to a hydroxyl group. The substrate is generally rendered biologically inactive and more water soluble, facilitating its elimination from the body. In humans, there are four families of cytosolic SULTs. The SULT1 and SULT2 families are responsible for the sulfonation of a wide variety of phenolic compounds and hydroxysteroids, respectively (Tibbs et al., 2015). Together, the catalytically active SULTs account for approximately 20% of conjugative phase II xenobiotic metabolism in humans (Evans and Relling, 1999).
In 2000, our laboratory identified a novel SULT isoform (SULT4A1) in human and rat brain cDNA libraries (Falany et al., 2000). Initially named “brain sulfotransferase-like,” it was later renamed SULT4A1 in the established SULT nomenclature based on sequence homology to other cytosolic SULTs (Blanchard et al., 2004). SULT4A1 is expressed throughout the central nervous system, with especially strong expression in the neurons of the choroid plexus, cerebral cortex, cerebellum, thalamus, pituitary, medial temporal lobe, and lentiform nucleus (Liyou et al., 2003). In zebrafish, SULT4A1 is also expressed in the retina (Crittenden et al., 2014). SULT4A1 shares several conserved sequence similarities with the other SULTs: the active site histidine, the KXXFTVXXXE dimerization domain common among SULTs, and the PAPS binding domain (Falany et al., 2000). Comparison of the SULT4A1 crystal structure with that of the other SULTs reveals conservation of tertiary structures, such as the β sheet backbone found in all SULTs, further justifying its classification as a SULT. SULT4A1 is the most conserved member of the SULT gene family, having been identified in every vertebrate species investigated to date. Humans and zebrafish, two species who share no other homologous SULT genes, have SULT4A1 genes that are 87% identical and 92% similar in amino acid sequence (Crittenden et al., 2014). SULT4A1’s singular conservation of protein sequence across vertebrate species sets it apart from the other cytosolic SULTs.
However, despite its high conservation and the sequence and structural similarity to the other cytosolic SULTs, no function has yet been described for SULT4A1. Initial studies failed to identify a substrate, and subsequent studies have suggested that SULT4A1 does not bind the cofactor, PAPS, as a purified protein. This is possibly due to the absence of a key tryptophan residue present in the catalytically active SULTs that is thought to pi-stack with the adenosine ring of PAPS (Falany et al., 2000; Allali-Hassani et al., 2007). Previous work has suggested that SULT4A1’s lack of activity in vitro could also be due to the protein’s post-translational modification in vivo (Mitchell and Minchin, 2009; Mitchell et al., 2011).
Our laboratory recently reported that morpholino knockdown of SULT4A1 expression in larval zebrafish leads to an upregulation of several genes known to be involved in phototransduction, specifically in cone photoreceptors (Crittenden et al., 2014). Zebrafish represent an excellent model organism for the investigation of SULT4A1, due to the highly conserved nature of human SULT4A1 and zebrafish SULT4A1, which suggests an equally conserved function. Furthermore, recent advances in genomic editing technology have allowed researchers to quickly generate heritable gene-specific mutations in the zebrafish genome using transcription activator-like effector nucleases (TALENs) (Bedell et al., 2012; Dahlem et al., 2012; Thomas et al., 2014). In this work, we detail the generation and characterization of a strain of zebrafish containing an 8-nucleotide deletion (Δ8) in the SULT4A1 gene sequence that results in a frameshift mutation after 89 amino acids (AA) and premature stop codon after 132 AA. Early in the adulthood of these fish, several observers noted that the mutant zebrafish were exhibiting excessively sedentary behaviors during the day, inconsistent with published reports on diurnal activity levels and sleep in zebrafish (Zhdanova et al., 2001; Yokogawa et al., 2007; Zhdanova, 2011). Zebrafish are largely diurnal organisms that are markedly active during the day and sedentary at night during their sleep period (Yokogawa et al., 2007). Therefore, an increase in sedentary behavior during the day represents an abnormal behavior and warrants further systematic analysis. This study describes the generation of SULT4A1 mutations in AB strain zebrafish and a detailed assessment of the activity patterns of wild-type (WT) and SULT4A1-deficient (SULT4A1Δ8/Δ8) zebrafish.
Materials and Methods
Zebrafish Lines and Maintenance.
WT AB strain and SULT4A1 mutant zebrafish were housed in a recirculation aquaria system (Aquaneering, San Diego, CA) in the University of Alabama Zebrafish Research Facility. Light cycle was maintained at 14 hours light/10 hours dark. All animals were cared for in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee of the University of Alabama.
TALEN Target Site Selection and Assembly.
TALENs were designed to target the zebrafish SULT4A1 gene. SULT4A1 gene exon sequences (GenBank accession number NP_001035334) were analyzed for potential TALEN targeting sites using the Old TALEN Targeter program at https://tale-nt.cac.cornell.edu/node/add/talen-old (Cermak et al., 2011; Doyle et al., 2012; Thomas et al., 2014). TALEN targeting sites within the second exon were chosen with the following parameters: left target sequence, 5′-ATTGATGAGCAGCTTCCAGT-3′; left repeat array sequence, NI, NG, NG, NN, NI, NG, NN, NI, NN, HD, NI, NN, HD, NG, NG, HD, HD, NI, NN, NG; right target sequence, 5′-AGCCGGGATTGGAGATTATCC-3′; right repeat array sequence, NN, NN, NI, NG, NI, NI, NG, HD, NG, HD, HD, NI, NI, NG, HD, HD, HD, NN, NN, HD; spacer length, 14 nucleotides. Target sequences were analyzed via BLAST to ensure that no identical sequences exist in the zebrafish genome. The Golden Gate TALEN and TAL Effector Kit 2.0 was purchased from Addgene (Cambridge, MA), and TALE repeats were assembled as previously described in the Addgene protocol (Cermak et al., 2011). Briefly, the TALENs were constructed by combining the desired TAL repeat plasmids and performing several cycles of digestion and ligation. These recombined vectors were transformed into Mach1 chemically competent cells (Life Technologies Grand Island, NY) to obtain plasmids that could then be digested and ligated into TALEN expression vectors pCS2TAL3DD and pCS2TAL3RR (Dahlem et al., 2012). These plasmids contained the Tal constant region, golden gate cloning region, and the left or right subunit of the Fok1 obligate heterodimer enzyme. Golden gate cloned plasmids were used for mRNA synthesis. TALEN mRNA was transcribed using the mMessage mMachine SP6 Kit (Life Technologies) and purified using the RNeasy Kit (Qiagen Valencia, CA). RNA concentrations were quantified using a NanoDrop ND-1000 spectrophotometer.
Microinjection of Zebrafish Embryos.
AB strain one-cell stage embryos were collected from natural breedings, and TALEN mRNA was injected into the yolk of the embryos using a regulated air-pressure microinjector (Harvard Apparatus, Holliston, MA; PL1-90). For TALEN mRNA injections, equal amounts of the left and right mRNAs were mixed to a final concentration of 100 ng/µL and injected at a volume of 0.5 nl into each embryo.
Extraction of Genomic DNA from TALEN-Injected Embryos and Adult Mutant Zebrafish.
TALEN-injected embryos were placed into individual wells of a 96-well plate containing a solution of 95% embryo lysis buffer (10 mM Tris, pH 8.3, 50 mM KCl, 0.3% Tween 20, 0.3% Nonidet P40) and 0.05 mg/ml proteinase K (Fisher Scientific, Fair Lawn, NJ). Embryos were incubated at 55°C overnight, and the proteinase K was then deactivated by incubation at 95°C for 10 minutes. The resultant solution was centrifuged at 2000g for 1 minute and used for subsequent high resolution melting analysis (HRMA). For extraction of DNA from adult fish, tail clippings were used in lieu of whole embryos.
High Resolution Melting Analysis.
To genotype fish, HRMA was performed, as described previously, using digested embryos or tail clippings as the source of template DNA (Parant et al., 2009). Each 10 µl reaction contained 1 µl LC Green Plus (BioFire Diagnostics, Salt Lake City, UT), 0.05 µl Ex Taq (TaKaRa, Ōtsu, Japan), 1 µl Ex Taq buffer, 0.4 mM dNTP (0.1 mM each), 0.1 µM forward primer, 0.1 µM reverse primer, and 1 µl DNA template. For screening of individual chimeric fish, the following primers were used: forwardchim (5′-ATGAGATCGGGCTCATGAAT-3′) and reversechim (5′-TGCGATATGCATGTGATAAAGA-3′). For screening of SULT4A1Δ8/Δ8 individuals, the following primers designed to sequences closer to the mutation site were used: forwardΔ8 (5′-TTGATGAGCAGCTTCCAGTG-3′) and reverseΔ8 (5′-TAATCTCCAATCCCGGCTGT-3′). Reaction solutions were covered with 20 µl mineral oil, and reactions were carried out in 96-well plates. After 40 polymerase chain reaction cycles (98°C for 10 seconds, 59°C for 20 seconds, 72°C for 15 seconds), the reactions were heated to 95°C for 10 seconds and then cooled to 4°C. Plates were analyzed for HRMA using a HR-1 96 LightScanner (Idaho Technology).
Immunoblot Analyses.
Cell lysates for immunoblot analysis were prepared from adult WT and SULT4A1Δ8/Δ8 zebrafish brains. Samples of brain tissue were dissected and placed in sterile phosphate-buffered saline with Complete Mini EDTA-free Protease Inhibitor Cocktail Tablets (Roche Indianapolis, IN) and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich, St. Louis, MO). Samples were disrupted by pipetting, vortexing 5 minutes at 4°C, and sonicating twice for 10 seconds with 30-second cooling on ice between sonications. The cycle of pipetting, vortexing, and sonicating was repeated, and lysate was collected after centrifugation at 15,000g for 20 minutes at 4°C. Lysate protein concentration was determined using a Bio-Rad protein analysis kit with γ globulin as a standard (Bradford, 1976), and 86 µg total protein was loaded onto the gel from each sample for SDS-PAGE. After transfer to nitrocellulose, immunoblot analyses were carried out using a rabbit polyclonal antibody raised to human SULT4A1 (Proteintech Chicago, IL) or a mouse polyclonal antibody raised to human α-tubulin (Abcam Cambridge, MA). The SULT4A1 antibody was detected using a goat anti-rabbit horseradish peroxidase–conjugated secondary antibody (Southern Biotech Birmingham, AL). The α-tubulin antibody was detected using a goat anti-mouse horseradish peroxidase–conjugated secondary (Southern Biotech). The immunoblots were developed with SuperSignal West Chemiluminescent Substrate (Thermo Scientific Waltham, MA) and exposed to autoradiograph film.
Behavioral Testing.
All behavioral tests (excluding activity analyses) were carried out between the hours of 10:00 AM and 2:00 PM. The water used in each apparatus was taken directly from the home aquaria system in which the fish had been housed and was changed between each trial. Each behavioral test (including activity analyses) was performed on a separate cohort of naive fish between 6 and 8 months of age. Test cohorts were an equal mix of male and female. All videos were recorded using an Ultra 720+ Resolution DSP True Day/Night Color Camera (EverFocus, Taipei, Taiwan) with near infrared recording abilities and analyzed using EthoVision XT (Noldus, Wageningen, The Netherlands) to track fish movement and activity levels. Genetically WT siblings of the SULT4A1Δ8/Δ8 fish being tested were used as controls in each behavioral test.
Novel Tank Test.
A standard novel tank test was used as a means to gauge stress and anxiety levels as well as locomotion in a novel tank environment (Levin et al., 2007; Bencan et al., 2009; Egan et al., 2009). For this experiment, the novel tank used was a narrow 1.8 L polycarbonate tank (Aquaneering), which restricted lateral movement, but allowed horizontal and vertical movement. The tank was filled with 1.4 L water from the same system as the home tank of the fish being tested. Test animals were moved into the recording room 1 hour prior to the beginning of testing. Fish were recorded individually in the novel tank for a total of 6 minutes, each using a camera positioned 60 cm away. Fish movement was tracked over the course of the experiment, and the following endpoints were determined: latency to enter upper half of tank, transitions to upper half, time in upper half, distance traveled, freezing bouts, and freezing bout duration. A freezing bout was defined as a lack of movement lasting at least 2 seconds.
Social Preference Test.
To examine zebrafish social behavior, a social preference test was used, as previously described by Grossman et al. (2010). Briefly, a clear acrylic open-top box was constructed with the following dimensions: 50 cm (length) × 10 cm (width) × 10 cm (height). The resulting 50-cm corridor was filled with home system water and divided into five separate, water-tight compartments through the use of four evenly spaced sliding doors. An unfamiliar target fish was placed in the conspecific compartment at one end of the corridor, and the compartment at the other end of the apparatus remained empty (empty compartment). To avoid lateral bias, the conspecific and empty compartments were switched between each trial for the same cohort. After acclimating the test cohorts to the recording room for 1 hour, WT or SULT4A1Δ8/Δ8 fish were introduced individually into the center compartment. After a period of 30 seconds (to minimize handling stress), the two center dividers were gently lifted to allow the test fish to swim freely along the 30-cm corridor comprised of the center zone, conspecific zone (adjacent to the conspecific compartment), and empty zone (adjacent to the empty compartment). Movement of the fish was recorded for 6 minutes using a camera positioned 80 cm above the tank, and the following end points were determined: empty zone entries, conspecific zone entries, time in empty zone, time in conspecific zone, and time in center zone.
Activity Analysis.
All fish used in the activity analyses were between 7 and 8 months of age. On day 1, a total of four fish (2 WT and 2 SULT4A1Δ8/Δ8) was transferred from home tanks and individually housed in 1.8 L polycarbonate tanks on an Aquaneering model 330B stand-alone housing rack. Tanks were backlit using two 8 W infrared (850 nm) light sources (Axton, North Salt Lake, UT). Two white, translucent, 0.4” plastic screens were placed behind the tanks to provide a uniform background and diffuse the infrared light. Two cameras were set up 75 cm from the tanks, with each camera set to record two tanks: one on top and one on bottom. Fish were allowed to habituate to the new tanks for a total of 96 hours on a light/dark cycle (14 hours light/10 hours dark) before recordings began. After habituation was complete, 48-hour recordings were initiated at zeitgeber time (ZT) 12. In the first recording, the two top tanks were occupied by WT fish, and the two bottom tanks were occupied by SULT4A1Δ8/Δ8 fish. In each subsequent recording, this arrangement was reversed so that there was an equal number of videos with each arrangement. Throughout the habituation and recording period, fish were fed twice daily at ZT12 and ZT19 with dry fish food. Temperature, pH, and conductivity of the system were maintained at 28°C, 7.4, and 1380–1450 µS, respectively. Day time illuminance on the front surface of the tanks was measured at 2.7 lux.
For each single-fish 48-hour trial, the arena was defined in EthoVision as the total area enclosed by the perimeter of the tank. The activity analysis function in EthoVision was used to determine activity levels within the arena over the course of each trial. All inactivity bouts lasting less than 1 second were excluded from analysis. Data output from the activity analysis was used to measure the following endpoints: mean activity (pixels/frame), mean activity length (s), inactive time percentage, inactivity bout frequency, and mean inactivity bout length (s). Data for each endpoint were grouped into 1-hour time bins, and JTK_CYCLE analyses (Hughes et al., 2010) were performed on the activity, inactive time percentage, inactivity bout frequency, and inactivity bout length data for each 48-hour trial to assess data rhythmicity and determine phase lag and amplitude. Period length was set at 24 hours across all data series.
Results
SULT4A1Δ8/Δ8 Mutant Generation.
The zebrafish SULT4A1 gene consists of seven exons and is located on chromosome 9 (ZDB-GENE-060421-2705). Using the Old TALEN Targeter program at https://tale-nt.cac.cornell.edu/node/add/talen-old, left and right TALEN sequences were designed to target sequences located within the second exon. The selected left and right TALEN sequences were 20 and 21 nucleotides long, respectively, separated by a spacer region of 14 nucleotides (Fig. 1A). By design, both the left and right TALEN sequences were coupled to complimentary subunits of a Fok1 heterodimer endonuclease. This minimizes the chance off-target effects by requiring that both halves of the obligate heterodimer are brought together by the binding of the genomic targeting domains to their respective target sequences within the genome before the endonuclease can become catalytically active. mRNA for the left and right TALENs was generated in vitro and injected into type AB zebrafish larvae at the one-cell stage. This founder generation (F0) was raised to adulthood and screened for the presence of SULT4A1 mutations by HRMA (Parant et al., 2009; Dahlem et al., 2012; Thomas et al., 2014). Mutants in this founder generation were chimeric with the potential for multiple mutations. To isolate singular mutations, F0 fish were crossed with WT fish and the progeny (F1) were raised to adulthood and screened for SULT4A1 mutations by HRMA (Fig. 1B). SULT4A1 gene sequencing of these heterozygous F1 fish revealed a mutation (SULT4A1Δ8) consisting of an 8-nucleotide deletion at the TALEN targeting site (Fig. 1C). Deletion of these 8 nucleotides results in a frameshift at AA 89 and premature stop codon after 132 AA (Fig. 2). Once identified, SULT4A1Δ8/WT fish were crossed with one another to generate fish that were homozygous for the SULT4A1Δ8 mutation. Immunoblot analysis of lysate from the brains of these SULT4A1Δ8/Δ8 individuals using a polyclonal antibody to human SULT4A1 revealed a lack of detectable immunoreactive SULT4A1 (Fig. 1D)
Fig. 1.
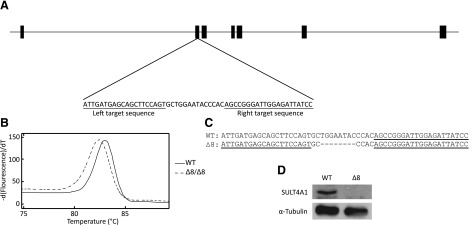
SULT4A1Δ8/Δ8 mutant generation. (A) Schematic of the zebrafish SULT4A1 gene with exons depicted as thick bars. Left and right TALEN target sequences are underlined. (B) SULT4A1Δ8/Δ8 fish were screened by HRMA analysis. WT and SULT4A1Δ8/Δ8 DNA showed a Tm difference of 0.6°C. (C) WT and SULT4A1Δ8 DNA sequence at mutation site. Left and right TALEN target sequences are underlined. Dashes represent single-nucleotide deletions. (D) Immunoblot analysis of WT (left) and SULT4A1Δ8/Δ8 (right) brain lysate. Wells were loaded with 86 µg protein, and membrane was probed with an anti-hSULT4A1 or anti–α-tubulin antibody.
Fig. 2.
WT and mutant SULT4A1Δ8 sequence alignment. Underlined sequence indicates divergence of SULT4A1Δ8 sequence from WT. Asterisks indicate a stop codon.
Anxiety in the Novel Tank Test.
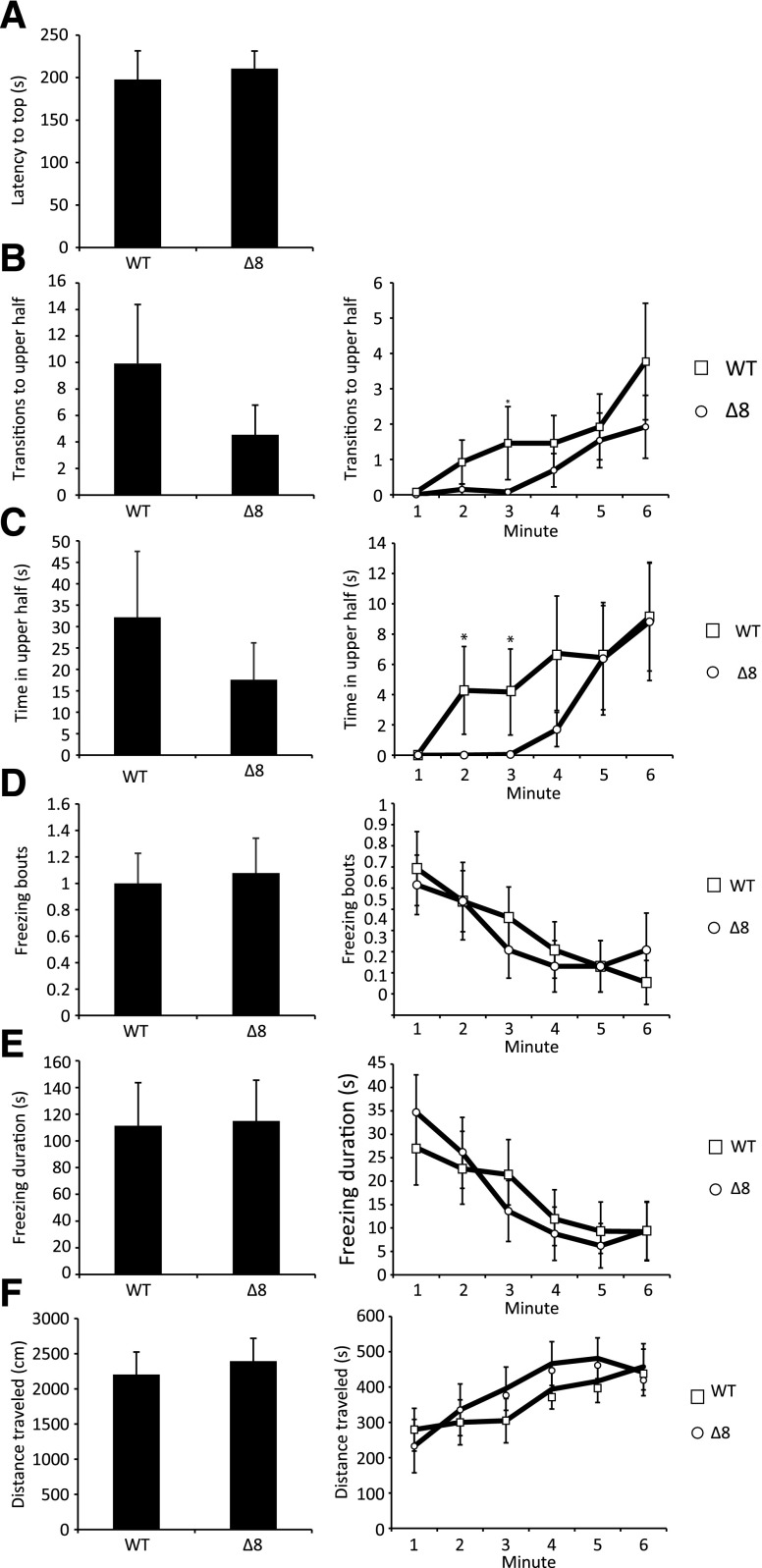
Anxiety has been shown to lead to prolonged periods of inactivity in zebrafish similar to those observed in the SULT4A1 mutants (Egan et al., 2009). Furthermore, natural variations in anxiety levels among zebrafish populations ensure that certain strains have higher anxiety levels than others. Such is the case with the leopard skin mutant, a strain of zebrafish that presents spots instead of stripes and that also exhibits increased anxiety-like behavior (Egan et al., 2009; Cachat et al., 2011; Maximino et al., 2013). To determine whether SULT4A1Δ8/Δ8 fish exhibited increased anxiety-like behavior, WT and mutant fish were subjected to a standard novel tank test, a test used to assess anxiety levels and locomotion in zebrafish when introduced into a novel environment (Levin et al., 2007; Bencan et al., 2009; Egan et al., 2009; Grossman et al., 2010). Over the course of the 6-minute experiment, SULT4A1Δ8/Δ8 fish did not exhibit any significant differences in latency to enter the upper half of the test chamber (Fig. 3A), freezing bouts (Fig. 3D), freezing duration (Fig. 3E), or total distance traveled (Fig. 3F). During the first 3 minutes of the experiment, the mutant fish displayed a decreased propensity to enter the upper half, but this did not translate into significant differences in either time in upper half or transitions to upper half (Fig. 3, B and C).
Fig. 3.
Standard 6-minute novel tank test. Error bars represent S.E.M. n = 13. (A) Latency to enter the upper half of the tank. (B) Left: total transitions to the upper half of the tank. Right: transitions to upper half per minute. (C) Left: cumulative time spent in the upper half of the tank. Right: time spent in the upper half per minute. (D) Left: total number of freezing bouts. Right: number of freezing bouts per minute. A freezing bout was defined as a total lack of movement lasting longer than 2 seconds. (E) Left: cumulative freezing duration. Right: freezing duration per minute. (F) Left: total distance traveled. Right: distance traveled per minute (*P < 0.05 in Student’s t test).
Social Preference.
WT and mutant fish were subjected to a social preference test designed to assess social behavior and motility (Fig. 4A). In accordance with previous studies (Grossman et al., 2010), WT fish spent significantly more time in the conspecific zone and entered the conspecific zone more frequently than the other zones (Fig. 4, B and D). SULT4A1Δ8/Δ8 fish also spent significantly more time in the conspecific zone than both the empty and center zones (Fig. 4E), but did not enter the conspecific zone more frequently (Fig. 4C). No significant differences were observed in the total distance traveled by either fish throughout the experiment (Fig. 4F).
Fig. 4.
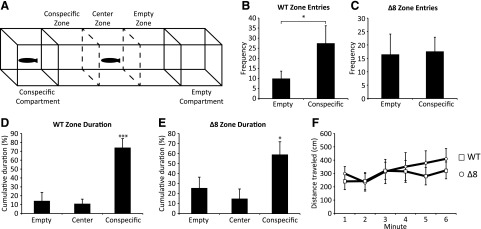
SULT4A1Δ8/Δ8 fish behavior in a social preference test. Error bars represent S.E.M. n = 10. (A) The apparatus consisted of a 50 cm × 10 cm × 10 cm clear polycarbonate tank with an open top filled maximally with water. Two 10 cm × 10 cm × 10 cm compartments at either end of the tank were separated from the rest of the tank by water-tight dividers. Two more sliding dividers separated the center area into three equal-volume zones. (B) WT fish frequency of entry into the empty and conspecific zones. (C) SULT4A1Δ8/Δ8 fish frequency of entry into the empty and conspecific zones. (D) WT fish cumulative duration in the empty, center, and conspecific zones. (E) SULT4A1Δ8/Δ8 fish cumulative duration in the empty, center, and conspecific zones. (F) Distance traveled per minute (*P < 0.05 in Student’s t test).
Activity Analysis.
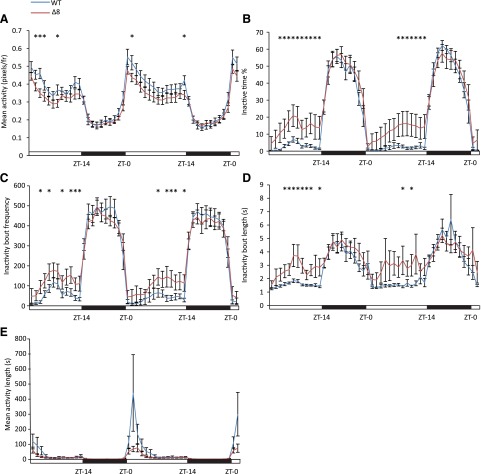
Anecdotal reports of the SULT4A1Δ8/Δ8 fish behavior suggested that the fish were less active during daytime hours. Consistent with this, EthoVision activity analysis showed a slight decrease in activity from ZT-1 to ZT-14 with no discernible difference from ZT-15 to ZT-24 (Fig. 5A). During daylight hours, SULT4A1Δ8/Δ8 spent a larger percentage of time in an inactive state (Fig. 5B) and displayed a higher inactivity bout frequency (Fig. 5C). Mutant fish also displayed increases in mean inactivity bout length during daylight hours (Fig. 5D). These data are summarized in Table 1. Due to the cyclic nature of these data, JTK_CYCLE analyses (Hughes et al., 2010) were used to calculate phase lag and amplitude of oscillations for each trial, and a significant decrease in the amplitude of these oscillations was observed in the mean activity level (Fig. 5A) as well as inactive time percentage (Fig. 5B) and inactivity bout length (Fig. 5D) of SULT4A1Δ8/Δ8 fish. In both WT and mutant fish, a sharp spike in mean activity bout length was observed within 2 hours of light onset. This peak in mean activity bout length was significantly shorter in mutant fish (Fig. 5E).
Fig. 5.
Suppressed activity in SULT4A1Δ8/Δ8 zebrafish. Error bars represent S.E.M. n = 14. (A) Mean activity levels. JTK_CYCLE analysis showed a significant drop in oscillatory amplitude in SULT4A1Δ8/Δ8 fish compared with WT fish. WT amplitude = 0.122 pixels (frame)−1 ± 0.017. SULT4A1Δ8/Δ8 amplitude = 0.084 pixels (frame)−1 ± 0.009. (B) Inactivity time percentage. JTK_CYCLE analysis showed a significant drop in oscillatory amplitude in SULT4A1Δ8/Δ8 fish compared with WT fish. WT amplitude = 11.83% ± 2.87%. SULT4A1Δ8/Δ8 amplitude = 6.38% ± 1.53%. (C) Inactivity bout frequency. JTK_CYCLE analysis did not show a significant drop in oscillatory amplitude in SULT4A1Δ8/Δ8 fish compared with WT fish. SULT4A1Δ8/Δ8 amplitude = 111.6 bouts (h)−1 ± 19.3. WT amplitude = 111.2 bouts (h)−1 ± 23.3. (D) Inactivity bout length. JTK_CYCLE analysis showed a significant drop in oscillatory amplitude in SULT4A1Δ8/Δ8 fish compared with WT fish. WT amplitude = 1.07 seconds ± 0.18 seconds. SULT4A1Δ8/Δ8 amplitude = 0.65 seconds ± 0.09. (E) Mean activity bout length. WT peak = 256 seconds ± 81.8. SULT4A1Δ8/Δ8 peak = 75.2 seconds ± 15.3.
TABLE 1.
Activity analysis in WT and SULT4A1Δ8/Δ8 fish
Asterisks indicate statistical significance in Student’s t test (P < 0.05). n = 14.
| Genotype |
|||
|---|---|---|---|
| Light/Dark | WT | Δ8 | |
| Activity (pixels/frame) | Light | 0.414 ± 0.023 | 0.354 ± 0.021* |
| Dark | 0.214 ± 0.018 | 0.209 ± 0.010 | |
| Inactive time % | Light | 2.27 ± 0.19 | 12.15 ± 1.13* |
| Dark | 44.97 ± 1.18 | 46.28 ± 1.28 | |
| Inactivity bout frequency (bouts/h) | Light | 45.01 ± 17.13 | 106.33 ± 36.2* |
| Dark | 419.85 ± 45.47 | 402.49 ± 39.84 | |
| Inactivity bout length (s) | Light | 1.65 ± 0.23 | 2.85 ± 0.78* |
| Dark | 4.14 ± 0.71 | 4.20 ± 0.54 | |
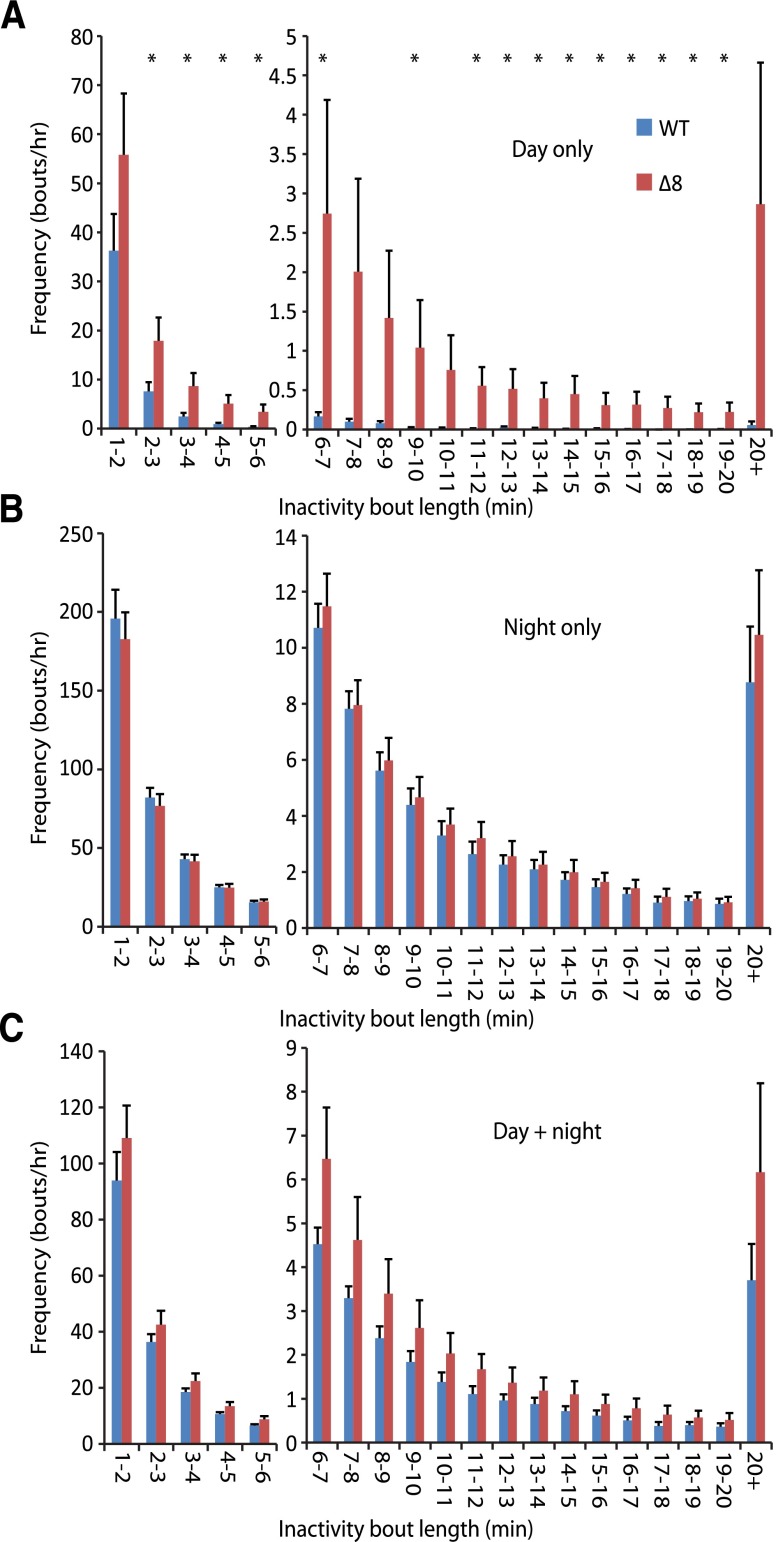
Previous reports have described a sleep-like behavior in zebrafish characterized by place preference (at the top or bottom of the tank), reversible immobility, and increased arousal threshold that peak during the night-time hours (Zhdanova, 2006, 2011; Yokogawa et al., 2007; Zhdanova et al., 2008; Appelbaum et al., 2009). Like humans, zebrafish display diurnal sleep patterns. But, unlike the consolidated sleep bouts seen in humans, zebrafish undergo many sleep bouts throughout the night. One defining characteristic of sleep in zebrafish is an increased arousal threshold. Yokogawa et al. (2007) used this arousal threshold increase to define the minimum epoch of immobility to distinguish sleep from simple immobility as 6 seconds. Thus, if the decreased activity seen in the SULT4A1Δ8/Δ8 mutants was attributable to abnormal sleep patterns, then a selective increase in inactivity bouts greater than 6 seconds would be expected. However, mutant fish displayed increased daytime inactivity bout frequency for bouts lasting less than 6 seconds as well as those lasting greater than 6 seconds (Fig. 6A). No changes were seen in night time or cumulative (day + night) inactivity bout frequency (Fig. 6, B and C).
Fig. 6.
Frequency of different inactivity bout lengths. Error bars represent S.E.M. n = 14 (*P < 0.05 in Student’s t test). (A) Day time inactivity bout frequency. (B) Night time inactivity bout frequency. (C) Cumulative (day + night) inactivity bout frequency.
Discussion
Since its identification in 2000, SULT4A1 has presented an enigma in the field of cytosolic SULT biology. Despite its very high level of conservation, no substrate or function has been identified. The characterization of activity suppression in SULT4A1 mutant fish represents a novel behavioral phenotype, the molecular mechanism of which remains unclear. Identification of that mechanism will allow pharmacologic intervention to study SULT4A1’s pathway in the future.
One possible explanation for the suppressed activity observed in SULT4A1Δ8/Δ8 fish is the disruption of normal sleep cycles. Zebrafish are diurnal animals whose sleep is markedly inhibited by light (Yokogawa et al., 2007). If sleep cycle dysregulation is responsible for the suppressed daytime activity seen in the mutant fish, this would provide a unique opportunity to elucidate SULT4A1’s biologic function. Much of the molecular machinery and effector molecules of sleep regulation, such as hypocretin and melatonin, are conserved among vertebrates (Chen et al., 2015). Zebrafish, however, are unique from most other vertebrates in that their circadian clock is decentralized (Whitmore et al., 1998; Cermakian et al., 2000). Most vertebrates (including humans) possess a small population of “pacemaker cells” within the suprachiasmatic nucleus (SCN) that are responsible for maintaining diurnal rhythms (Mistlberger, 2005). Zebrafish possess a SCN, but evidence suggests that it is not required for the normal development of circadian rhythms (Noche et al., 2011). Instead, the zebrafish brain has been shown to be globally rhythmic and light-sensitive (Whitmore et al., 1998; Moore and Whitmore, 2014). Given the diffuse nature of SULT4A1 expression in the retina and brain outside the SCN and pineal gland in mammals (Liyou et al., 2003), it is unlikely that SULT4A1 is involved in the maintenance of diurnal rhythms. However, it is possible that SULT4A1 may be involved in regulating the neuronal response to circadian input from effector molecules such as hypocretins or melatonin. As in other diurnal vertebrates, hypocretins and melatonin play a central role in the regulation of sleep and wakefulness in zebrafish (Appelbaum et al., 2009; Mieda et al., 2013). Furthermore, hypocretin deficiency has been shown to cause narcolepsy in nocturnal as well as diurnal animals, including humans (Chemelli et al., 1999; Lin et al., 1999; Peyron et al., 2000; Thannickal et al., 2000). If SULT4A1 is involved in the regulation of hypocretin signaling at the postsynaptic level, then that may help explain an increase in sleep-like behavior during the day. In order for the observed inactivity bouts to be conclusively characterized as sleep, an increase in arousal threshold will need to be demonstrated during the inactivity bouts. Due to the brevity and unpredictable timing of these inactivity bouts, demonstrating an increased arousal threshold is exceedingly difficult. So at this point, the possibility of sleep dysregulation remains.
Previous studies have shown that anxiety in zebrafish can lead to increased inactivity bouts (Egan et al., 2009; Cachat et al., 2010, 2011; Grossman et al., 2010). However, the results of this study suggest that the increase in inactivity bouts is most likely not attributable to anxiety. In the novel tank test, designed to induce and analyze anxiety in zebrafish, SULT4A1Δ8/Δ8 fish did not show any increase over WT fish in freezing bout frequency, freezing duration, or total distance traveled. SULT4A1Δ8/Δ8 fish did show a decreased propensity to enter the upper half of the tank in the novel tank test, but only during the second and third minutes of the experiment, after which they did not behave differently from WT fish (Fig. 3).
In 2014, our laboratory reported an upregulation of several cone-specific phototransduction genes in SULT4A1 knockdown zebrafish larvae (Crittenden et al., 2014). This dysregulation of cone genes was observed at 72 hours postfertilization and was not accompanied by any overt morphologic or developmental defects. It is possible that the dysregulation of cone phototransduction genes may carry over into adulthood in the SULT4A1 mutant fish. If such is the case, however, it does not appear as though this results in blindness in the fish. SULT4A1Δ8/Δ8 fish were able to see and identify the conspecific fish in a social preference test. Given the water-tight nature of the boundary between the center and conspecific compartments in the test, this preference is more likely attributable to vision than another social stimulus such as olfaction. Although the SULT4A1Δ8/Δ8 fish are most likely not blind, the possibility that they may have impaired color vision remains to be investigated.
Elucidation of SULT4A1’s role in activity regulation will require a comprehensive inquiry into the biochemical activity of SULT4A1 within the central nervous system. Recent work has described the post-translational modification of SULT4A1 via phosphorylation/dephosphorylation as well as a possible interaction with the peptidyl-prolyl cis-trans isomerase, Pin1 (Mitchell and Minchin, 2009; Mitchell et al., 2011). Yet little is known about the exact biologic function that SULT4A1 plays on a molecular level, and understanding its role in the regulation of activity will require extensive work. Although it does not resolve and address the question of the enzymatic activity or biochemical function of SULT4A1, this study represents a major step forward in the search for this protein’s function in its identification of a behavioral phenotype associated with SULT4A1 mutation.
Acknowledgments
The University of Alabama–Heflin sequencing core and the University of Alabama Zebrafish Research Facility were strongly used. The authors thank Karen Gamble and Lauren Hablitz for statistical expertise, and Patty Oden for diligent observation of the SULT4A1Δ8/Δ8 zebrafish.
Abbreviations
- Δ8
8-nucleotide deletion
- AA
amino acid
- HRMA
high resolution melting analysis
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- SCN
suprachiasmatic nucleus
- SULT
sulfotransferase
- TALEN
transcription activator-like effector nuclease
- WT
wild type
- ZT
zeitgeber time
Authorship Contributions
Participated in research design: Crittenden, Parant, Falany.
Conducted experiments: Crittenden, Thomas.
Contributed new reagents or analytic tools: Parant.
Performed data analysis: Crittenden, Falany.
Wrote or contributed to the writing of the manuscript: Crittenden, Falany.
Footnotes
This work was supported by the National Institutes of Health [Grants R21MH095946 and P30NS47466].
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L, Wang GX, Maro GS, Mori R, Tovin A, Marin W, Yokogawa T, Kawakami K, Smith SJ, Gothilf Y, et al. (2009) Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc Natl Acad Sci USA 106:21942–21947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencan Z, Sledge D, Levin ED. (2009) Buspirone, chlordiazepoxide and diazepam effects in a zebrafish model of anxiety. Pharmacol Biochem Behav 94:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14:199–211. [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- Cachat J, Stewart A, Grossman L, Gaikwad S, Kadri F, Chung KM, Wu N, Wong K, Roy S, Suciu C, et al. (2010) Measuring behavioral and endocrine responses to novelty stress in adult zebrafish. Nat Protoc 5:1786–1799. [DOI] [PubMed] [Google Scholar]
- Cachat J, Stewart A, Utterback E, Hart P, Gaikwad S, Wong K, Kyzar E, Wu N, Kalueff AV. (2011) Three-dimensional neurophenotyping of adult zebrafish behavior. PLoS One 6:e17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Whitmore D, Foulkes NS, Sassone-Corsi P. (2000) Asynchronous oscillations of two zebrafish CLOCK partners reveal differential clock control and function. Proc Natl Acad Sci USA 97:4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. (1999) Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98:437–451. [DOI] [PubMed] [Google Scholar]
- Chen Q, de Lecea L, Hu Z, Gao D. (2015) The hypocretin/orexin system: an increasingly important role in neuropsychiatry. Med Res Rev 35:152–197. [DOI] [PubMed] [Google Scholar]
- Crittenden F, Thomas H, Ethen CM, Wu ZL, Chen D, Kraft TW, Parant JM, Falany CN. (2014) Inhibition of SULT4A1 expression induces up-regulation of phototransduction gene expression in 72-hour postfertilization zebrafish larvae. Drug Metab Dispos 42:947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. (2012) Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8:e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. (2012) TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40:W117-W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan RJ, Bergner CL, Hart PC, Cachat JM, Canavello PR, Elegante MF, Elkhayat SI, Bartels BK, Tien AK, Tien DH, et al. (2009) Understanding behavioral and physiological phenotypes of stress and anxiety in zebrafish. Behav Brain Res 205:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV. (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286:487–491. [DOI] [PubMed] [Google Scholar]
- Falany CN, Xie X, Wang J, Ferrer J, Falany JL. (2000) Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem J 346:857–864. [PMC free article] [PubMed] [Google Scholar]
- Grossman L, Utterback E, Stewart A, Gaikwad S, Chung KM, Suciu C, Wong K, Elegante M, Elkhayat S, Tan J, et al. (2010) Characterization of behavioral and endocrine effects of LSD on zebrafish. Behav Brain Res 214:277–284. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ED, Bencan Z, Cerutti DT. (2007) Anxiolytic effects of nicotine in zebrafish. Physiol Behav 90:54–58. [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. (1999) The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98:365–376. [DOI] [PubMed] [Google Scholar]
- Liyou NE, Buller KM, Tresillian MJ, Elvin CM, Scott HL, Dodd PR, Tannenberg AE, and McManus ME (2003) Localization of a brain sulfotransferase, SULT4A1, in the human and rat brain: an immunohistochemical study. J Histochem Cytochem 51:1655-1664. [DOI] [PubMed]
- Maximino C, Puty B, Matos Oliveira KR, Herculano AM. (2013) Behavioral and neurochemical changes in the zebrafish leopard strain. Genes Brain Behav 12:576–582. [DOI] [PubMed] [Google Scholar]
- Mieda M, Tsujino N, Sakurai T. (2013) Differential roles of orexin receptors in the regulation of sleep/wakefulness. Front Endocrinol 4:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. (2005) Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus. Brain Res Brain Res Rev 49:429–454. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Butcher NJ, Minchin RF. (2011) Phosphorylation/dephosphorylation of human SULT4A1: role of Erk1 and PP2A. Biochim Biophys Acta 1813:231–237. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Minchin RF. (2009) Cytosolic Aryl sulfotransferase 4A1 interacts with the peptidyl prolyl cis-trans isomerase Pin1. Mol Pharmacol 76:388–395. [DOI] [PubMed] [Google Scholar]
- Moore HA, Whitmore D. (2014) Circadian rhythmicity and light sensitivity of the zebrafish brain. PLoS One 9:e86176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noche RR, Lu PN, Goldstein-Kral L, Glasgow E, Liang JO. (2011) Circadian rhythms in the pineal organ persist in zebrafish larvae that lack ventral brain. BMC Neurosci 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant JM, George SA, Pryor R, Wittwer CT, and Yost HJ (2009) A rapid and efficient method of genotyping zebrafish mutants. Dev Dyn 238:3168-3174. [DOI] [PMC free article] [PubMed]
- Peyron C, Faraco J, Rogers W, Ripley B, Overeem S, Charnay Y, Nevsimalova S, Aldrich M, Reynolds D, Albin R, et al. (2000) A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med 6:991–997. [DOI] [PubMed] [Google Scholar]
- Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, Cornford M, Siegel JM. (2000) Reduced number of hypocretin neurons in human narcolepsy. Neuron 27:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas HR, Percival SM, Yoder BK, Parant JM. (2014) High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS One 9:e114632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbs ZE, Rohn-Glowaki KJ, Crittenden F, Guidry AL, Falany CN. (2015) Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab Pharmacokinet 30:3–20. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strähle U, Sassone-Corsi P. (1998) Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci 1:701–707. [DOI] [PubMed] [Google Scholar]
- Yokogawa T, Marin W, Faraco J, Pézeron G, Appelbaum L, Zhang J, Rosa F, Mourrain P, Mignot E. (2007) Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol 5:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova IV. (2006) Sleep in zebrafish. Zebrafish 3:215–226. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV. (2011) Sleep and its regulation in zebrafish. Rev Neurosci 22:27–36. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV, Wang SY, Leclair OU, Danilova NP. (2001) Melatonin promotes sleep-like state in zebrafish. Brain Res 903:263–268. [DOI] [PubMed] [Google Scholar]
- Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S, Guelin E. (2008) Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res Bull 75:433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]